Abstract
Multiple strains of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis isolated from animal, clinical, or food samples have been analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Whole bacterial cells were harvested from colonies or confluent growth on agar and transferred directly into solvent and then to a spot of dried 3-methoxy-4-hydroxycinnamic acid (matrix). Multiple ions in the 5,000- to 15,000-Da mass range were evident in spectra for each strain; one or two ions in the 9,500- to 11,000-Da range were consistently high intensity. “Species-identifying” biomarker ions (SIBIs) were evident from analyses of multiple reference strains for each of the six species, including the genome strains C. jejuni NCTC 11168 and C. jejuni RM1221. Strains grown on nine different combinations of media and atmospheres yielded SIBI masses within ±5 Da with external instrument calibration. The highest-intensity C. jejuni SIBIs were cytosolic proteins, including GroES, HU/HCj, and RplL. Multiple intraspecies SIBIs, corresponding probably to nonsynonymous nucleotide polymorphisms, also provided some intraspecies strain differentiation. MALDI-TOF MS analysis of 75 additional Campylobacter strains isolated from humans, poultry, swine, dogs, and cats revealed (i) associations of SIBI type with source, (ii) strains previously speciated incorrectly, and (iii) “strains” composed of more than one species. MALDI-TOF MS provides an accurate, sensitive, and rapid method for identification of multiple Campylobacter species relevant to public health and food safety.
Thermophilic strains of Campylobacter account for a high percentage of the estimated 76 million yearly incidents of food-borne illness in the United States (49). Although many outbreaks of Campylobacter disease have been reported in the last 25 years, most illnesses occur sporadically and are caused by C. jejuni (51). Of the 16 species of the genus Campylobacter identified to date, at least eight have been identified as potential human gastrointestinal pathogens: C. jejuni, C. coli, C. lari, C. fetus, C. upsaliensis, C. sputorum, C. concisus, and C. curvus (1, 7, 15, 22, 23, 29, 37, 38, 47, 48, 53, 60, 62, 64, 66). Approximately 0.1% of C. jejuni cases are associated with a serious paralytic disease, Guillain-Barré syndrome (55). Also, C. rectus and possibly C. showae are associated with periodontal disease (44).
There are a variety of existing assays for confirming Campylobacter by genus and species. These include biochemical (2), genetic (PCR) (3, 6, 20, 32, 71), immunochemical (34, 36, 45), chemotaxonomic fatty acid profiling (9), protein one-dimensional gel electrophoresis methods (67) and, recently, a microarray-based method (69). Some of the limitations in these methods are hippuricase-negative C. jejuni strains (24, 30, 43), the necessity for multiple hybridization steps and/or primers with multispecies PCR-based assays (3, 6, 20, 25, 31, 43, 59, 68, 71), specificity issues with 16S rRNA PCR (42, 43; unpublished observations), and chemotaxonomic methods (9, 63), and only limited information (e.g., species) is obtained. Details of the fine specificities of immunochemical reagents and assay methods and comparisons of results with multiple strains of C. jejuni, C. coli, and emerging Campylobacter species have been limited (34, 36, 56).
Mass spectrometric methods for analyzing whole bacterial cells for intact proteins, including the identification of protein biomarker ions, have been reported previously (5, 11, 16, 28, 35, 70). In a preliminary study most relevant to this work, Campylobacter and Helicobacter strains were analyzed by matrix-assisted laser desorption-time of flight mass spectrometry (MALDI-TOF MS), and biomarker ions in the 10- to 20-kDa range (presumably proteins) were reported to be the most discriminatory of those observed (73).
The major advantages of MALDI-TOF MS for analyzing bacteria are (i) ease and speed of analysis, (ii) identification of mixed cultures, and (iii) rapid identification of candidate biomarkers, even when minimal genetic data are available. We have developed an expanded MALDI-TOF MS method for analyzing multiple Campylobacter species and applied this method to determine species, and limited subspecies classes, of suspected Campylobacter strains isolated from a variety of food and animal sources.
MATERIALS AND METHODS
Bacterial strains.
The RM strain number is the designation for strains in the Produce Safety and Microbiology Research Unit strain file. Sources and other information about the strains are noted in Table 1.
TABLE 1.
Strains used in the study
| PSM strain no. | Putative species | Source | Location | Additional strain info.a | Species by MALDI-TOF MSb |
|---|---|---|---|---|---|
| RM1048 | C. jejuni | Human | Canada | ATCC 43432, HS4 | C. jejuni |
| RM1050 | C. jejuni | Human | Canada | ATCC 43449, HS23 | C. jejuni |
| RM1051 | C. coli | Human | Canada | ATCC 43479, HS30 | C. coli |
| RM1155 | C. jejuni | Human | Canada | HL1 | C. jejuni |
| RM1156 | C. jejuni | Human | Canada | HL2 | C. jejuni |
| RM1158 | C. jejuni | Human | Canada | HL5 | C. jejuni |
| RM1159 | C. jejuni | Human | Canada | HL6 | C. jejuni |
| RM1161 | C. coli | Human | Canada | HL8 | C. coli |
| RM1163 | C. jejuni | Human | Canada | HL11 | C. jejuni |
| RM1164 | C. jejuni | Human | Canada | HL15 | C. jejuni |
| RM1165 | C. jejuni | Chicken | Canada | HL17 | C. jejuni |
| RM1166 | C. coli | Chicken | Canada | HL21 | C. coli |
| RM1167 | C. jejuni | Human | Canada | HL28 | C. jejuni |
| RM1168 | C. jejuni | Human | Canada | HL36 | C. jejuni |
| RM1169 | C. coli | Human | Canada | HL55 | C. coli |
| RM1199 | C. jejuni | Chicken, processing water | United States | This study | C. jejuni |
| RM1216 | C. jejuni | Chicken, retail | United States | This study | C. jejuni |
| RM1221 | C. jejuni | Chicken, retail | United States | This study and reference 50 | C. jejuni |
| RM1244 | C. jejuni | Human | United States | CDHS, 90A2737 | C. jejuni |
| RM1246 | C. jejuni | Human | United States | CDHS, 92A3120 | C. jejuni |
| RM1247 | C. jejuni | Human | United States | CDHS, 96A11074 | C. jejuni |
| RM1292 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1327 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1343 | C. coli | Turkey | United States | This study | C. coli |
| RM1352 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1357 | C. coli | Turkey | United States | This study | C. coli |
| RM1367 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1372 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1373 | C. coli | Turkey | United States | This study | C. coli |
| RM1374 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1376 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1399 | C. coli | Turkey | United States | This study | C. coli |
| RM1420 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1421 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1438 | C. jejuni | Turkey | United States | This study | C. jejuni |
| RM1462 | C. lari | Human | Not known | ATCC 35223 | C. lari |
| RM1485 | C. sputorum biovar fecalis | Ovine | Not known | ATCC 33709 | C. sputorum bv. fecalis |
| RM1488 | C. upsaliensis | Human | United States | ATCC 49815 | C. upsaliensis |
| RM1495 | C. sputorum biovar bulbulus | Not determined | Not known | ATCC 33491 | C. sputorum bv. bulbulus |
| RM1862 | C. jejuni | Human | United Kingdom | HL4, NCTC 11168 (58) | C. jejuni |
| RM1875 | C. coli | Swine | Not known | ATCC 33559 | C. coli |
| RM1876 | C. coli | Human | Belgium | ATCC 43473, HS14 | C. coli |
| RM1877 | C. coli | Human | Canada | ATCC 43482, HS46 | C. coli |
| RM1878 | C. coli | Human | Canada | ATCC 43474, HS20 | C. coli |
| RM1879 | C. coli | Human | Canada | ATCC 43479, HS30 | C. coli |
| RM1880 | C. coli | Human | United States | ATCC 43485, HS49 | C. coli |
| RM1881 | C. jejuni | Human | United States | ATCC 33291 | C. jejuni |
| RM1882 | C. jejunic | Human | United States | ATCC 43429, HS1 | C. coli +C. jejuni |
| RM1883 | C. jejuni | Calf | Canada | ATCC 43430, HS2 | C. jejuni |
| RM1885 | C. jejuni | Not known | Not known | ATCC 43463, HS44 | C. jejuni |
| RM1886 | C. jejuni | Bovine | Not known | ATCC 33560 | C. jejuni |
| RM1887 | C. lari | Human | United Kingdom | ATCC 35223, NCTC 11458 | C. lari |
| RM1888 | C. lari | Avian | Northern Europe | ATCC 35221, NCTC 11352 | C. lari |
| RM1889 | C. lari | Dog | Northern Europe | ATCC 35222, NCTC 11457 | C. lari |
| RM1890 | C. lari | Human | Canada | ATCC 43675 | C. lari (+C. jejuni?) |
| RM1891 | C. coli | Chicken farm | United States | K20 | C. coli |
| RM1892 | C. jejuni | Chicken farm | United States | K21 | C. jejuni |
| RM1893 | C. jejuni | Chicken farm | United States | K22 | C. jejuni |
| RM1894 | C. jejuni | Chicken farm | United States | K23 | C. jejuni |
| RM1895 | C. jejuni | Chicken farm | United States | K24 | C. jejuni |
| RM1896 | C. coli | Swine farm | United States | 1921 | C. coli |
| RM1897 | C. coli | Swine farm | United States | 1926 | C. coli |
| RM1898 | C. coli | Swine farm | United States | 1931 | C. coli |
| PSM strain no. | Putative species | Source | Location | Additional strain info.a | Species by MALDI-TOF MSb |
| RM1899 | C. coli | Swine farm | United States | 1936 | C. coli |
| RM1900 | C. coli | Swine farm | United States | 1941 | C. coli |
| RM1901 | C. coli | Swine farm | United States | 2151 | C. coli |
| RM1902 | C. coli | Swine farm | United States | 2156 | C. coli |
| RM1904 | C. coli | Swine farm | United States | 2171 | C. coli |
| RM1905 | C. coli | Swine farm | United States | 2176 | C. coli |
| RM1906 | C. coli | Swine farm | United States | 2181 | C. coli |
| RM1907 | C. coli | Swine farm | United States | 2191 | C. coli |
| RM1908 | C. coli | Swine farm | United States | 2196 | C. coli |
| RM1909 | C. jejuni | Human | Canada | ATCC 43431, TGH9011, HS3 | C. jejuni |
| RM1910 | C. jejuni | Chicken | United States | This study | C. jejuni |
| RM2089 | C. sputorumbv.fecalis | Ovine | Not known | CDC D1137 | C. upsaliensis |
| RM2090 | C. sputorum bv. fecalis | Ovine | United States | CDC D1169, NCTC 11415 | C. sputorum bv. fecalis |
| RM2091 | C. sputorum bv. fecalis | Ovine | Not known | CDC D1138 | C. sputorum bv. fecalis |
| RM2092 | C. upsaliensis | Human | United States | CDC D1673 | C. upsaliensis |
| RM2093 | C. upsaliensis | Human | United States | CDC D1178 | C. upsaliensis |
| RM2095 | C. jejuni subsp. doylei | Human (blood) | United States | CDC 2722 | C. jejuni subsp. doylei |
| RM2096 | C. jejuni subsp. doylei | Human | United States | CDC D2781 | C. jejuni subsp. doylei |
| RM2098 | C. lari | Human | United States | CDC D70 | C. lari |
| RM2099 | C. lari | Human | United States | CDC D71 | C. lari |
| RM2100 | C. lari | Human | United States | CDC D67 | C. lari |
| RM2225 | C. jejuni | Chicken | United States | MDR | C. coli |
| RM2228 | C. jejuni | Chicken | United States | MDR | C. coli |
| RM2230 | C. coli | Chicken | United States | MDR | C. coli |
| RM2231 | C. coli | Chicken | United States | MDR | C. coli |
| RM2236 | C. coli | Chicken | United States | MDR | C. coli |
| RM2241 | C. jejuni | Chicken | United States | MDR | C. coli |
| RM2243 | C. jejuni | Chicken | United States | MDR | C. coli |
| RM3195 | C. upsaliensis | Human | South Africa | 300.94 | C. upsaliensis |
| RM3228 | C. helveticus | Feline | Switzerland | ATCC 51209, NCTC 12470 | C. helveticus |
| RM3776 | C. upsaliensis | Human | South Africa | 365.96 | C. upsaliensis |
| RM3777 | C. upsaliensis | Human | South Africa | 3.97 | C. upsaliensis |
| RM3778 | C. upsaliensis | Human | South Africa | 5.97 | C. upsaliensis |
| RM3807 | C. upsaliensis | Feline | United States | This study | C. helveticus |
| RM3808 | C. upsaliensis | Canine | United States | This study | C. upsaliensis |
| RM3809 | C. upsaliensis | Canine | United States | This study | C. upsaliensis |
| RM3810 | C. upsaliensis | Feline | United States | This study | C. upsaliensis |
| RM3812 | C. upsaliensis | Canine | United States | This study | C. upsaliensis |
| RM3937 | C. upsaliensis | Human | United States | CDHS-LA #3 | C. upsaliensis |
| RM3939 | C. upsaliensis | Human | United States | CDHS-LA #21 | C. upsaliensis |
| RM3940 | C. upsaliensis | Human | United States | CDHS-LA #54 | C. upsaliensis |
| RM3941 | C. upsaliensis | Human | United States | CDHS-LA #58 | C. upsaliensis |
| RM3942 | C. upsaliensis | Human | United States | CDHS-LA #675 | C. upsaliensis |
| RM3943 | C. upsaliensis | Human | United States | CDHS-LA #1419 | C. upsaliensis |
| RM3945 | C. upsaliensis | Canine | United States | CDHS-LA #3846 | C. upsaliensis |
| RM3946 | C. upsaliensis | Canine | United States | CDHS-LA #1883 | C. upsaliensis |
| RM3947 | C. upsaliensis | Canine | United States | CDHS-LA #1884 | C. upsaliensis |
| RM3948 | C. upsaliensis | Canine | United States | CDHS-LA #1885 | C. upsaliensis |
| RM3949 | C. upsaliensis | Canine | United States | CDHS-LA #2041 | C. upsaliensis |
| RM3950 | C. upsaliensis | Canine | United States | CDHS-LA #2042A | C. upsaliensis |
| RM4039 | C. upsaliensis | Human | South Africa | SA 181.00 | C. upsaliensis |
| RM4040 | C. upsaliensis | Human | South Africa | 101.01 | C. upsaliensis |
| RM4042 | C. upsaliensis | Human | South Africa | 106.01 | C. upsaliensis |
| RM4087 | C. helveticus | Feline | Switzerland | CCUG 30566 | C. helveticus+C. upsaliensis?d |
| RM4088 | C. helveticus | Feline | Sweden | CCUG 34016 | C. helveticus |
| RM4120 | C. sputorum bv. paraureolyticus | Human | Canada | LMG11764 | C. sputorum bv. paraureolyticus |
| RM4121 | C. sputorum bv. fecalis | Ovine | United Kingdom | CCUG 20703 | C. sputorum bv. fecalis |
| PSM strain no. | Putative species | Source | Location | Additional strain info.a | Species by MALDI-TOF MSb |
| RM4124 | C. upsaliensis | Canine | Switzerland | CCUG 19607 | C. upsaliensis |
| RM4244 | C. upsaliensis | Human | Belgium | LMG9104 | C. upsaliensis |
| RM4245 | C. upsaliensis | Human | Belgium | LMG9108 | C. upsaliensis |
| RM4246 | C. upsaliensis | Human | Belgium | LMG9114 | C. upsaliensis |
| RM4248 | C. upsaliensis | Human | Belgium | LMG9125 | C. upsaliensis |
| RM4249 | C. upsaliensis | Human | Belgium | LMG9129 | C. upsaliensis |
| RM4250 | C. upsaliensis | Human | Belgium | LMG9140 | C. upsaliensis |
| RM4252 | C. upsaliensis | Human | Belgium | LMG9226 | C. upsaliensis |
| RM4254 | C. upsaliensis | Human | Belgium | LMG9234 | C. upsaliensis |
| RM4257 | C. upsaliensis | Human | Belgium | LMG9265 | C. upsaliensis |
| RM4258 | C. upsaliensis | Human | Belgium | LMG9269 | C. upsaliensis |
| RM4385 | C. upsaliensis | Feline | United States | This study | C. upsaliensis |
| RM4386 | C. upsaliensis | Feline | United States | This study | C. upsaliensis |
| RM4387 | C. upsaliensis | Feline | United States | This study | C. helveticus |
| RM4388 | C. upsaliensis | Feline | United States | This study | C. helveticus |
| RM4389 | C. upsaliensis | Feline | United States | This study | C. helveticus |
| RM4390 | C. upsaliensis | Feline | United States | This study | C. helveticus |
| RM4391 | C. upsaliensis | Feline | United States | This study | C. helveticus |
| RM4392 | C. upsaliensis | Feline | United States | This study | C. helveticus |
The original strain numbers are noted also, if known. MDR, multidrug resistant; HS, Penner heat-stable serotype; HL, Lior heat-sensitive (labile) serotype; CDC, Centers for Disease Control and Prevention; ATCC, American Type Culture Collection, United States; NCTC, National Type Culture Collection, United Kingdom; CDHS-LA, California Dept. of Health Services—LA Laboratory; CCUG, Culture Collection of the University of Göteborg, Sweden; LMG, Laboratorium voor Microbiologie, Universiteit Gent, Belgium.
Boldface type indicates a “strain” with results inconsistent with original species designation.
MLST data indicate possible mixture of C. helveticus and C. upsaliensis or C. helveticus-C. upsaliensis chimeric alleles.
Bacterial isolation and culture conditions.
C. coli and C. jejuni strains were maintained on brucella agar (BA) containing, per liter, 28 g brucella broth (BD Biosciences, San Jose, CA), supplements (0.25 g FeSO4, 0.25 g sodium metabisulfite [anhydrous], 0.25 g sodium pyruvate [anhydrous]) and 15 g of Bacto agar (BD Biosciences) as described previously (2, 50). Other Campylobacter strains were grown on BA and/or brain heart infusion agar medium containing 37 g of brain heart infusion broth medium, 6 g yeast extract, 15 g Bacto agar (BD Biosciences), and 100 ml laked horse blood (Hema Resource and Supply, Aurora, OR) per liter. All Campylobacter strains were grown under microaerophilic conditions at 37°C or 42°C by placing plates in a sealable plastic bag with a gas mixture of 10% CO2, 5% H2, and 85% N2 (microaerophilic gas) or a commercial sachet (CampyPak; Oxoid, Inc.). In one experiment to compare the effects of different growth conditions on species-identifying biomarker ions (SIBIs), C. jejuni strain RM1221 was grown also on BA with 5% laked horse blood (BAB) and GC agar medium, which contains GC medium base and 1% Bacto agar, and supplemented with final concentrations of 0.3% glucose, 0.00084% ferric nitrate (nonahydrate), 0.01% l-glutamine, 0.001% thiamine pyrophosphate, and 0.005% l-cysteine, as described previously (46, 72). The media were incubated in a microaerophilic atmosphere as above or in 5% CO2, 5% O2, 90% N2 at either 37°C or 42°C.
Some C. upsaliensis and C. helveticus strains analyzed in this study were isolated by a membrane filtration method modified from one described previously (41). An approximately 20% suspension of cat or dog feces in saline was incubated at 37°C for 30 min, and then four 50-μl samples were applied at different locations on a 0.6-μm membrane filter (ME26; 47 mm; Schleicher & Schuell) placed on top of a BAB plate. The sample was incubated on the filter at room temperature for 15 min, and then any solution remaining was removed and the filter was removed with sterile tweezers. The plate was placed in a plastic sealable bag (Ziploc Freezer; Johnson and Son), and the bag was filled with 10% H2, 10% CO2, and 80% N2 and evacuated at least twice before final filling with gas; the bag and plate were incubated at 37°C for 48 to 72 h. Suspect Campylobacter colonies were subcultured on BAB for further analysis. The majority of the colonies were Campylobacter species.
Materials.
Acetonitrile, ferrous sulfate, sodium metabisulfite, sodium pyruvate, brucella agar, and Bacto agar were purchased from Fisher Scientific (Pittsburgh, PA). Trifluoroacetic acid (TFA) and 3-methoxy-4-hydroxycinnamic acid (ferulic acid) were purchased from Aldrich Chemical (Milwaukee, WI). Adrenocorticotropic hormone clip (18-39), angiotensin II, substance P, bovine insulin, and horse heart myoglobin were purchased from Sigma Chemical Company (St. Louis, MO). Purified water (Milli-Q system; Millipore, Bedford, MA) was used for all experiments.
Preparation of bacterial cell lysates and MALDI sample targets.
Cells were transferred from the plate to the extraction tube with a 1.0-μl disposable inoculating loop (NUNC Brand Products, Denmark); this volume corresponded to approximately 109 cells. The extraction tube was a 2.0-ml conical screw cap microtube (QSP; catalog no. 522; Porex Bio Products Inc.) with cap and O-ring. The extraction solution contained 0.5 ml of 33% HPLC-grade acetonitrile (Fisher Scientific, Fair Lawn, NJ), 67% HPLC-grade water (Burdick & Jackson, Muskegon, MI), 0.1% sequencing-grade TFA (Sigma-Aldrich, St. Louis, MO), and ∼40 mg of 0.1-mm zirconia-silica beads (catalog no. 11079101z; BioSpec Products Inc., Bartlesville, OK). Extraction tubes were then tightly capped, placed into a 96-well aluminum holder, and agitated for 60 s on a beadbeater (Mini-beadbeater-96+; BioSpec Products, Bartlesville, OK). Bead beating for longer than 60 s was avoided to minimize sample heating due to bead friction. The solution was frothy and opaque after this step. Tubes then were centrifuged for 4 to 5 min at 10,600 × g (model 4515C; Eppendorf, Hamburg, Germany). The undissolved cellular material and beads at the bottom of the tube were pelleted by centrifugation, after which the supernatant was clear (bacterial cell lysate).
A saturated solution of trans-4-hydroxy-3-methoxy-cinammic acid (Sigma- Aldrich, St. Louis, MO) also known as ferulic acid, was prepared in 0.5 ml of 33% acetonitrile, 67% water, and 0.1% TFA (saturated matrix solution). A working matrix solution was prepared by adding 100 μl of the saturated matrix solution to 200 μl of 33% acetonitrile, 67% water, and 0.1% TFA. The solution was then briefly vortexed, 0.5 μl was deposited on a sample square of a 7 by 7 spot homemade stainless steel MALDI target plate, and the matrix solution was allowed to air dry at room temperature. Finally, 0.5 μl of bacterial cell lysate-supernatant was deposited onto the dried matrix and allowed to dry. This was repeated for each bacterial lysate sample.
Mass spectrometry.
All experiments were run in the positive ion mode on a Bruker Daltonics (Billerica, MA) Reflex II instrument using the reflectron mode with delayed extraction; the delayed extraction setting was set to “medium.” Ionization of molecules in the samples was achieved with a nitrogen laser (337 nm). A minimum sum of 200 laser shots per sample was used to generate each ion spectrum used for analysis. The instrument was calibrated externally before each batch run of test samples using the adrenocorticotropic hormone clip (peptide 18-39), angiotensin II, substance P, bovine insulin, and horse heart myoglobin as standards.
Identification of SIBI proteins based on mass determined by MALDI-TOF MS.
The masses of SIBIs determined by MALDI-TOF MS were compared to the predicted molecular weights of proteins encoded by the annotated genes of C. jejuni strain NCTC 11168 (58) and C. jejuni strain RM1221 (65) for a tentative protein identification. The C. jejuni SIBI type 1 was confirmed by tryptic mass mapping (26). A tentative identification of other SIBIs was determined similarly from the partial genome sequences of C. coli strain RM2228, C. lari strain RM2100, and C. upsaliensis strain RM3195 (21).
MAbs.
Two monoclonal antibodies (MAbs) shown previously to be specific for C. jejuni (anti-Cj) or both C. jejuni and C. coli (anti-Cc/Cj) were used in some experiments in a whole-cell enzyme-linked immunosorbent assay to identify C. coli and C. jejuni strains. Strains were cultured on BA or BAB, and the bacteria were harvested and then suspended in phosphate-buffered saline (10 mM sodium phosphate, 150 mM NaCl, pH 7.4) to an optical density at 620 nm of 0.2 to 0.3. Seventy microliters of the suspension was added to polystyrene assay wells (Immulon II [Dynatech, Chantilly, Va.] or Maxisorp [Nunc, Roskilde, Denmark]). The plates were allowed to dry by incubating them for 18 to 24 h at 37°C, and they were washed three times with Tris-HCl-buffered saline containing 1% Tween 20 (TBS-Tween) or phosphate-buffered saline-Tween (diluents) and then with a final rinse of pyrogen-free water. Nonspecific binding sites on the wells were blocked by adding 200 μl of 1% casein blocking buffer and then incubating the plate at room temperature for 1 h. The plates were rinsed and washed as described above and used immediately, or they were stored in a desiccator at 4°C for later use. Dilutions of MAbs (100 μl) diluted in 1% bovine serum albumin in TBS-Tween were added to wells, and the wells were incubated with gentle shaking for 1 to 2 h at room temperature. Wells were emptied and then washed and rinsed with water. Bound MAb was detected by adding 100 μl (0.2 μg) of alkaline phosphatase-conjugated rabbit anti-mouse immunoglobulin (Zymed, South San Francisco, CA), incubating the wells for 1 h with shaking, and then washing and rinsing the wells with water. One hundred microliters of p-nitrophenylphosphate (1 mM in 10 mM diethanolamine, 0.5 mM MgCl2, pH 9.5) was added, and the absorbance at 405 nm was determined on a Spectramax 340 microplate reader (Molecular Devices, Sunnyvale, CA) (45).
Hippuricase activity.
Hippuricase activity was determined by a method described previously (24).
MLST.
Selected strains were analyzed by a multilocus sequence typing (MLST) method as described previously (52).
Fluorescent Campylobacter strains.
C. jejuni strain RM1221 carrying a plasmid encoding cyan fluorescent protein (CFP) has been described previously (50). C. lari strain RM2100 was transformed with the same plasmid encoding green fluorescent protein (GFP) by a similar method (50).
RESULTS
MALDI-TOF MS spectra obtained for Campylobacter species reference strains.
Initial MALDI-TOF MS studies were initiated with well-characterized reference C. coli and C. jejuni strains, and then the analysis was expanded to C. lari, C. upsaliensis, C. helveticus, and C. sputorum strains. In a typical analysis of Campylobacter strains by MALDI-TOF MS, 10 to 20 prominent ion peaks were noted in the spectra in the region between 6,000 and 15,000 Da, with the highest-intensity peaks consistently in the range of 9,000 to 10,300 Da. Figures 1 to 5, below, are representative generally of results obtained with multiple strains of each of the six species tested. Comparisons of spectra for multiple strains within a species revealed that multiple ions corresponded to a characteristic “fingerprint.” Samples analyzed at different times over the course of several hours after calibration yielded results that were reproducible to 0.03% (data not shown), indicating that the precision of the method was sufficient for rapid determination of Campylobacter species. Mass abundance was reflected by the height of a peak; however, peak height was not useful generally for species identification or accurate quantification of cells. Masses illustrated in Fig. 1 to 5, below, for C. jejuni, C. coli, C. lari, C. upsaliensis, C. helveticus and C. sputorum, respectively, are the values determined for an individual experiment. The molecular weights of putative SIBIs were determined by averaging the results for multiple strains tested in multiple experiments. The precision of the externally calibrated data was approximately ±10 Da.
FIG. 1.
MALDI-TOF MS spectra of whole-cell extracts of C. jejuni reference strains. Spectra representative of MS profile type 1 and profile type 2 were obtained from C. jejuni strains RM1862 (NCTC 11168) (A) and RM1438 (B), respectively. The relative intensities of the ions are shown on the y axis, and the masses (Da) of the ions are shown on the x axis.
FIG. 5.
MALDI-TOF MS spectra of whole-cell extracts of C. helveticus and C. sputorum reference strains. Spectra for C. helveticus reference strain RM3228 (A) and C. sputorum reference strain RM1485 (B) are shown. The relative intensities of the ions are shown on the y axis, and the masses (in Da) of the ions are shown on the x axis.
Designation of SIBIs from MALDI-TOF MS analysis of reference strains.
Ions detected for C. jejuni, C. coli, C. lari, C. upsaliensis, C. helveticus, and C. sputorum biovar fecalis reference strains by MALDI-TOF MS (see Fig. 1 to 5, below) were designated as a candidate initially if they occurred in an approximate relative abundance of >10% of the most abundant ion in an experiment, were at least 10 Da different from other SIBIs, and were relatively conserved for a species. For example, C. jejuni strain RM1862 (NCTC 11168) (Fig. 1A) had prominent ions at 10,275 and 13,728, whereas neither ion was present in spectra for C. coli strains RM1161 and RM1051 (Fig. 2A and B). It was observed that some ions appeared common between species, e.g., 7,035- (7,036-) and 8,153- (8,154-) Da ions occur in both C. coli and C. jejuni strains (Fig. 1 and 2), but not in C. lari and C. upsaliensis strains (Fig. 3 and 4). For C. lari and C. upsaliensis strains, minor-intensity ions nearest to the 7,035- (7,036-) and 8,153- (8,154-) Da ions occurred at ∼6,998 (C. lari) and 7,063 (C. upsaliensis) Da and at ∼8,000 Da (C. lari and C. upsaliensis), respectively.
FIG. 2.
MALDI-TOF MS spectra of whole-cell extracts of C. coli reference strains. Spectra representative of MS profile type 1 and profile type 2 were obtained from C. coli strains RM1161 (A) and RM1051 (B), respectively. The relative intensities of the ions are shown on the y axis, and the masses (in Da) of the ions are shown on the x axis.
FIG. 3.
MALDI-TOF MS spectra of whole-cell extracts of C. lari reference strains. Spectra representative of MS profile type 1 and profile type 2 were obtained from C. lari strains RM2100 (A) and RM2099 (B), respectively. The relative intensities of the ions are shown on the y axis, and the masses (in Da) of the ions are shown on the x axis.
FIG. 4.
MALDI-TOF MS spectra of whole-cell extracts of C. upsaliensis reference strains. Spectra for reference strains representative of SIBI types 1, 2, 3, and 4 were obtained with C. upsaliensis strains RM2092 (A), RM3776 (B), RM3195 (C), and RM4245 (D), respectively. The relative intensities of the ions are shown on the y axis, and the masses (in Da) of the ions are shown on the x axis. The SIBI types are designated above the singly charged ion for each strain.
MALDI-TOF analysis of C. coli, C. jejuni, and two strains of C. jejuni subsp. doylei.
Analysis of multiple reference strains of the same species, however, occasionally yielded spectra with a SIBI that varied slightly in mass from that identified initially. For example, a major peak was observed at ∼10,275 Da (designated SIBI type 1) in profiles for most C. jejuni strains (Fig. 1A), but a 10,307-Da ion (designated SIBI type 2) was the nearest mass ion observed for other strains (Fig. 1B). Similarly, a major ion of 10,031 Da (SIBI type 1), present in spectra for most C. coli strains (Fig. 2A), appeared at 10,061 Da (SIBI type 2) for at least one C. coli strain analyzed (Fig. 2B). Similar differences in ions were noted for C. lari strains at masses of 9,617 (SIBI type 1) and 9,650 (SIBI type 2) Da (Fig. 3A and B).
An ion at 13,729 Da was present in profiles for the majority of the C. jejuni reference strains and, thus, was designated a putative “common” ion (Table 2). However, this ion was missing in a few strains, and an ion of 12,887 or 12,888 Da was present instead (Table 2, RM1050, RM1158, and RM1159). It is not known whether these ions represent variant proteins or completely different proteins. However, subsequent analyses of two strains of C. jejuni subsp. doylei (RM2095 and RM2096) identified the 10,276-Da C. jejuni type 1 SIBI, but they identified a 12,888-Da ion in the absence of the 13,729-Da C. jejuni common ion (data not shown). These results suggest that some of the C. jejuni reference strains (Table 2, RM1050, RM1158, and RM1159) and animal isolates (see Table S1 in the supplemental material [RM1216, RM1420, and RM1886]) may be C. jejuni subsp. doylei. Additional well-characterized C. jejuni subsp. doylei strains would be required to clarify this subspecies assignment.
TABLE 2.
SIBIs identified for reference strains of C. coli, C. jejuni, C. lari, C. sputorum biovars and C. upsaliensis by MALDI-TOF MSa
| PSM strain no. | Species | Sourceb | Cc1 (10,032 Da) | Cc2 (10,060 Da) | Cc-C (12,855 Da) | Cj1 (10,276 Da) | Cj2 (10,303 Da) | Cj-C (13,729 Da) | Cl1 (9,619 Da) | Cl2 (9,651 Da) | Cl-C (12,972 Da) | Ch1 (10,189 Da) | Ch-C (9,504 Da) | Cu1 (10,171 Da) | Cu2 (10,197 Da) | Cu3 (10,272 Da) | Cu4 (10,229 Da) | Cu-C (9,504 Da) | Csp1 (9,797 Da) | Csp2 (9.817 Da) | Csp-C (12,786 Da) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RM1051 | C. coli | H-CN | X | Xc | X | ||||||||||||||||
| RM1161 | C. coli | H-CN | X | X | |||||||||||||||||
| RM1166 | C. coli | C-CN | X | X | |||||||||||||||||
| RM1169 | C. coli | H-CN | X | X | |||||||||||||||||
| RM2228 | C. coli | C-US | X | X | |||||||||||||||||
| RM2230 | C. coli | C-US | X | X | |||||||||||||||||
| RM2231 | C. coli | C-US | X | X | |||||||||||||||||
| RM2236 | C. coli | C-US | X | X | |||||||||||||||||
| RM1048 | C. jejuni | H-CN | X | X | |||||||||||||||||
| RM1050 | C. jejuni | H-CN | X | VRd | |||||||||||||||||
| RM1155 | C. jejuni | H-CN | X | X | |||||||||||||||||
| RM1156 | C. jejuni | H-CN | X | X | |||||||||||||||||
| RM1862 | C. jejuni | H-UK | X | X | |||||||||||||||||
| RM1158 | C. jejuni | H-CN | X | VRd | |||||||||||||||||
| RM1159 | C. jejuni | H-CN | X | VRd | |||||||||||||||||
| RM1163 | C. jejuni | H-CN | X | X | |||||||||||||||||
| RM1164 | C. jejuni | H-CN | X | X | |||||||||||||||||
| RM1165 | C. jejuni | C-CN | X | Xc | X | ||||||||||||||||
| RM1167 | C. jejuni | H-CN | X | X | |||||||||||||||||
| RM1168 | C. jejuni | H-CN | X | X | |||||||||||||||||
| RM1462 | C. lari | H-? | X | X | X | ||||||||||||||||
| RM2098 | C. lari | H-US | X | X | |||||||||||||||||
| RM2099 | C. lari | H-US | X | ||||||||||||||||||
| RM2100 | C. lari | H-US | X | X | |||||||||||||||||
| RM3228 | C. helveticus | F-SW | X | Xe | |||||||||||||||||
| RM4087 | C. helveticus | F-SW | X | X | |||||||||||||||||
| RM4088 | C. helveticus | F-S | X | X | |||||||||||||||||
| RM1485 | C. sputorum bv. fecalis | O-? | X | X | |||||||||||||||||
| RM1495 | C. sputorum bv. bulbulus | ND | X | X | |||||||||||||||||
| RM2089 | C. upsaliensis | O-? | X | X | |||||||||||||||||
| RM2090 | C. sputorum bv. fecalis | O-US | X | X | |||||||||||||||||
| RM2091 | C. sputorum bv. fecalis | O-? | X | X | |||||||||||||||||
| RM4120 | C. sputorum bv. paraureolyticus | H-CN | X | X | |||||||||||||||||
| RM4121 | C. sputorum bv. fecalis | O-UK | X | X | |||||||||||||||||
| RM1488 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM2092 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM2093 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM3937 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM3939 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM3940 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM3941 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM3942 | C. upsaliensis | H-US | X | X |
MALDI-TOF analysis of C. upsaliensis.
Four SIBIs differing by <60 Da in mass were identified for C. upsaliensis strains analyzed in this study. These C. upsaliensis SIBI types (designated 1 to 4) corresponded to ions at 10,171, 10,197, 10,272, and 10,229 Da, respectively (Fig. 4A to D). In addition, a common ion at 9,504 Da was observed for most of the C. upsaliensis reference strains (Table 2), regardless of SIBI type. Although this appeared to be a “common” C. upsaliensis SIBI, subsequent work indicated it was present also in C. helveticus strains (see below).
MALDI-TOF analysis of C. helveticus.
Three reference strains of C. helveticus, including the type strain RM3228 (ATCC 51209), were analyzed by MALDI. An ion at approximately 10,189 Da was designated the C. helveticus SIBI-1 ion, since it was identified in all 10 C. helveticus strains analyzed (a 10,189-Da ion with a range of precision of ±10 Da gives a 10,199-Da ion as shown in Fig. 5A; this spectrum is an example of the ∼10 Da difference between the mass calculated with external standardization and the actual mass). A common ion at 9,504 Da was noted for two of the three reference strains (RM4087 and RM4088) but was 28 Da lower than an ion at 9,532 Da observed consistently for the type strain of C. helveticus, RM3228 (9,541 Da − 10 Da = 9,531 Da [Fig. 5A]). Nevertheless, the C. helveticus common ion was designated 9,504 Da, since subsequent studies with additional C. helveticus strains revealed that this was the major C. helveticus common ion observed (see Table S1 in the supplemental material). As noted above, the Ch-C ion at 9,504 Da was also a common ion for C. upsaliensis SIBI types 1 to 4. Thus, observance of an ion at 9,504 Da indicated a putative C. helveticus or C. upsaliensis strain, necessitating accurate analysis of the C. helveticus SIBI at 10,189 Da and the type 2 C. upsaliensis SIBI at 10,197 Da. It is worth noting that multiple common ions at masses of <9,000 Da also were observed for all 10 C. helveticus strains analyzed (e.g., 5,094 ± 2 and 8,451 ± 8 Da), also facilitating species determination.
MALDI-TOF analysis of C. sputorum.
Four reference strains of C. sputorum biovar fecalis isolated from sheep and a single strain each of C. sputorum biovars bulbulus and paraureolyticus were analyzed by MALDI-TOF MS. A profile of the C. sputorum biovar bulbulus reference strain RM1495 is shown in Fig. 5B. Possible SIBIs and/or common ions at 9,796, 11,170, and 12,786 Da were noted in five of six C. sputorum reference strains (Table 2). The 9,796-Da ion was designated SIBI type 1 because it was present in spectra for each of the four C. sputorum biovar fecalis strains and the single C. sputorum biovar bulbulus strain, and it generally was of high intensity. A comparable ion at 9,817 Da (∼20-Da difference) was present for the single C. sputorum biovar paraureolyticus strain (RM4120). Similarly an ion at 11,203 Da was present for C. sputorum biovar paraureolyticus instead of the 11,170-Da ion (∼30-Da difference). Both of these ions may be useful as C. sputorum type 1 SIBIs.
Strain RM2089 was received for analysis as a C. sputorum biovar fecalis strain, but our analysis determined it to be C. upsaliensis based on a prominent C. upsaliensis SIBI at 10,181 Da and a C. upsaliensis/C. helveticus common ion at 9,509 Da. This strain was confirmed subsequently as C. upsaliensis in an expanded MLST system developed for genotyping Campylobacter (52).
Additional ions that appeared to be potential SIBIs were observed for each of the six species, e.g., 9,459 (or 9,460) Da for C. jejuni, 8,573 Da for C. coli, 10,212 (to 10,216) Da for C. lari, 11,135 to 11,139 Da for C. upsaliensis; 8,452 to 8,460 and 15,100 to 15,130 for C. helveticus, and 11,170 Da for C.sputorum (Fig. 1 to 5). These ions were not selected as prototype SIBIs because they were not as clearly a species-identifying ion, were of lesser abundance, and/or were not as conserved in a species as the ions selected as SIBIs.
MALDI-TOF analysis of putative or suspected C. coli and C. jejuni strains.
More than 50 strains suspected of being C. coli or C. jejuni, plus recent isolates from environmental sources (including poultry and swine carcasses and dog and cat feces), were analyzed by MALDI-TOF MS, and the results were analyzed to determine species (see Table S1 in the supplemental material). For the C. coli and C. jejuni strains, the MS results were compared also to results obtained with anti- C. coli and anti-C. jejuni specific MAbs and a hippuricase biochemical assay (see Table S1). Either a C. coli or a C. jejuni SIBI was identified for all suspected C. coli or C. jejuni strains, resulting in unambiguous identification of the species.
Identification of mixed C. coli and C. jejuni “strains.”
For most experiments, data were consistent with the immunochemical and biochemical results (see Table S1 in the supplemental material). However, inconsistent results were observed in the spectrum for “C. jejuni” strain RM1882 (Fig. 6). Close examination of the profile revealed that both a C. coli and a C. jejuni SIBI were present in the sample assumed to be a pure culture of C. jejuni (Fig. 6). Eight single-colony picks were obtained for “C. jejuni” RM1882 and tested separately for hippuricase activity. Six of the eight single-colony picks were positive for hippuricase activity, suggesting that “strain” RM1882 was a mixture of C. jejuni and C. coli or that hippuricase-negative C. jejuni was present. The hippuricase-positive and hippuricase-negative isolates were analyzed, and the immunochemical and MALDI-TOF MS results were consistent with the presence of both C. coli and C. jejuni colonies: the isolated strains expressed either a C. coli or a C. jejuni SIBI, but not both (data not shown).
FIG. 6.
MALDI-TOF MS spectra of a whole-cell extract of C. coli and C. jejuni cells in a sample expected to be a pure culture of either C. coli or C. jejuni. For strain RM1882, both C. coli and C. jejuni SIBIs (*) and common ions (**) were evident in the spectrum (also see Table S1 in the supplemental material).
MALDI-TOF MS of C. lari strains.
Four strains received as either C. coli or C. jejuni (RM1887 to RM1890) (Table 1; see also Table S1 in the supplemental material) were of particular interest because they lacked the characteristic C. coli and C. jejuni SIBI. The spectrum for strain RM1889 contained prominent ions at 6,995, 9,648, 10,212, 11,250, and 12,970 Da (see spectra in Fig. 3 for other C. lari). In subsequent studies, all four strains were confirmed as predominately C. lari by comparison with MS data obtained with C. lari reference strains (Fig. 3). A minor C. jejuni SIBI (10,278-Da) peak was observed in the spectra for C. lari RM1890 (see Table S1). No attempt was made to isolate a possible C. jejuni from this sample, since MLST results did not reveal evidence of any other species contaminating RM1890 (data not shown).
MALDI-TOF MS of C. helveticus and C. upsaliensis strains from cats and dogs.
Thirteen strains of suspected C. helveticus or C. upsaliensis isolated in our laboratory from dog and cat feces by a membrane filtration method were analyzed with MALDI-TOF MS. Initial results suggested that at least six of the strains had a prominent type 1 C. upsaliensis SIBI (10,171 Da); the remaining seven strains had a prominent ion ranging from 10,188 to 10,194 Da, a mass closest to the type 2 C. upsaliensis SIBI (10,197 Da). In addition, the C. upsaliensis common ion was identified for 12 of 13 strains (see Table S1 in the supplemental material). A 16S rRNA analysis indicated that seven strains with the 10,188- to 10,194-Da ions were either C. upsaliensis or C. helveticus, but the exact species could not be determined based on comparisons to C. helveticus and C. upsaliensis 16S data in GenBank (4). However, analyses of these strains by an MLST method developed by our laboratory confirmed them as C. helveticus (see Table S1) (52). The common ion identified for C. upsaliensis strains (9,504 Da) was expressed also by C. helveticus strains and, fortuitously, was not observed in profiles for other Campylobacter species. Therefore, the 9,504-Da ion (C. upsaliensis/C. helveticus) and accurate determination of the C. upsaliensis and C. helveticus SIBIs (±1 Da with internal standard calibration) allowed differentiation of these two closely related species. All of the C. helveticus strains in this set were isolated from cats; three C. upsaliensis strains each were isolated from cats and dogs (Table 1; see also Table S1). A C. upsaliensis type 1 SIBI (10,171 Da) was present also in profiles of multiple analyses of putative C. helveticus strain RM3807, indicating either that it was composed of a mixture of C. upsaliensis and C. helveticus cells or that both types of SIBIs can be expressed by this strain (e.g., a chimera). Analyses of additional C. helveticus strains will be required to determine whether this C. upsaliensis-specific SIBI can occur in other C. helveticus strains.
Detection sensitivity of C. coli and C. jejuni in a mixture.
To determine the detection sensitivity of C. coli or C. jejuni in mixed cultures under conditions similar to those encountered for analyses of unknown strains, C. jejuni strain RM1221 and C. coli strain RM2228 were analyzed as a mixture at different proportions of each strain. Cells were removed from agar, combined in proportions varying from 10 to 100% for each strain, and then tested by MALDI-TOF MS. The results shown in Fig. 7 illustrate that the intensity of the C. jejuni and C. coli SIBI (at 10,277 Da [C. jejuni SIBI type 1] and 10,063 Da [C. coli SIBI type 2]) corresponded directly to the concentration of each strain. These results suggest that, at least under these ideal experimental conditions, different species of Campylobacter with SIBIs distinguishable by MALDI-TOF MS can be detected when the strain in lowest concentration is at least 5 to 10% of the mixture.
FIG. 7.
Detection of a C. jejuni biomarker ion in a mixture of C. jejuni strain RM1221 and C. coli strain RM2228 whole cells. C. coli and C. jejuni cells were suspended in different proportions ranging from 0 to 100% of the total. The results are plotted as the percentage of the intensity of the C. jejuni biomarker ion measured by MS compared to the percentage of C. jejuni in the mixture.
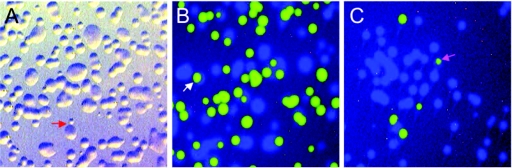
It is worth noting that a single sample of bacteria harvested from agar plates likely will not reflect exactly the overall proportion of different species and strains in a mixed culture due to heterogeneity in the distribution on a plate. Different Campylobacter species and strains mixed in complex environmental samples can be difficult, if not impossible, to distinguish on selective or nonselective media. To illustrate the potential colony morphology encountered on plates sampled for analysis, a C. jejuni strain and a C. lari strain expressing CFP (C. jejuni-CFP) or GFP (C. lari-GFP), respectively, were mixed in different proportions (1:1 C. lari-GFP/C. jejuni-CFP [Fig. 8B]; 1:10 C. lari-GFP/C. jejuni-CFP [Fig. 8C]), plated, and incubated, and then growth was examined under a fluorescence stereomicroscope. C. jejuni (Fig. 8) often grew as large and heterogeneously shaped colonies that result from swarming motility. In contrast, C. lari growth was present predominantly in small, round colonies (Fig. 8A to C). C. lari-GFP colonies were observed often surrounded, but not covered, by confluent C. jejuni-CFP growth, or on top of confluent C. jejuni growth. Colonies of C. lari and C. jejuni growing next to one another and indistinguishable under regular light were sometimes observed (Fig. 8C, top). No sectored colonies corresponding to a mixed strain “CFU” were observed in this experiment. These results reflect the importance of proper sampling and analysis of multiple samples to obtain a thorough assessment of the diversity of Campylobacter strains and species that may be present in either clinical or environmental samples.
FIG. 8.
Mixed culture of C. jejuni-CFP and C. lari-GFP on BAB medium. C. jejuni-CFP and C. lari-GFP cells were harvested from BAB medium and suspended in broth in proportions of 1:1 (B) or 10:1 (C), respectively, mixed, and then plated on BAB. The plates were incubated for 30 h, and growth was observed under a fluorescence stereomicroscope (Leica MZIII). (A) No filters; the image shows growth of a 1:1 proportion of C. jejuni and C. lari. Note the larger colonies of C. jejuni-CFP (blue) compared to C. lari-GFP (green) (B and C). The red arrow points to a C. lari colony next to a C. jejuni colony (A); the white arrow in panel B points to a C. lari-GFP colony on top of a C. jejuni-CFP colony; the violet arrow in panel C points to a small C. lari-GFP colony on top of C. jejuni-CFP growth.
Consistency of spectra of C. jejuni strain RM1221 after growth on different media and under different atmospheric conditions.
To determine whether the SIBI expression was stable under different culture conditions, C. jejuni strain RM1221 was subcultured on three different media under two atmospheric conditions at two different temperatures, and samples of bacteria were analyzed by MALDI-TOF MS. The masses determined for the C. jejuni SIBI type 1 (10,276 Da), the C. jejuni common ion (13,729 Da), and a third ion at ∼9,551 Da (probable 30S ribosomal protein S17) were very reproducible for samples of C. jejuni RM1221 grown under each of nine combinations of conditions (see Table S2 in the supplemental material). No significant differences were noted in the mass of high-intensity ions, regardless of the media (e.g., blood), gas mixture (5% or 10% CO2 and 5% H2 or 5% O2), or incubation temperature (37°C versus 42°C), as shown in the following results: C. jejuni SIBI type 1 = 10,276.0 ± 1.3; C. jejuni common SIBI = 13,729.0 ± 1.6; “ion 2” = 9,551.0 ± 1.4. In addition, eight replicate samples from a plate of C. coli RM2228, C. lari RM2100, and C. upsaliensis RM3195 grown under a single condition (BAB medium, 10% CO2-5% H2, 37°C) were analyzed (see Table S2). The standard deviations for the replicates for the three ions for the three strains ranged from 0.9 (C. coli SIBI type 1) to 3.1 (C. upsaliensis ion 2), reflecting good intraexperimental reproducibility. A similar experiment with three different strains of C. helveticus (RM3228, RM4087, and RM4088) grown under the same conditions resulted in average values of 10,188.3 ± 1.4, 9,522.1, and 9,504.0 ± 0.7 for the C. helveticus SIBI, C. helveticus common SIBI for RM4087 and 4088, and C. helveticus common SIBI for RM3228, respectively, for six samples total (two for each strain) for each ion (data not shown). Two strains of C. sputorum biovar fecalis (RM1495 and RM4121) and C. sputorum biovar paraureolyticus strain RM4120 were analyzed in a second experiment with replicate samples. The average values for the SIBI and common SIBI for the two C. sputorum biovar fecalis strains were 9,797.1 ± 1.26 and 11,167 ± 2.61, respectively; for the single C. sputorum biovar paraureolyticus strain the values were 9,813.7 ± 0.1 and 11,193 ± 0.4 (data not shown). These values reflect the potential difference between C. sputorum biovar fecalis/C. sputorum biovar bulbulus and C. sputorum biovar paraureolyticus described above (Table 2), although additional strains of each biovar would need to be tested to confirm this biovar-specific difference.
These results indicate that conventional growth conditions do not influence markedly the mass of ions detected. It is worth noting, however, that horse blood-related proteins apparently incorporated by the cells or into CFU from the underlying blood agar medium were identified in some profiles. This was confirmed in control experiments with samples of BAB medium extracted directly from plates without bacteria (e.g., 14,890, 15,050, 15,100, and 16,075 Da). We suspected that the 16,075-Da ion corresponded to the β-chain protein of horse hemoglobin (61). This was exploited in later experiments as a useful internal protein standard of an appropriate mass for calibration, thus facilitating extremely accurate mass assignments of SIBIs (±1 Da) for identification of C. helveticus, C. upsaliensis, and C. sputorum biovar fecalis strains (see Table S1 in the supplemental material).
DISCUSSION
We have presented data that multiple Campylobacter species grown on solid medium under a variety of conditions can be analyzed by MALDI-TOF MS to yield high-intensity and, in most cases, intact protein ions in the 9- to 14-kDa range that are diagnostic of Campylobacter species. Pure cultures of Campylobacter can be analyzed rapidly with less ambiguous results, compared to many methods used currently for confirming species. An added advantage of MALDI-TOF MS is that multiple species of Campylobacter in mixed cultures can be identified more easily by MS than by conventional methods; reproducibility of the MALDI-TOF MS protein profiles for C. coli and C. jejuni strains was sufficient to allow species identification of isolates.
Summary of Campylobacter SIBI types.
The different types of SIBIs, their approximate masses, and the candidate proteins identified for each ion for six species of Campylobacter are summarized in Table 3. A comparison of the SIBI masses illustrates that in most cases there are sufficient differences between SIBIs (>12 to 15 Da) for accurate identification of isolates and mixed species cultures by MALDI-TOF MS. However, there are three sets of SIBIs that are either identical (C. helveticus and C. upsaliensis common ions) or very similar (<10-Da difference for C. helveticus type 1 and C. upsaliensis type 2 and for C. jejuni type 1 and C. upsaliensis type 3) that require very accurate mass assignments. The level of accuracy required to differentiate these species was achieved by internal calibration with standard proteins or blood proteins from the medium.
TABLE 3.
Summary of SIBIs for six Campylobacter species
| Species | SIBI type | Observed mass (Da)a | Confirmed, or possible, protein identityb | Predicted mass (Da) |
|---|---|---|---|---|
| C. coli | Common | 12,855 | 50S ribosomal, L7/L12 (possible), posttranslationally modifiedc | 12,985 |
| C. coli | 1 | 10,032 | DNA binding, HU homolog | 10,030 |
| C. coli | 2 | 10,060 | DNA binding, HU homolog | 10,060j |
| C. jejuni | Common | 13,729 | 30S ribosomal, S13 subunit | 13,735 |
| C. jejuni | 1 | 10,276d | DNA binding, HU homolog | 10,274 |
| C. jejuni | 2 | 10,303 | DNA binding, HU homologe | 10,304j |
| C. lari | Common | 12,972 | 50S ribosomal, L7/L12 or L18 (possible), posttranslationally modifiedf | 13,099/13,100 |
| C. lari | 1 | 9,619 | GroES chaperonin (cpn10) | 9,617 |
| C. lari | 2 | 9,651 | Conserved hypothetical | 9,655 |
| C. sputorum biovars | Common | 12,786 | Not determined | Unknown |
| C. sputorum biovars | 1 | 9,796g | Not determined | Unknown |
| C. helveticus | Common | 9,504 | Not determined | Unknown |
| C. helveticus | 1 | 10,189h | DNA binding, HU homolog | 10,187j |
| C. upsaliensis | Common | 9,504 | Conserved hypothetical | 9,503 |
| C. upsaliensis | 1 | 10,171 | DNA binding, HU homolog | 10,172j |
| C. upsaliensis | 2 | 10,197 | DNA binding, HU homolog | 10,199j |
| C. upsaliensis | 3 | 10,272i | DNA binding, HU homolog | 10,271 |
| C. upsaliensis | 4 | 10,229 | DNA binding, HU homolog | 10,227j |
Masses shown in boldface type denote ions of <10 Da different from a SIBI for another species: C. helveticus common and C. upsaliensis common (0 Da), C.helveticus type 1 and C. upsaliensis type 2 (8 Da), C. jejuni type 1 and C. upsaliensis type 3 (4 Da).
The potential protein was determined from the predicted protein masses for genes annotated for the completed genomes of C. jejuni strains NCTC 11168 (see Parkhill et al. [58]) and partial genome sequences of C. coli RM2228 (C. coli SIBI type 1), C. lari RM2100 (C. lari SIBI type 1), and C. upsaliensis RM3195 (C. upsaliensis SIBI type 3) (65).
A predicted protein mass was present in the Cc RM2228 database at 12,851 Da. However, the best candidate protein for this Cc SIBI is a ribosomal protein minus the N-terminal methionine (12,985 Da minus 131 Da = 12,854 Da).
The putative gene encoding this protein is designated Cj0913c for C. jejuni strain NCTC 11168. The identity of this protein was confirmed by tryptic mass mapping analysis (data not shown).
A cytochrome c oxidase protein (cbb3 type, subunit IV) of predicted mass 10,303 Da is annotated in C. jejuni RM1221; however, a high-copy-number HU protein with one or more single amino acid changes is a more probable candidate for the type 2 C. jejuni SIBI.
No predicted protein mass was present in the C. lari RM2100 database within ±10 Da of 12,972. Two candidates for this C. lari SIBI are two C. lari ribosomal proteins with the loss of the N-terminal methionine (13,099 or 13,100 Da minus 131 Da = 12,968/12,969 Da).
The ion nearest in mass for a single strain of C. sputorum bv. paraureolyticus (RM4120) was 9,817 Da.
The C. helveticus strains were presumed to be C. upsaliensis strains (see Table S1 in the supplemental material). The predicted mass for this SIBI type is an actual mass for strain RM3807 determined by tryptic mass mapping analysis (18).
The identity of this protein was confirmed by tryptic mass mapping analysis.
Determined by hup sequencing (18).
The identification of Campylobacter species based on the presence or absence of biomarker proteins in the selected mass range from 9 to 14 kDa was 100% accurate for the 76 samples analyzed in our semiblinded study (see Table S1 in the supplemental material). Generally, the species of the strain could be determined by the presence of a single SIBI (e.g., Table 2, SIBI types). However, additional slightly higher mass ions were usually present to assist in confirming the species (e.g., “common” ions).
The approach to species identification for Campylobacter strains presented in this study is based on the presence or absence of SIBIs. However, we have noted variation in the SIBI masses for certain strains, arising from possibly nonsynonymous mutations in genes encoding the SIBIs and resulting in protein mass shifts that allow some limited differentiation of the strains. For example, a 10,276-Da ion was present in 13 of 14 strains (93%) of the reference strain set of C. jejuni (Table 2) used to identify potential SIBIs (a 10,303-Da ion occurred in one strain, C. jejuni RM1165). However, 6 of 31 (19%) putative C. jejuni strains tested in the semiblinded samples (see Table S1 in the supplemental material) expressed the ∼10,303-Da ion (SIBI type 2). Six of the seven SIBI type 2 C. jejuni strains were isolated from chickens (Canada, Texas, and California), possibly indicating an association of SIBI type with source.
Table 2 and also Table S1 in the supplemental material include only a subset of the ions detected in both C. coli and C. jejuni strains. Although the C. jejuni 13,729-Da ion occurred at approximately the same mass in most of the C. jejuni strains, it was absent in six human and animal C. jejuni strains and was replaced by an ∼12,884- to 12,888-Da ion (∼30 Da higher than the C. coli common ion) (Table 2; see also Table S1 in the supplemental material). The “12,888 ion” was identified also in the only C. jejuni subsp. doylei strains we had available for analysis (both human clinical strains). Similarly, the C. coli SIBI at 12,857 Da was absent in a few strains tested, and an ion at 12,914 Da was observed (57-Da difference). These results indicate the importance of identifying multiple SIBIs for accurate analysis and identification and the potential value of MALDI-TOF analysis for identifying subspecies/biovar differences in Campylobacter.
In a previous study of a few C. coli and C. jejuni strains by MALDI-TOF MS, biomarker ions were identified at 10,074 Da for C. coli strain ATCC 43474 and at 10,285 Da for C. jejuni strain ATCC 43464 (73). It is probable that these ions correspond to the C. coli type 2 (10,060 Da) and C. jejuni type 1 (10,276 Da) SIBIs identified in our study. The 9- and 14-Da mass differences for these ions in the two studies may reflect an actual difference (e.g., amino acid polymorphism or posttranslational modification) or differences in measurement precision. However, our analysis of the same C. coli strain as the previous study (RM1878) (Table 1; see also Table S1 in the supplemental material) suggests the difference is due to precision.
Identities or putative identities of some SIBIs.
Many of the SIBIs identified in this study are putatively abundant, cytosolic proteins, of relatively low mass compared to the majority of proteins, and apparently conducive to ionization in MALDI-TOF MS (Table 3, HU-homolog, ribosomal subunit, and chaperonin proteins). A C. jejuni HU homolog encoded by the hup gene was characterized previously by Konkel et al. and designated HCj (33). The calculated molecular mass of HCj was 10,267 Da, with a pI of 10.1. Presumably, this small, highly basic, histone-like protein has properties similar to other HU proteins with regards to abundance (30,000 to 50,000 HU dimers per cell in E. coli) and DNA binding and condensing activity (14). It is probable that the high intensity of HU/HCj and possible HCj-homologs in other Campylobacter species is due to a combination of similar high abundance plus efficient ionization of these highly basic proteins. In addition, we speculate that cytosolic proteins whose functions and sequences have been highly conserved through evolution are good candidates for species identification, although of limited utility for strain differentiation. It will be interesting to determine whether these same high-intensity ions identified by MALDI-TOF MS (e.g., the HU protein) are good predictors of other Campylobacter species (18) and bacterial species of other genera.
The possible identities for some of the SIBIs shown in Table 3 must be considered speculative until confirmed by tryptic mass mapping experiments. For example, the C. coli and C. lari common SIBIs have been designated as 50S ribosomal L7/L12 proteins (RplL), even though the predicted protein is ∼130 Da larger than that observed by MALDI-TOF MS. This is because no predicted protein mass closer than 10 Da was present in the genome database, suggesting that post- or cotranslational modification had occurred, resulting in removal of the N-terminal methionine (131 Da) by a specific aminopeptidase (13, 27).
Polymorphisms and variation of SIBIs.
Ongoing genetic and mass mapping studies in our laboratory indicate that the mass differences between SIBI types, in fact, are due to nonsynonymous single or multiple nucleotide polymorphisms, rather than differences due to sensitivities of methods (18). These SIBI types representing authentic nonsynonymous mutations will be useful for population analyses of strains. Strains related epidemiologically (e.g., from an outbreak or prevalent in a single source) can be identified based on a pattern of small mass differences reflecting single or multiple amino acid changes that are different from other strains. For example, all of the C. upsaliensis strains from the United States were C. upsaliensis SIBI type 1 (∼10,171 Da), in contrast to six of seven C. upsaliensis strains from South Africa, which were C. upsaliensis SIBI type 2 (∼10,197 Da). However, of the 11 C. upsaliensis reference strains from Belgium, two, three, and six strains represented SIBI types 1, 2, and 4, respectively. The common ion at 9,504 Da was observed in all of the C. upsaliensis MALDI-TOF MS profiles. The four C. upsaliensis SIBI types are consistent with the genomic heterogeneity reported previously for animal and human (8, 40, 57) and, specifically, dog and cat C. upsaliensis strains (40, 54). Twelve of 13 dog and cat C. upsaliensis strains in our study were from the United States, and all were C. upsaliensis SIBI type 1. Although SIBI types provide probably only a minor discrimination of strains, it will be interesting eventually to compare SIBI types with sequence types determined by a new MLST method for C. coli, C. lari, C. helveticus, and C. upsaliensis (52).
As we expand our MALDI-TOF MS analyses to include other species of Campylobacter, it is anticipated that protein homologs to the SIBIs described in this study will be identified as biomarkers. For example, we have identified recently a 10.5-kDa protein as the HU protein, and a potential SIBI, for C. concisus strains (18). The probability will increase that SIBIs in different species with similar masses (∼10 Da) will be identified, again emphasizing the need for accurate mass assignments. As noted above, the identification of medium-derived horse hemoglobin β-chain in some experiments permitted recalibration of data, yielding ±1 Da accuracy that proved to be essential for differentiating the C. helveticus SIBI (∼10,189 Da) from a C. upsaliensis type 2 SIBI (∼10,197 Da). Thus, media proteins distinct from bacterial proteins, or addition of a reference protein to each sample, can be exploited for internal calibration to improve precision and accuracy of mass measurement.
The correct species designations for >130 strains analyzed in this study were identified usually by identifying only two SIBIs: a single SIBI among multiple variant SIBIs (e.g., C. upsaliensis), plus a SIBI common among all or most strains of a species. However, it is worth emphasizing that additional SIBIs are observed in the mass range of 7 to 15 kDa that permit confirmation of species when SIBIs for different species are of similar mass (±10 Da) or are variable among environmental strains of the same species. Therefore, identifying a set of SIBIs would be advantageous for unambiguous assignments of species and important intraspecies strain differences.
As more organisms are sequenced and the data are entered into public databases (e.g., NCBI GenBank), the accuracy in identification and analysis of unknown isolates by proteomics approaches also will increase (12, 13, 19). In a single study reporting species identification by querying ion masses against protein databases (19), organisms could be identified only to the genus level. In our study, and other studies, isolates are classified tentatively as members of a particular genus and/or species through both sample source and the special selection and enrichment conditions involved in obtaining them. MALDI-TOF MS has proved to be useful for rapid confirmation of species.
Environmental samples and mixed species cultures.
Selective media for isolation of C. coli and C. jejuni from clinical or environmental (animals, food, and water) samples commonly include antibiotics for minimizing other microbial flora. The unintended consequence of this approach is that antibiotic-sensitive strains of C. coli and C. jejuni and other Campylobacter species are not culturable. MALDI-TOF MS speciation is very efficient with pure cultures of Campylobacter. However, we have shown also that multiple Campylobacter species can be identified with a preparation of Campylobacter cells growing confluently, or as single colonies, on common agar media (Fig. 8). We are developing an approach currently to analyze multiple colonies of suspected Campylobacter species obtained following passage through 0.6-μm filters and growth on nonselective culture medium in the appropriate atmosphere (22, 39). Since multiple strains of a majority of Campylobacter, Helicobacter, and other spiral-form bacteria could be present and difficult to distinguish, MALDI-TOF MS analysis provides a simple method for identifying potential Campylobacter species, and also potential mixed-species colonies (Fig. 8), as has been reported previously (17, 50). We confirmed numerous times during this study that “isolates” received from other sources were either speciated incorrectly or contained multiple species.
Intraspecies variability of the Campylobacter SIBIs, potentially due to single or multiple amino acid sequence changes, has been identified; these data are not attainable rapidly in most other assays. MALDI-TOF MS may be especially useful for characterizing non-C. coli and non-C. jejuni strains because of the minimal genetic and biochemical data available for emerging Campylobacter species (10). The recent increase in genomic sequence data available for Campylobacter species will enhance MALDI-TOF MS as a method of analysis for Campylobacter (21, 65), since accurate protein masses yield gene and protein identifications for strain differentiation and further analysis.
ADDENDUM IN PROOF
We have recently created a database of all of the MALDI-TOF MS data acquired in our laboratory for Campylobacter species and other species of bacteria, as well as a program (M2MASS) for comparing all of the ions detected for a strain with ions obtained for every other strain represented in the database. This analysis yields a score for determining the similarities between mass data among strains and assigning species. Within the program, the masses of selected ions can be compared to predicted masses for genes in public and PSMRU sequence databases, facilitating comparisons of large amounts of data and, thus, increasing the accuracy of the conclusions.
Supplementary Material
Table 2a.
| RM3943 | C. upsaliensis | H-US | X | X | |||||||||||||||||
| RM3945 | C. upsaliensis | D-US | X | X | |||||||||||||||||
| RM3946 | C. upsaliensis | D-US | X | X | |||||||||||||||||
| RM3947 | C. upsaliensis | D-US | X | X | |||||||||||||||||
| RM3948 | C. upsaliensis | D-US | X | X | |||||||||||||||||
| RM3949 | C. upsaliensis | D-US | X | X | |||||||||||||||||
| RM3950 | C. upsaliensis | D-US | X | X | |||||||||||||||||
| RM4124 | C. upsaliensis | D-SW | X | X | |||||||||||||||||
| RM4249 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4252 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM3776 | C. upsaliensis | H-SA | X | X | |||||||||||||||||
| RM3777 | C. upsaliensis | H-SA | X | X | |||||||||||||||||
| RM4039 | C. upsaliensis | H-SA | X | X | |||||||||||||||||
| RM3778 | C. upsaliensis | H-SA | X | X | |||||||||||||||||
| RM4040 | C. upsaliensis | H-SA | X | X | |||||||||||||||||
| RM4042 | C. upsaliensis | H-SA | X | X | |||||||||||||||||
| RM4244 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4246 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4254 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM3195 | C. upsaliensis | H-SA | X | X | |||||||||||||||||
| RM4245 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4248 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4258 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4250 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4257 | C. upsaliensis | H-BE | X | X | |||||||||||||||||
| RM4258 | C. upsaliensis | H-BE | X | X |
SIBIs identified for each strain are designated with an X. Multiple SIBIs were identified for most species. SIBI type 1 (e.g., Cc1) refers to the biomarker ion observed consistently among the strains tested of the designated species and the ion mass; SIBI type 2 (e.g., Cc2) designates a second high-intensity SIBI. The same designations apply for the other SIBIs.
Abbreviations: H, human; C, chicken; F, feline; O, ovine; D, canine; CN, Canada; US, United States; SW, Switzerland; S, Sweden; BE, Belgium; SA, South Africa; UK, United Kingdom; ND, not determined.
Two biomarker ions were present for this “strain.”
VR, variant result. No C. jejuni common ion was identified; a 12,887- or 12,888-Da ion was noted.
Ch-C ion for RM3228 (C. helveticus type strain) was 9,532 Da, ∼28 Da higher than the Ch-C observed for other C. helveticus strains.
Acknowledgments
Strains used in this study were provided generously by Sharon Abbott, Paula Cray, Mark Englen, Patricia Guerry, Roger Harvey, Al Lastovica, Rick Meinersmann, Mabel Nicholson, Stephen L. On, Larry Stanker, Peter Vandamme, Irene Wesley, David Woodward, and the California Dept. of Health Services—Los Angeles Microbiology Laboratory. Andrew Lieberman wrote the software for comparing and correlating data from multiple spectra (BACMASS). We thank Brandon Garbus for creating a database of MS data acquired by the PSMRU and software for web-based access and data analysis. We thank William Vensel for assistance in MS, Feli Bautista for help in preparing samples of bacteria, and David Brandon for collaboration in production of MAbs.
This work was supported by the USDA Agricultural Research Service CRIS project 5325-42000-041 and supports a U.S. collaboration in the European Commission Fifth Framework Project QLK1-CT-2002-02201, “CAMPYCHECK.”
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbott, S. L., M. Waddington, D. Lindquist, J. Ware, W. Cheung, J. Ely, and J. M. Janda. 2005. Description of Campylobacter curvus and C. curvus-like strains associated with sporadic episodes of bloody gastroenteritis and Brainerd's diarrhea. J. Clin. Microbiol. 43:585-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acuff, G. R. 1992. Media, reagents, and stains, p. 1093-1208. In C. Vanderzant and D. F. Splittstoesser (ed.), Compendium of methods for the microbiological examination of foods. American Public Health Association, Washington, D.C.
- 3.Al Rashid, S. T., I. Dakuna, H. Louie, D. Ng, P. Vandamme, W. Johnson, and V. L. Chan. 2000. Identification of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, Arcobacter butzleri, and A. butzleri-like species based on the glyA gene. J. Clin. Microbiol. 38:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, R. J., J. A. Karty, A. D. Ellington, and J. P. Reilly. 1999. Monitoring the growth of a bacteria culture by MALDI-MS of whole cells. Anal. Chem. 71:1990-1996. [DOI] [PubMed] [Google Scholar]
- 6.Bang, D. D., A. Wedderkopp, K. Pedersen, and M. Madsen. 2002. Rapid PCR using nested primers of the 16S rRNA and the hippuricase (hipO) genes to detect Campylobacter jejuni and Campylobacter coli in environmental samples. Mol. Cell. Probes 16:359-369. [DOI] [PubMed] [Google Scholar]
- 7.Bourke, B., V. L. Chan, and P. Sherman. 1998. Campylobacter upsaliensis: waiting in the wings. Clin. Microbiol. Rev. 11:440-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourke, B., P. M. Sherman, D. Woodward, H. Lior, and V. L. Chan. 1996. Pulsed-field gel electrophoresis indicates genotypic heterogeneity among Campylobacter upsaliensis strains. FEMS Microbiol. Lett. 143:57-61. [DOI] [PubMed] [Google Scholar]
- 9.Brondz, I., and I. Olsen. 1991. Multivariate analyses of cellular fatty acids in Bacteroides, Prevotella, Porphyromonas, Wolinella, and Campylobacter spp. J. Clin. Microbiol. 29:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CAMPYCHECK. 2003. Improved physiological, immunological and molecular tools for the recovery and identification of emerging Campylobacteraceae (CAMPYCHECK): a European Commission research project (QLK1 CT 2002 02201). [Online.] www.campycheck.org.
- 11.Claydon, M. A., S. N. Davey, V. Edwards-Jones, and D. B. Gordon. 1996. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 14:1584-1586. [DOI] [PubMed] [Google Scholar]
- 12.Demirev, P. A., Y. P. Ho, V. Ryzhov, and C. Fenselau. 1999. Microorganism identification by mass spectrometry and protein database searches. Anal. Chem. 71:2732-2738. [DOI] [PubMed] [Google Scholar]
- 13.Demirev, P. A., J. S. Lin, F. J. Pineda, and C. Fenselau. 2001. Bioinformatics and mass spectrometry for microorganism identification: proteome-wide post-translational modifications and database search algorithms for characterization of intact H. pylori. Anal. Chem. 73:4566-4573. [DOI] [PubMed] [Google Scholar]
- 14.Drlica, K., and J. Rouviere-Yaniv. 1987. Histone-like proteins of bacteria. Microbiol. Rev. 51:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dronda, F., I. Garcia-Arata, E. Navas, and L. de Rafael. 1998. Meningitis in adults due to Campylobacter fetus subspecies fetus. Clin. Infect. Dis. 27: 906-907. [DOI] [PubMed] [Google Scholar]
- 16.Easterling, M. L., C. M. Colangelo, R. A. Scott, and I. J. Amster. 1998. Monitoring protein expression in whole bacterial cells with MALDI time-of-flight mass spectrometry. Anal. Chem. 70:2704-2709. [DOI] [PubMed] [Google Scholar]
- 17.Englen, M. D., and P. J. Fedorka-Cray. 2002. Evaluation of a commercial diagnostic PCR for the identification of Campylobacter jejuni and Campylobacter coli. Lett. Appl. Microbiol. 35:353-356. [DOI] [PubMed] [Google Scholar]
- 18.Fagerquist, C. K., W. G. Miller, L. A. Harden, A. H. Bates, W. H. Vensel, G. Wang, and R. E. Mandrell. 2005. Genomic and proteomic identification of a DNA-binding protein used in the “fingerprinting” of Campylobacter species and strains by MALDI-TOF-MS protein biomarker analysis. Anal. Chem. 77:4897-4907. [DOI] [PubMed]
- 19.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 20.Fermer, C., and E. O. Engvall. 1999. Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis.J. Clin. Microbiol. 37:3370-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:72-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goossens, H., B. Pot, L. Vlaes, C. Van den Borre, R. Van den Abbeele, C. Van Naelten, J. Levy, H. Cogniau, P. Marbehant, J. Verhoef, et al. 1990. Characterization and description of “Campylobacter upsaliensis” isolated from human feces. J. Clin. Microbiol. 28:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goossens, H., L. Vlaes, M. De Boeck, B. Pot, K. Kersters, J. Levy, P. De Mol, J. P. Butzler, and P. Vandamme. 1990. Is “Campylobacter upsaliensis” an unrecognised cause of human diarrhoea? Lancet 335:584-586. [DOI] [PubMed] [Google Scholar]
- 24.Hani, E. K., and V. L. Chan. 1995. Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase (hippuricase) gene in Escherichia coli. J. Bacteriol. 177:2396-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmon, K. M., G. M. Ransom, and I. V. Wesley. 1997. Differentiation of Campylobacter jejuni and Campylobacter coli by multiplex polymerase chain reaction. Mol. Cell. Probes 11:195-200. [DOI] [PubMed] [Google Scholar]
- 26.Henzel, W. J., T. M. Billeci, J. T. Stults, S. C. Wong, C. Grimley, and C. Watanabe. 1993. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc. Natl. Acad. Sci. USA 90:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirel, P. H., M. J. Schmitter, P. Dessen, G. Fayat, and S. Blanquet. 1989. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. USA 86:8247-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland, R. D., C. R. Duffy, F. Rafii, J. B. Sutherland, T. M. Heinze, C. L. Holder, K. J. Voorhees, and J. O. Lay, Jr. 1999. Identification of bacterial proteins observed in MALDI TOF mass spectra from whole cells. Anal. Chem. 71:3226-3230. [DOI] [PubMed] [Google Scholar]
- 29.Ichiyama, S., S. Hirai, T. Minami, Y. Nishiyama, S. Shimizu, K. Shimokata, and M. Ohta. 1998. Campylobacter fetus subspecies fetus cellulitis associated with bacteremia in debilitated hosts. Clin. Infect. Dis. 27:252-255. [DOI] [PubMed] [Google Scholar]
- 30.Jauk, V. 2003. Phenotypic and genotypic differentiation of Campylobacter spp. isolated from Austrian broiler farms: a comparison. Avian Pathol. 32:33-37. [DOI] [PubMed] [Google Scholar]
- 31.Kirk, R., and M. T. Rowe. 1994. A PCR assay for the detection of Campylobacter jejuni and Campylobacter coli in water. Lett. Appl. Microbiol. 19:301-303. [DOI] [PubMed] [Google Scholar]
- 32.Klena, J. D., C. T. Parker, K. Knibb, J. C. Ibbitt, P. M. L. Devane, S. T. Horn, W. G. Miller, and M. E. Konkel. 2004. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis using a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J. Clin. Microbiol. 42:5549-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konkel, M. E., R. T. Marconi, D. J. Mead, and W. Cieplak, Jr. 1994. Cloning and expression of the hup encoding a histone-like protein of Campylobacter jejuni. Gene 146:83-86. [DOI] [PubMed] [Google Scholar]
- 34.Kosunen, T. U., B. E. Bang, and M. Hurme. 1984. Analysis of Campylobacter jejuni antigens with monoclonal antibodies. J. Clin. Microbiol. 19:129-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnamurthy, T., P. L. Ross, and U. Rajamani. 1996. Detection of pathogenic and non-pathogenic bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:883-888. [DOI] [PubMed] [Google Scholar]
- 36.Lamoureux, M., A. MacKay, S. Messier, I. Fliss, B. W. Blais, R. A. Holley, and R. E. Simard. 1997. Detection of Campylobacter jejuni in food and poultry viscera using immunomagnetic separation and microtitre hybridization. J. Appl. Microbiol. 83:641-651. [DOI] [PubMed] [Google Scholar]
- 37.La Scola, B., S. Chambourlier, and P. Bouillot. 1998. Campylobacter fetus ssp. fetus brain abscess. J. Infect. 37:309-310. [DOI] [PubMed] [Google Scholar]
- 38.Lastovica, A., E. Le Roux, R. Warren, and H. Klump. 1993. Clinical isolates of Campylobacter mucosalis. J. Clin. Microbiol. 31:2835-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lastovica, A. J., and E. le Roux. 2000. Efficient isolation of campylobacteria from stools. J. Clin. Microbiol. 38:2798-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lentzsch, P., B. Rieksneuwohner, L. H. Wieler, H. Hotzel, and I. Moser. 2004. High-resolution genotyping of Campylobacter upsaliensis strains originating from three continents. J. Clin. Microbiol. 42:3441-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.le Roux, E., and A. J. Lastovica. 1998. The Cape Town Protocol: how to isolate the most campylobacters for your dollar, pound, franc, yen, etc., p. 30-33. In A. J. Lastovica, D. G. Newell, and E. E. Lastovica (ed.), Campylobacter, Helicobacter and related organisms, 9th international workshop. Institute of Child Health, University of Cape Town, Cape Town, South Africa.
- 42.Linton, D., F. E. Dewhirst, J. P. Clewley, R. J. Owen, A. P. Burnens, and J. Stanley. 1994. Two types of 16S rRNA gene are found in Campylobacter helveticus: analysis, applications and characterization of the intervening sequence found in some strains. Microbiology 140:847-855. [DOI] [PubMed] [Google Scholar]
- 43.Linton, D., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macuch, P. J., and A. C. Tanner. 2000. Campylobacter species in health, gingivitis, and periodontitis. J. Dent. Res. 79:785-792. [DOI] [PubMed] [Google Scholar]
- 45.Mandrell, R. E., A. H. Bates, and D. L. Brandon. May 2002. Monoclonal antibodies against Campylobacter jejuni and Campylobacter coli outer membrane antigens. U.S. patent 6,395,879 B1.
- 46.Mandrell, R. E., A. J. Lesse, J. V. Sugai, M. Shero, J. M. Griffiss, J. A. Cole, N. J. Parsons, H. Smith, S. A. Morse, and M. A. Apicella. 1990. In vitro and in vivo modification of Neisseria gonorrhoeae lipooligosaccharide epitope structure by sialylation. J. Exp. Med. 171:1649-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinot, M., B. Jaulhac, R. Moog, S. De Martino, P. Kehrli, H. Monteil, and Y. Piemont. 2001. Campylobacter lari bacteremia. Clin. Microbiol. Infect. 7:96-97. [DOI] [PubMed] [Google Scholar]
- 48.Matsheka, M. I., A. J. Lastovica, and B. G. Elisha. 2001. Molecular identification of Campylobacter concisus. J. Clin. Microbiol. 39:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller, W. G., and R. E. Mandrell. 2005. Campylobacter in the food and water supply: prevalence, outbreaks, isolation, and detection, p. 101-163. In J. Ketley and M. E. Konkel (ed.), Campylobacter jejuni: new perspectives in molecular and cellular biology. Horizon Scientific Press, Norfolk, United Kingdom.
- 52.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montero, A., X. Corbella, J. A. Lopez, M. Santin, and I. H. Ballon. 1997. Campylobacter fetus-associated aneurysms: report of a case involving the popliteal artery and review of the literature. Clin. Infect. Dis. 24:1019-1021. [DOI] [PubMed] [Google Scholar]
- 54.Moser, I., B. Rieksneuwohner, P. Lentzsch, P. Schwerk, and L. H. Wieler. 2001. Genomic heterogeneity and O-antigenic diversity of Campylobacter upsaliensis and Campylobacter helveticus strains isolated from dogs and cats in Germany. J. Clin. Microbiol. 39:2548-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newell, D. G. 1986. Monoclonal antibodies directed against the flagella of Campylobacter jejuni: cross-reacting and serotypic specificity and potential use in diagnosis. J. Hyg. (London) 96:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owen, R. J., and J. Hernandez. 1990. Genotypic variation in “Campylobacter upsaliensis” from blood and faeces of patients in different countries. FEMS Microbiol. Lett. 60:5-10. [DOI] [PubMed] [Google Scholar]
- 58.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 59.Rasmussen, H. N., J. E. Olsen, K. Jorgensen, and O. F. Rasmussen. 1996. Detection of Campylobacter jejuni and Camp. coli in chicken faecal samples by PCR. Lett. Appl. Microbiol. 23:363-366. [DOI] [PubMed] [Google Scholar]
- 60.Simor, A. E., and L. Wilcox. 1987. Enteritis associated with Campylobacter laridis.J. Clin. Microbiol. 25:10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, D. B. 1968. Amino acid sequences of some tryptic peptides from the beta-chain of horse hemoglobin. Can. J. Biochem. 46:825-843. [DOI] [PubMed] [Google Scholar]
- 62.Soderstrom, C., C. Schalen, and M. Walder. 1991. Septicaemia caused by unusual Campylobacter species (C. laridis and C. mucosalis). Scand. J. Infect. Dis. 23:369-371. [DOI] [PubMed] [Google Scholar]
- 63.Steinbrueckner, B., G. Haerter, K. Pelz, and M. Kist. 1999. Routine identification of Campylobacter jejuni and Campylobacter coli from human stool samples. FEMS Microbiol. Lett. 179:227-232. [DOI] [PubMed] [Google Scholar]
- 64.Tauxe, R. V., C. M. Patton, P. Edmonds, T. J. Barrett, D. J. Brenner, and P. A. Blake. 1985. Illness associated with Campylobacter laridis, a newly recognized Campylobacter species. J. Clin. Microbiol. 21:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.The Institute for Genomic Research. 2004. TIGR microbial database, unfinished microbial genomes. [Online.] http://www.tigr.org.
- 66.Vandamme, P., E. Falsen, B. Pot, B. Hoste, K. Kersters, and J. De Ley. 1989. Identification of EF group 22 campylobacters from gastroenteritis cases as Campylobacter concisus. J. Clin. Microbiol. 27:1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandamme, P., B. Pot, and K. Kersters. 1991. Differentiation of campylobacters and Campylobacter-like organisms by numerical analysis of one-dimensional electrophoretic protein patterns. Syst. Appl. Microbiol. 14: 57-66. [Google Scholar]
- 68.van Doorn, L. J., A. Verschuuren-van Haperen, A. Burnens, M. Huysmans, P. Vandamme, B. A. Giesendorf, M. J. Blaser, and W. G. Quint. 1999. Rapid identification of thermotolerant Campylobacter jejuni, Campylobacter coli, Campylobacter lari, and Campylobacter upsaliensis from various geographic locations by a GTPase-based PCR-reverse hybridization assay. J. Clin. Microbiol. 37:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Volokhov, D., V. Chizhikov, K. Chumakov, and A. Rasooly. 2003. Microar-ray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis.J. Clin. Microbiol. 41:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wall, D. B., D. M. Lubman, and S. J. Flynn. 1999. Rapid profiling of induced proteins in bacteria using MALDI-TOF mass spectrometric detection of nonporous RP HPLC-separated whole cell lysates. Anal. Chem. 71:3894-3900. [DOI] [PubMed] [Google Scholar]
- 71.Wang, G., C. G. Clark, T. M. Taylor, C. Pucknell, C. Barton, L. Price, D. L. Woodward, and F. G. Rodgers. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40:4744-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White, L. A., and D. S. Kellogg. 1965. Neisseria gonorrhoeae identification in direct smears by a fluorescent antibody-counterstain method. Appl. Microbiol. 13:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winkler, M. A., J. Uher, and S. Cepa. 1999. Direct analysis and identification of Helicobacter and Campylobacter species by MALDI-TOF mass spectrometry. Anal. Chem. 71:3416-3419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.