Abstract
The microeukaryotic community in Zodletone Spring, a predominantly anaerobic sulfide and sulfur-rich spring, was examined using an 18S rRNA gene cloning and sequencing approach. The majority of the 288 clones sequenced from three different locations at Zodletone Spring belonged to the Stramenopiles, Alveolata, and Fungi, with members of the phylum Cercozoa, order Diplomonadida, and family Jakobidae representing a minor fraction of the clone library. No sequences suggesting the presence of novel kingdom level diversity were detected in any of the three libraries. A large fraction of stramenopile clones encountered were monophyletic with either members of the genus Cafeteria (order Bicosoecida) or members of the order Labyrinthulida (slime nets), both of which have so far been encountered mainly in marine habitats. The majority of the observed fungal clone sequences belonged to the ascomycetous yeasts (order Saccharomycetales), were closely related to yeast genera within the Hymenobasidiomycetes (phylum Basidiomycetes), or formed a novel fungal lineage with several previously published or database-deposited clones. To determine whether the unexpected abundance of fungal sequences in Zodletone Spring clone libraries represents a general pattern in anaerobic habitats, we generated three clone libraries from three different anaerobic settings (anaerobic sewage digester, pond sediment, and hydrocarbon-exposed aquifer sediments) and partially sequenced 210 of these clones. Phylogenetic analysis indicated that clone sequences belonging to the kingdom Fungi represent a significant fraction of all three clone libraries, an observation confirmed by phospholipid fatty acid and ergosterol analysis. Overall, this work reveals an unexpected abundance of Fungi in anaerobic habitats, describes a novel, yet-uncultured group of Fungi that appears to be widespread in anaerobic habitats, and indicates that several of the previously considered marine protists could also occur in nonmarine habitats.
The utilization of culture-independent approaches for describing microbial assemblages in various ecosystems has altered our view on the breadth of prokaryotic diversity. For example, by extensive 16S rRNA gene-based analysis of various marine and terrestrial ecosystems, the number of bacterial divisions and candidate divisions has increased from 12 in 1987 (68) to at least 52 (53). Similarly, numerous novel archaeal lineages, many of which have a global distribution, have been detected in both marine (11) and terrestrial (7) ecosystems.
Recently, a similar approach utilizing small subunit (SSU) rRNA gene amplification, cloning, and sequencing has been applied to study eukaryotic diversity. The few available studies primarily examined the microeukaryotic community in different marine environments, including the photic and aphotic zones of pelagic oceans (12, 13, 25, 39, 43, 61), anoxic marine salt marsh sediments (60), and hydrothermal vents (17, 38). Collectively, these studies suggest an unexpected level of diversity with the detection of sequences representing potential novel lineages within known eukaryotic groups, as well as sequences suggesting the presence of novel kingdom level diversity within the microeukaryotic community. The use of 18S rRNA gene-based analysis to study freshwater anaerobic communities has received relatively less attention than marine surveys. However, the work of Dawson and Pace (10) suggests the presence of a similar broad diversity at the kingdom level within freshwater lake sediments.
An argument has recently been presented stating that SSU rRNA surveys overestimate the level of eukaryotic diversity at the megaevolution level (6, 9). The reasoning is that there is an inadequate number of available 18S rRNA gene sequences representing some of these novel fast-evolving groups, chimeric sequences have been incorporated into phylogenetic analysis, long branch attraction artifacts may have occurred, and there are a relatively high number of described eukaryotic taxa with no available sequence data (6).
The present work examines the microeukaryotic communities in Zodletone Spring in southwestern Oklahoma. Zodletone Spring has a high dissolved sulfide concentration in the emergent water (8 to 10 mM) and maintains hypoxic conditions in the water and anoxia in the underlying sediments throughout the course of the spring. Another important characteristic of the spring is the continuous bubbling of short-chain alkanes (methane, ethane, and propane) from the source of the spring, resulting in high hydrocarbon levels, especially in the source area. Recent surveys of the bacterial and archaeal communities in Zodletone Spring revealed an extremely diverse microbial community with several novel bacterial and archaeal lineages (18, 19). The study of the microeukaryotic community in the spring will add to our knowledge regarding microeukaryotes that can thrive under anaerobic conditions in general and under extreme conditions of high sulfide concentrations and hydrocarbon exposure in particular. The resulting increase in the 18S rRNA gene sequences in databases will lead to better phylogenetic resolution for both known and novel eukaryotic lineages that suffer from low sampling. Also, extensive 18S rRNA gene sampling in Zodletone Spring will allow us to practically test the two previously outlined hypotheses regarding the level of eukaryotic diversity at the kingdom level. Finally, comparing the overall patterns of eukaryotic diversity in Zodletone Spring with patterns observed in other anaerobic environments examined in this and other studies will contribute to our understanding of the global patterns of microeukaryotic diversity within anaerobic habitats. Our results support the view that the number of novel high-level eukaryotic lineages is much lower than previously inferred from other SSU rRNA surveys, and we document the presence of several eukaryotic groups previously thought to be restricted to marine environments. We also show an unexpected abundance of fungi in all studied ecosystems and suggest a wide distribution of a novel fungal lineage in anaerobic environments.
MATERIALS AND METHODS
Site description.
Zodletone Spring emerges near Zodletone Mountain in the Anadarko basin in southwestern Oklahoma. Water emerges at the spring source at a rate of 8 liters/s and flows for about 20 m before discharging at a neighboring creek (Stinking Creek). The source is mainly an anaerobic area of sulfide-saturated sediments which approximately 50 cm of water overlies. As a result of light exposure and constant high sulfide concentrations, microbial mats of differing structures are visible throughout the spring. It has been shown that due to phototrophically driven sulfide oxidation mediated by the spring microbial community, barite and calcite are formed throughout the spring and as mineral crusts located on the bank of an adjacent creek (55). These crusts appear to precipitate from sulfide-laden groundwater that percolates out of the creek banks.
Sampling.
Eukaryotic diversity was examined in three locations in Zodletone Spring: the sulfide-saturated, hydrocarbon-exposed spring source; the microbial mat community; and the bacterial crust formations on the banks of the creek. Samples were collected using a sterile spatula and frozen immediately on dry ice. They were transferred to the laboratory within 3 h of sampling, where they where stored at −20°C. In addition to samples obtained in Zodletone Spring, samples were also obtained from three different anaerobic locations: an anaerobic sewage digester from a sewage treatment plant in Norman, OK (clone library ww), anaerobic sediments from a gas-condensate-contaminated site near Ft. Lupton, CO (clone library ftlp), and anoxic sediment (5 cm deep) from a freshwater pond (duck pond) in Norman, OK (clone library dp). All samples were collected in August 2003, frozen immediately, and stored at −20°C for a maximum of 1 week prior to DNA extraction. Subsurface sediment used for constructing the fl library was collected in June 2001 and stored frozen for 2 years.
DNA extraction, PCR amplification, cloning, and sequencing.
DNA isolation was carried out using a lysis-bead-beating protocol (15). The custom primers (Invitrogen Corp., Carlsbad, CA) used for amplifying the 18S rRNA gene were 82f and 1520r (10). We checked the primers for specificity by confirming the length of the amplification product using agarose gel electrophoresis. As well, all sequences obtained from the cloning procedure were 18S rRNA gene sequences. 18S rRNA genes were amplified from the bulk community DNA in a 50-μl reaction mixture containing (final concentration): 2 μl of 1:100 dilution of extracted DNA, 1× PCR buffer (Invitrogen), 1.5 mM MgSO4, 0.2 mM (each) deoxynucleoside triphosphate mixture, 2.5 U of platinum Taq DNA polymerase (Invitrogen), and 10 μM (each) forward and reverse primers. PCR amplification was carried out on a Gene Amp PCR system 9700 thermocycler according to the following protocol: denaturation for 5 min at 94°C, 30 cycles of 94°C for 30 s, 55°C for 15 s, and 72°C for 2 min, followed by a final extension at 72°C for 10 min. PCR products were cloned into a TOPO-TA cloning vector and sequenced as previously described (19).
Phylogenetic analysis.
Shannon-Weiner Index, species evenness, average nucleotide diversity (θ), and percentage of coverage were determined according to previously described procedures (32, 42). For phylogenetic placement, sequences initially were compared to the GenBank nr database and checked using BLAST (1). Sequences with more than 98% similarity were considered to be of the same operational taxonomic unit (OTU). The generation of chimeric sequences during PCR-based diversity studies has been frequently observed (36), and several studies have demonstrated the accumulation of chimeric bacterial, archaeal (31), and eukaryotic (6) rRNA gene sequences in the databases. The presence of chimeric sequences in our data set was checked by screening all sequences with the CHECK-CHIMERA program, available through the ribosomal database website (40). Also, sequences with low (<92%) database similarity were manually inspected to detect the presence of universally conserved regions. In addition, we used a partial treeing analysis approach (31), in which trees were generated using various segments of the 18S rRNA gene sequence and the resulting tree topologies were compared to each other and to the full-length tree. An example of partial treeing analysis for the fungal data set is provided as supplementary material (see Fig. S1a-c in the supplemental material). Overall, 12 chimeric sequences (10 from Zeuk and 1 each from the dp and ftlp clone libraries) were detected and removed from the data set. Zodletone sequences and GenBank-downloaded sequences were aligned using the ClustalX program (63). The program ModelTest (52) was used to choose the optimum model of DNA substitution for each data set. Phylogenetic trees were constructed using representatives of closely related reference sequences to highlight the phyogenetic affiliation of clones obtained in this study. Evolutionary distance (neighbor-joining algorithm) and maximum-parsimony trees were constructed using PAUP (version 4.01b10; Sinauer Associates, Sunderland, Mass.). Bayesian analysis for the fungal data set were performed with MrBayes version 3.0b4 (30), using the GTR+G model of evolution with 500,000 generations run and 5,000 trees sampled. The first 100 trees were discarded, and the consensus tree was computed from the remaining trees using PAUP 4.01b. All phylogenetic trees show the frequencies of occurrence of specific OTUs in the source, mat, and crust clone libraries in parentheses from Zodletone samples (designated Zeuk), as well as in ww, dp, or ftlp libraries.
Phospholipid fatty acid (PLFA) and ergosterol analysis.
Sediment or solids were extracted with the single-phase chloroform-methanol buffer system of Bligh and Dyer (8) as modified by White et al. (67). The total lipid extract was fractionated into neutral lipids, glycolipids, and polar lipids by silicic acid column chromatography (28). The neutral lipids (sterols) were saponified and then derivatized with N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) (46), while the polar lipids were transesterified to the fatty acid methyl esters by a mild alkaline methanolysis (28).
Ergosterol and PLFA methyl esters were analyzed by gas chromatography/mass spectroscopy using a Hewlett-Packard 6890 series gas chromatograph interfaced to a Hewlett-Packard 5973 mass selective detector using a 50-m DB-1 column (0.1-mm inner diameter, 0.1-μm film thickness). The injector and detector were maintained at 290°C and 300°C, respectively. The column temperature was programmed at 60°C for 1 min, then ramped at 20°C min−1 to 150°C, held for 4 min, then ramped at 7°C min−1 to 230°C, held for 2 min, and finally ramped at 10°C min−1 to 300°C and held for 3 min. Mass spectra were determined by electron impact at 70 eV. Methyl nonadecanonate was used as the internal standard, and the PLFA and ergosterol were expressed as the equivalent peak response to the internal standard.
Nucleotide sequence accession numbers.
Sequences obtained in this study were deposited in GenBank under accession numbers AY916560 through AY916665.
RESULTS
Composition and comparative diversity of Zodletone source, mat, and crust clone libraries.
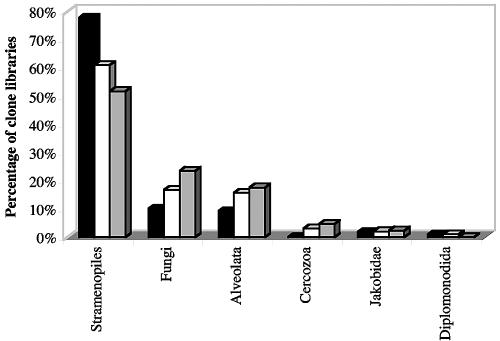
A library of 108, 95, and 85 clones was constructed and sequenced from the source, mat, and crust clone libraries, respectively. Figure 1 summarizes the group level affiliation of clone sequences. Clone sequences belonging to the Stramenopiles, Fungi, and Alveolata collectively represented 97, 94, and 93% of the source, mat, and crust clone libraries, respectively, while those belonging to the Cercozoa, Diplomonadida, and Jakobidae represented a minor fraction of the clone libraries. In spite of sequencing a total of 288 clones from Zodletone Spring, no novel kingdom level diversity was detected. Selection for a few dominant species was evident, as five OTUs belonging to the orders Bicosoecida, Labyrinthulida, and Bacillariophyceae made up 54% of the total number of clones sequenced. Various diversity indices (see Table S1 in the supplemental material) suggested a lower level of diversity within the source clone library, compared to the mat and crust clone libraries. This lower diversity is probably a reflection of the harsher conditions (higher sulfide and hydrocarbon concentration) at the spring source.
FIG. 1.
Distribution of Zodletone source (black columns), mat (white columns), and crust (gray columns) clones among various eukaryotic groups.
Phylogenetic placement. (i) Stramenopiles.
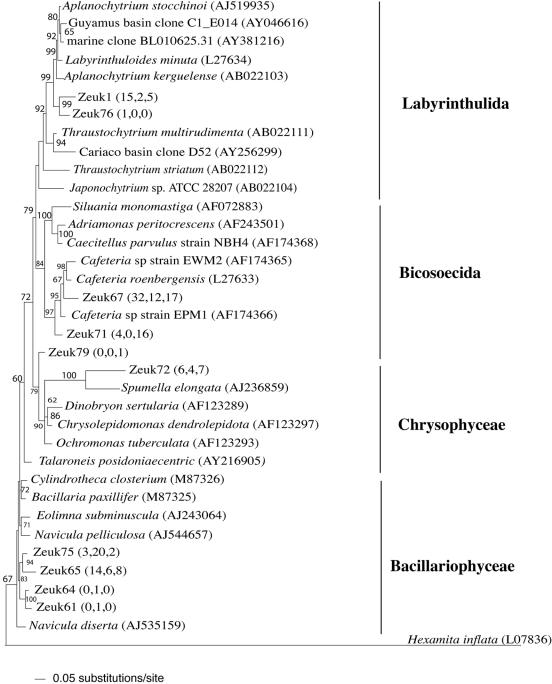
Within the three Zodletone clone libraries, Stramenopiles clone sequences belonged to four different groups: the pinnate diatoms (class Bacillariophyceae), golden algae (class Chrysophyceae), order Bicosoecida, and slime nets (order Labyrinthulida) (Fig. 2). Bicosoecida clones (2 OTUs, 81 clones) were monophyletic, with strong bootstrap support, to members of the genus Cafeteria. All currently known Cafeteria spp. had been obtained from marine sources such as marine plankton and hydrothermal vents (2, 5, 23, 34). Labyrinthulida clone sequences (2 OTUs, 11 clones) were only 93% similar to their closest relatives (Aplanochytrium kerguelense) (Fig. 2). Similar to Cafeteria, all members of the Labyrinthulida have so far been isolated from marine and estuarine environments (51), and culture-independent studies have often encountered this group in anaerobic, marine habitats (17, 39, 43, 61). Therefore, this work shows that this group of microeukaryotes could also occur in other aquatic habitats.
FIG. 2.
Distance dendrogram based on the 18S sequences of stramenopile OTUs encountered in Zodletone source, mat, and crust clone libraries. The tree was constructed using a general time reversible substitution model, with a proportion of invariable site value of 0.13128 and a variable site gamma distribution shape parameter at 0.3493. Hexamita inflata (L07836) was used as the outgroup. Bootstrap values (in percentages) are based on 1,000 replicates and are shown for branches with more than 50% bootstrap support. Numbers in parentheses represent the frequencies of occurrence of a specific OTU in the source, mat, and crust clone libraries, respectively.
(ii) Fungi.
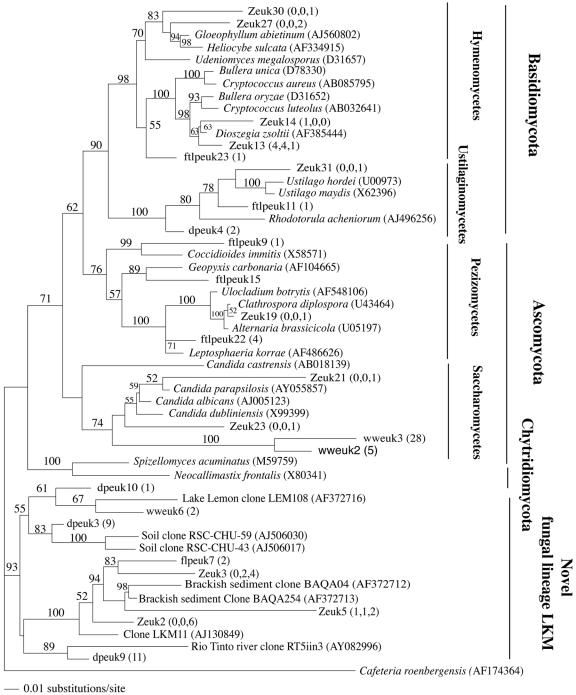
Fungal clones represented a significant fraction of the clone libraries in Zodletone Spring (16% of the total number of clones) (Fig. 1). Fungal sequences are often observed (10) in clone libraries during surveys of eukaryotes from anaerobic environments (10, 17, 60, 61) but are rarely discussed. In Zodletone clone libraries, most fungal clones belonged to four main groups (Fig. 3). (i) Several clone sequences (3 OTUs, 16 clones) had no affiliation with any of the known fungal phyla and, together with several published or database-deposited clones (10, 66), form a novel lineage, which we named novel fungal lineage LKM, after clone LKM11 (AJ130849), the first published sequence belonging to this group (66). (ii) Members of the tremallales clade within the Hymenomycetes (phylum Basidiomycetes), with their closest relatives being members of known yeast genera, e.g., Bullera, Fellomyces, and Kockovaella. (iii) Members of the plant pathogenic class Ustilaginomycetes within the Basidiomycetes with close (99%) similarity to the smut fungi Ustilago hordei and Ustilago maydis. (iv) Members of the class Saccharomycetes within the Ascomycota. The source clone library had a high percentage (55% of the total fungal clones) of Hymenobasidiomycetes, clone sequences related to yeast genera. In contrast, the majority of the fungal clone sequences in the mat and crust clone libraries belonged to the novel LKM group.
FIG. 3.
Distance dendrogram based on the 18S sequences of fungal OTUs encountered in Zodletone source, mat, and crust clone libraries as well as in ww, ftlp, and dp clone libraries. The tree was constructed using a Tamura-Nei substitution model, with a proportion of invariable site value of 0.1218 and a variable site gamma distribution shape parameter at 0.4114. Cafeteria roenbergensis was used as the outgroup. Bootstrap values (in percentages) are based on 1,000 replicates and are shown for branches with more than 50% bootstrap support. Numbers in parentheses represent the frequencies of occurrence of a specific OTU in the source, mat, and crust clone libraries, respectively.
(iii) Alveolates.
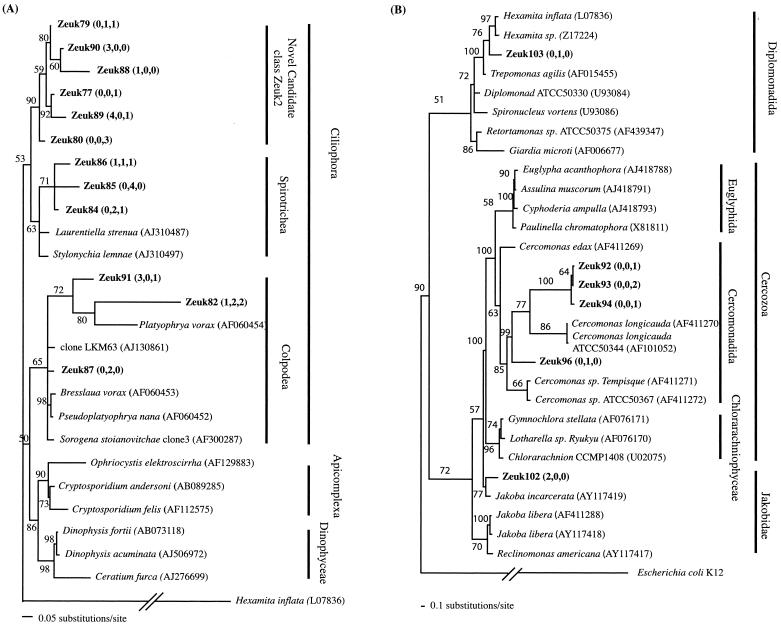
All alveolates from Zodletone belonged to the class Ciliophora (ciliates), the presence of which has long been established in anaerobic environmments (22). Zodletone ciliates either belonged to the classes Spirotrichea or Colpodea or formed a novel lineage within the ciliates that is designated candidate ciliate class Zeuk (Fig. 4A)
FIG. 4.
(A) Distance dendrogram based on the 18S sequences of Alveolata OTUs encountered in Zodletone source, mat, and crust clone libraries. The tree was obtained under a Tamura-Nei substitution model, with a variable-site gamma distribution parameter at 0.4022. Hexamita inflata (L07836) was used as the outgroup. (B) Distance dendrogram based on the 18S sequences of Cercozoa, Diplomonadida, and Jakoba encountered in Zodletone source, mat, and crust clone libraries. The tree was obtained under a Tamura-Nei substitution model, with a variable-site gamma distribution parameter at 0.5402. Escherichia coli strain K-12 16S rRNA gene was used as an outgroup. Bootstrap values (in percentages) are based on 1,000 replicates and are shown for branches with more than 50% bootstrap support. Numbers in parentheses represent the frequencies of occurrence of a specific OTU in the source, mat, and crust clone libraries, respectively.
Jakoba, Diplomonadida, and Cercozoa.
Zodletone Jakoba clones were closely related to Jakoba incarcerata, isolated from intertidal sediments in Quibray Bay, Australia (Fig. 4B). Two Diplomonadida clones were detected from the Zodletone source, and mat samples and were only 91 to 93% similar to their closest relative, the free-living amitochondrial flagellate Hexamita inflata (Fig. 4B). These Diplomonadida clones were the only sequences encountered in this study that belonged to one of the early branching amitochondrial eukaryotic groups. Cercozoa clones belonged to the order Cercomonadida that has been often encountered in anoxic habitats (5, 10).
Comparison of the Zodletone Spring microeukaryotic community to other anaerobic environments.
Samples from an anaerobic sewage digester (ww sequences), a gas-condensate-impacted aquifer (ftlp sequences), and freshwater pond sediments (dp sequences) were used to construct clone libraries, and a total of 210 clones were partially sequenced. The composition of all three libraries is given in Table 1. Interestingly, fungus-affiliated clone sequences represented a significant fraction in all three clone libraries (Fig. 3) (63% in the ww library, 35% of the fl library, and 34% of the dp library). Moreover, clone sequences belonging to the novel fungal LKM group were present in all three environments, suggesting a widespread distribution of this group in anaerobic ecosystems. This group represented 82% of the fungal clones (n = 27) and 28% of the total number of clones (n = 79) in the dp clone library. The remaining fungal clone sequences in the dp clone libraries were members of the phyla Glomeromycota and Chytridiomycota and the class Ustilaginomycetes. Fungal clone sequences from the anaerobic digestor were either saccharomycetous yeasts or members of the fungal LKM group (Fig. 3). Anaerobic aquifer fungal clone sequences were ascomycetous or basidiomycetous yeasts and belonged to the LKM group as well as the orders Pezizales and Ustilaginomycetes (Fig. 3).
TABLE 1.
Phylogenetic affiliation of the microeukaryotic community encountered in wastewater, hydrocarbon-contaminated, and pond sediment clone librariesa
| Sample | Fungi
|
Stramenopiles
|
Alveolates
|
Green algae
|
Metazoa (%) | Higher plants (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| % | Componentsb | % | Componentsb | % | Componentsb | % | Componentsb | |||
| Wastewater (ww) | 63 | Saccharomycetales (34) | 0 | NA | 0 | NA | 0 | NA | 37 | 0 |
| LKM group (3) | ||||||||||
| Gas condensate-contaminated aquifer (ftlp) | 35 | Saccharomycetales (7) | 13 | Labyrinthulida (1) | 7 | Suessiales (1) | 0 | NA | 24 | 22 |
| LKM group (1) | Oomycetes (6) | Stichotrichia (2) | ||||||||
| Pezizomycetes (14) | Bacillariophyceae (2) | Cyrtophorida (1) | ||||||||
| Hymenobasidiomycetes (2) | Cyrtolophosidida (1) | |||||||||
| Ustilaginomycetes (1) | ||||||||||
| Freshwater pond (dp) | 34 | LKM group (22) | 24 | Labyrinthulida (1) | 25 | Armophorida (4) | 1 | Volvocales (7) | 4 | 0 |
| Chytridomycota (2) | Oomycetes (1) | Eimeriida (1) | 4 | Chlorococcales (2) | ||||||
| Glomeromycetes (1) | Bacillariophycease (16) | Halteriidae (3) | Chlorosarcinales (1) | |||||||
| Ustilaginomycetes (2) | Raphidophyceae (1) | Peniculida (3) | Prasiolales (1) | |||||||
| Gymnodiniales (1) | ||||||||||
| Euplotida (5) | ||||||||||
| Pleuronematida (3) | ||||||||||
Detailed phylogenetic placement of fungal clones is given in Fig. 2.
Number of clones encountered belonging to each phylogenetic group is given in parentheses. NA, not applicable.
PLFA and sterol analysis.
To estimate the relative abundance of eukaryotes in general and fungi in particular in anaerobic environments, we quantified the total, prokaryotic, and eukaryotic PLFA, two fungus-specific PLFAs (18:2w6 and 18:3ω3), and ergosterol in Zodletone Spring as well as in ww, dp, and ftlp samples. Ergosterol, a fungus-specific sterol used as an indicator of living fungal biomass (27), was detected in all environments except Zodletone crust samples and ftlp samples (Table 2). However, fungus-specific PLFAs were detected in all samples analyzed, confirming the presence of fungi in all of the anaerobic ecosystems examined. These fungus-specific PLFAs (18:2ω6 and 18:3ω3) represented 4 to 6% of the total PLFAs in the dp, ww, and ftlp samples and represented a significant fraction (between 35% in Zodletone crust samples and 79% in Zodletone source samples) of the total eukaryotic PLFAs in all samples (Table 2). It is interesting to note that among all three Zodletone Spring samples, the highest total eukaryotic PLFA and lowest prokaryotic/eukaryotic ratios were observed in the source sample. Also, the source samples contained more fungus-specific PLFAs (226 pmol/g [dry weight]) than the mat and crust samples (Table 2). The fact that the source has a higher sulfide concentration and lower exposure to light and atmospheric oxygen compared to the mat and crust attests to the ability of eukaryotic groups detected in Zodletone Spring, including Fungi, to survive and grow under strict anaerobic, highly reduced conditions.
TABLE 2.
Biomass, ergosterol, and PLFA analysis of sediments from samples examined using 18S rRNA gene analysis in this studya
| Total
|
Ergosterol biomassb | Fungus-specific PLFA (18:2ω6 and 3ω3)b
|
|||||
|---|---|---|---|---|---|---|---|
| PLFA biomassb | Prokaryoticb | Eukaryoticb | Prokaryotic/eukaryotic ratio | Biomass | mol% | ||
| Zodletone source | 14,755 (3,222) | 14,469 (3,135) | 286 (86) | 52 (4) | 0.8 (0.2) | 226 | <1 |
| Zodletone mat | 9,246 (3,116) | 9,194 (3,095) | 52 (20) | 177 (11) | 0.9 (0.8) | 19 | <1 |
| Zodletone crust | 11,557 (415) | 11,392 (359) | 165 (55) | 74 (22) | 0 | 58 | <1 |
| Freshwater pond (dp) | 22,756 (597) | 20,722 (718) | 2,034 (121) | 11 (1) | 3.3 (1.5) | 1,315 | 6 |
| Wastewater (ww) | 1,198,626 (299,952) | 1,137,662 (279,713) | 60,964 (20,239) | 20 (2) | 27.6 (1.5) | 44,726 | 4 |
| Gas condensate-contaminated sediment (ftlp)c | 3,053 | 2,882 | 171 | 18 | 0 | 143 | 5 |
Standard deviation values are given in parentheses.
Values are expressed as pmol/g (dry weight).
Only one sample was analyzed; for all other samples, n = 2.
DISCUSSION
In this study, we surveyed the microeukaryotic community in Zodletone Spring to determine the identity of eukaryotes in this anaerobic, sulfide and sulfur-rich, hydrocarbon-impacted environment. Although this study did not reveal the presence of any novel kingdom level diversity, it suggests the presence of novel lineages within known groups (e.g., novel fungal lineage LKM within the fungi or novel Zeuk lineage within the ciliates) and expands the geographical presence of some groups (genus Cafeteria, order Labyrinthulida, and genus Jakoba) previously thought to be restricted to marine habitats.
The presence of ciliates within the anoxic community in Zodletone Spring is not surprising since ciliates have long been known to be inhabitants of anaerobic environments (22). However, several Zodletone clone sequences have low similarity to all GenBank-deposited ciliate 18S rRNA genes and potentially represent a novel uncultured candidate class within the ciliates.
Cafeteria species have been shown to be abundant in a variety of marine habitats. They have been isolated from anaerobic tidal waves (2), pelagic ocean, and hydrothermal vents (2). Here, we show that members of Cafeteria spp. are not restricted to marine settings but could also be encountered in other aquatic ecosystems. This conclusion is also true for Jakoba spp. and the order Labyrinthulida, both of which have previously been detected using culture-dependent and -independent analyses solely in marine environments (17, 39, 43, 51, 61). It is interesting to note that all three previously mentioned groups have been shown to be an integral component of hydrothermal vent ecosystems. Similarly, many clone groups within the prokaryotic community in Zodletone Spring have been observed in hydrothermal vent prokaryotic communities (18, 19, 62). The resemblance between a deep-sea hydrothermal vent and a mesophilic spring is quite unexpected. However, both environments have several geochemical characteristics in common, including the low redox potential, high sulfide content, and high levels of short-chain gaseous alkanes. Therefore, it appears that these previously mentioned factors could play an important role in shaping the microbial communities in both ecosystems.
In Zodletone Spring, samples with sequences suggesting the presence of two different phototrophic groups (within the Stramenopiles) were observed: the classes Chrysophyceae (golden algae) and Bacillariophyceae (diatoms). The fact that almost all Chrysophyceae species described so far are oxygenic phototrophs suggests these organisms are introduced into this anaerobic environment with the input of exogenous organic matter (e.g., wood, leaves) to the stream. Alternatively, in the case of the mat and crust, the Chrysophyceae might be localized on the uppermost, air-exposed layers and hence directly exposed to atmospheric air and sunlight. These assumptions could also be true with members of the Bacillariophyceae, especially since they represent a larger fraction of the clone libraries of the light-exposed microbial mats compared with the less light-exposed source. It should be noted, however, that some of the closest relatives of Zodletone Chrysophyceae clone sequences are closely related to the genus Spumela (Fig. 2), all of which are obligately phagocytic (33, 37).
Perhaps the most interesting finding of this study is the unexpected abundance and the novel fungal diversity in Zodletone Spring as well as in all other surveyed anaerobic environments (Fig. 1 and 3; Tables 1 and 2). Fungal clones were previously detected and often represented a significant fraction of the 18S rRNA sequences in clone libraries from anaerobic environments (10, 17, 39, 60). Apart from members of the class Neocallimasticaceae within the Chytridiomycota, no strict anaerobic fungal species has yet been described and fungi have always been thought to play a minor role in ecosystem processes in anaerobic ecosystems (14, 41). Within the various groups encountered in all six clone libraries examined, fermentative metabolism and anaerobic growth abilities are known to exist among yeast species of the order Saccharomycetales. It had been shown that Candida albicans and Saccharomyces cerevisiae could be grown and maintained under strict anaerobic conditions (16, 58). Also, basidiomycetous yeasts are known to be saprophytes, thriving in high-organic-content environments (20). Although fermentation occurs in only very few of the basidiomycetous yeasts (20), some species had been isolated from seemingly anaerobic environments like subseafloor habitats (45). These fermentative capabilities could explain the presence of these two groups of fungi in Zodletone Spring as well as in other clone libraries. A third group of fungal clones in the libraries is characterized by their close similarity to well known aerobic, wood- and plant-associated fungi e.g., Ustilago sp. and Pezizales, and their presence is probably a reflection of plant input to the system.
The final group of fungal sequences encountered in Zodletone Spring as well as other environments is the novel fungal lineage LKM (Fig. 3). Since no pure culture representative of this group is yet available, we can only speculate on their metabolic capabilities based on the environments where they have been encountered. A database search revealed the presence of clones belonging to this group in anaerobic brackish sediments and anaerobic freshwater lake sediments (10). This group has also been encountered in soil and lake sediments where no definite information regarding the redox potential is available (66) (GenBank accession numbers AJ506017 and AJ506030), as well as in extremely acidic river sediments (69). However, the fact that several studies using the 18S rRNA gene sequence to study the fungal communities in highly oxic systems, including soil (65), the rhizosphere (57, 64), and freshwater habitats (47), did not detect this group, coupled with their presence in all clone libraries tested in this study as well as several other anaerobic habitats, provides strong evidence for the indigenous nature of this group and hence the ability to thrive under anaerobic conditions. However, it is important to note that some of the previous studies examining fungal diversity utilizing the large ribosomal subunit (rRNA) as a biomarker have detected several unique yet uncultured fungal lineages (54). Comparing the phylogeny of the novel fungal lineage obtained in this study to that of novel lineages detected in large-subunit rRNA-based studies is not feasible.
PLFA and ergosterol determinations confirmed the presence of fungi in all ecosystems studied. The fact that ergosterol and fungal PLFA levels were highest in anaerobic digester samples is in agreement with the observation that Fungi constituted the majority of clone sequences (63%) in ww clone library. Assuming a conversion factor of 5.4 mg ergosterol g−1 fungal biomass (35), fungal concentrations were 0.06 to 0.07 mg fungal biomass per gram of sediment in the Zodletone source and mat samples, respectively, 0.24 mg per gram in the pond, and 2.02 mg per gram in sewage digester samples. The lower fungal biomass in Zodletone Spring is in agreement with previous studies showing that ergosterol content decreases considerably with depth-associated change from aerobic to anaerobic redox potential in salt marsh sediments (41). However, fungal biomass values in Zodletone Spring are on average only 2 orders of magnitude less than values obtained in studies examining fungal biomass in aerobic, fungus-rich habitats such as compost (35), fungus-infested sludge (24), leaf litter (26, 59), and biocontaminated building materials (50). Moreover, it has been suggested that ergosterol measurements might underestimate the fungal biomass of yeast cells (50). The fact that several Zodletone clones are closely related to yeast lineages suggests that ergosterol analysis might have underestimated the fungal biomass in Zodletone Spring.
The fact that many of the clones in dp, ftlp, and ww clone libraries are derived from the metazoa and Viridiplantae (Table 1), coupled with the unexpected abundance of fungi in these settings and relatively small number of clones sequenced renders this study far from a comprehensive survey of the microeukaryotic community in common anaerobic freshwater settings. Studies examining similar environments using culturing techniques targeting a single group, e.g., ciliates or flagellates, have isolated several microeukaryotes, many of which appear to be numerous but that we failed to detect (5, 21, 22, 29, 48, 49, 56). Nevertheless, the 18S rRNA gene-based study and PLFA analysis collectively suggest that culture-based studies generally underestimate fungal levels in anaerobic ecosystems.
Finally, the lack of novel kingdom level diversity or even clones affiliated with recently described novel kingdoms in all six clone libraries analyzed (a total of 496 clones) is surprising. Previous studies collectively suggest the presence of a large number of yet-undescribed eukaryotic kingdoms (10, 17, 44, 60). However, a recent reanalysis of sequences proposed to represent these kingdoms led the authors to propose only five novel lineages, three of which were detected in more than one study (6). These five lineages may represent new eukaryotic kingdoms; however, additional studies are likely needed to validate them. The view that we may be overestimating the level of eukaryotic diversity at the kingdom level based upon 18S rRNA gene analysis is supported by recent studies which used multiple gene analysis to show that most of the eukaryotic diversity could be grouped into eight supergroups and that many of the lineages previously thought to be early diverging branches of the eukaryotic tree based on the 18S sequence are related to groups belonging to the crown radiation eukaryotes (3, 4).
Supplementary Material
Acknowledgments
We thank Marielle Hoefnagels and David A. Caron for critical evaluation of the manuscript.
This work has been supported by the National Science Foundation microbial observatories program (no. MCB_0240683).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkins, M. S., A. P. Teske, and O. R. Anderson. 2000. A survey of flagellate diversity at four deep-sea hydrothermal vents in the Eastern Pacific ocean using structural and molecular approaches. J. Eukaryot. Microbiol. 47: 400-411. [DOI] [PubMed] [Google Scholar]
- 3.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, C., A. G. Simpson, and D. J. Patterson. 2000. Some free living flagellates (Protista) from anoxic habitats. Ophelia 52:113-142. [Google Scholar]
- 6.Berney, C., J. Fahrni, and J. Pawlowski. 2004. How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bligh, E. G., and W. J. Deyer. 1954. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 9.Cavalier-Smith, T. 2004. Only six kingdoms of life. Proc. R. Soc. Lond. B 271:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diez, B., C. Pedros-Alio, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diez, B., C. Pedros-Alio, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dighton, J. 2003. Fungi in ecosystem processes, 1st ed., vol. 17. Marcel Dekker, Inc., New York, N.Y.
- 15.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumitru, R., J. M. Hornby, and K. W. Nickerson. 2004. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob. Agents Chemother. 48:2350-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elshahed, M. S., F. Z. Najar, B. A. Roe, A. Oren, T. A. Dewers, and L. R. Krumholz. 2004. Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide- and sulfur-rich spring. Appl. Environ. Microbiol. 70:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elshahed, M. S., J. M. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, J. R. Spear, and L. R. Krumholz. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fell, J. W., T. Boekhout, A. Fonseca, and J. P. Sampaio. 2001. Basidiomycetous yeast, p. 3-35. In E. G. McLaughlin and P. A. Lemke (ed.), The mycota: systemics and evolution, 1st ed., vol. 7. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 21.Fenchel, T., C. Bernard, G. Esteban, B. J. Finlay, P. J. Hansen, and N. Iversen. 1995. Microbial diversity and activity in a Danish fjord with anoxic deep water. Ophelia 45:45-100. [Google Scholar]
- 22.Fenchel, T., and B. J. Finlay. 1995. Ecology and evolution in anoxic worlds, 1st ed. Oxford University Press, Oxford, United Kingdom.
- 23.Fenchel, T., and D. J. Patterson. 1988. Cafeteria roenbergensis nov. gen., nov. sp., a heterotrophic microflagellate from marine plankton. Mar. Microb. Food Webs 3:9-19. [Google Scholar]
- 24.Gardner, R. M., G. W. Tindall, S. M. Cline, and K. L. Brown. 1993. Ergosterol determination in activated sludge and its application as a biochemical marker for monitoring fungal biomass. J. Microbiol. Methods 17:49-60. [Google Scholar]
- 25.Gast, R. J., M. R. Dennett, and D. A. Caron. 2004. Characterization of protistan assemblages in the Ross Sea, Antarctica, by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2028-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gessner, M. O., and E. Chauvet. 1993. Ergosterol-to-biomass conversion factors for aquatic hyphomycetes. Appl. Environ. Microbiol. 59:502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gessner, M. O., and S. Y. Newell. 1997. Bulk quantitative methods for the examination of eukaryotic organoosmotrophs in plant litter, p. 295-308. In C. J. Hrust, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 28.Guckert, J. B., C. P. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid, ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 29.Harvey, R. W., N. Mayberry, N. E. Kinner, D. W. Metge, and F. Novarino. 2002. Effect of growth conditions and staining procedure upon the subsurface transport and attachment behaviors of a groundwater protist. Appl. Environ. Microbiol. 68:1872-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 31.Hugenholtz, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 54:289-293. [DOI] [PubMed] [Google Scholar]
- 32.Humayoun Shaheen, B., N. Bano, and T. Hollibaugh James. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnes, R. I. 2000. Mixotrophy in planktonic protists: an overview. Freshwater Biol. 45:219-226. [Google Scholar]
- 34.Karpov, S. A. 2000. Ultrastructure of the aloricate bicosoecid Pseudobodo tremulans with revision of the order Bicosoecida. Protistology 1:101-109. [Google Scholar]
- 35.Klamer, M., and E. Baath. 2004. Estimation of conversion factors for fungal biomass determination in compost using ergosterol and PLFA 18:2w6,9. Soil Biol. Biochem. 36:57-65. [Google Scholar]
- 36.Kopczynski, E. D., M. M. Bateson, and D. M. Ward. 1994. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl. Environ. Microbiol. 60:746-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristiansen, J. 1990. Phylum Chrysophyta. In L. Margulis, J. O. Corliss, M. Melkonian, and D. J. Chapman (ed.), Handbook of protocista. Jones and Bartlett Publishers, Boston, Mass.
- 38.Lopez-Garcia, P., H. Philippe, F. Gail, and D. Moreira. 2003. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl. Acad. Sci. USA 100: 697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Allo, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 40.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. T. Olsen, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansfield, S. D., and F. Barlocher. 1993. Seasonal variation of fungal biomass in the sediment of a salt marsh in New Brunswick. Microb. Ecol. 26:37-45. [DOI] [PubMed] [Google Scholar]
- 42.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68: 3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon-van der Staay, S. Y., R. De Wachtert, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 44.Moreira, D., and P. Lopez-Garcia. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 45.Nagahama, T., M. Hamamoto, T. Nakase, Y. Takaki, and K. Horikoshi. 2003. Cryptococcus surugaensis sp. nov., a novel yeast species from sediment collected on the deep-sea floor of Suruga Bay. Int. J. Syst. Evol. Microbiol. 53:2095-2098. [DOI] [PubMed] [Google Scholar]
- 46.Nichols, P. D., J. K. Volkman, and R. B. Johns. 1983. Sterols and fatty acids of the marine unicellular alga, FCRG 51. Phytochemistry 22:1447-1452. [Google Scholar]
- 47.Nikolcheva, L. G., A. M. Cockshutt, and F. Barlocher. 2002. Determining diversity of freshwater fungi on decaying leaves: comparison of traditional and molecular approaches. Appl. Environ. Microbiol. 69:2548-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novarino, G., A. Warren, H. Butler, G. Lambourne, A. Boxshall, J. Bateman, N. E. Kinner, R. W. Harvey, R. A. Mosse, and B. Teltsch. 1997. Protistan communities in aquifers: a review. FEMS Microbiol. Rev. 20:261-275. [DOI] [PubMed] [Google Scholar]
- 49.Novarino, G., A. Warren, N. E. Kinner, and R. W. Harvey. 1994. Protists from a sewage-contaminated aquifer on Cape-Cod, Massachusetts. Geomicrobiology 12:23-36. [Google Scholar]
- 50.Pasanen, A. L., K. Y. Pietila, P. Pasanen, P. Kalliokoski, and J. Tarhanen. 1999. Ergosterol content in various fungal species and biocontaminated building materials. Appl. Environ. Microbiol. 65:138-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porter, D. 1990. Phylum labyrinthulomycota, p. 388-398. In L. Margulis, J. O. Corliss, M. Melkonian, and D. J. Chapman (ed.), Handbook of Protocista, 1st ed. Jones and Bartlett Publishers, Boston, Mass.
- 52.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 53.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 54.Schadt, C. W., A. P. Martin, D. A. Lipson, and S. K. Schmidt. 2003. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301:1359-1361. [DOI] [PubMed] [Google Scholar]
- 55.Senko, J. M., B. S. Campbell, J. R. Henriksen, M. S. Elshahed, T. A. Dewers, and L. R. Krumholz. 2004. Barite deposition resulting from phototrophic sulfide-oxidizing bacterial activity. Geochim. Cosmochim. Acta 68:773-780. [Google Scholar]
- 56.Sleigh, M. 1989. Protozoa and other protists. Edward Arnolds, New York, N.Y.
- 57.Smit, E., P. Leeflang, B. Glandorf, J. D. Van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonderegger, M., M. Jeppsson, B. Hahn-Haegerdal, and U. Sauer. 2004. Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Appl. Environ. Microbiol. 70:2307-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sridhar, K. R., and F. Barlocher. 2000. Initial colonization, nutrient supply, and fungal activity on leaves decaying in streams. Appl. Environ. Microbiol. 66:1114-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoeck, T., G. T. Taylor, and S. S. Epstein. 2003. Novel eukaryotes from the permanently anoxic Cariaco basin (Caribbean Sea). Appl. Environ. Microbiol. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teske, A., K.-U. Hinrichs, V. P. Edgcomb, A. D. V. Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vandenkoornhuyse, P., S. L. Baldauf, C. Leyval, J. Straczek, and J. P. W. Young. 2002. Extensive fungal diversity in plant roots. Science 295:2051. [DOI] [PubMed] [Google Scholar]
- 65.Van Elsas, J. D., G. F. Duarte, A. Keijzer-Wolters, and E. Smit. 2000. Analysis of fungal-specific PCR of soil DNA followed by denaturing gradient gel eletrophoresis. J. Microbiol. Methods 43:133-151. [DOI] [PubMed] [Google Scholar]
- 66.Van Hannen, E. J., W. Mooij, M. P. Van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White, D. C., R. J. Bobbie, J. S. Herron, J. D. King, and S. J. Morrison. 1979. Biochemical measurements of microbial mass and activity from environmental samples, p. 69-81. In J. W. Costerton and R. R. Colwell (ed.), Native aquatic bacteria: enumeration, activity, and ecology, ASTM STP 695. American Society for Testing and Materials, Philadelphia, Pa.
- 68.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zettler, L. A. A., F. Gomez, E. Zettler, B. K. Keenan, R. Amils, and M. L. Sogin. 2002. Microbiology: eukaryotic diversity in Spain's River of Fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.