Abstract
Listeria monocytogenes is a significant food-borne human and veterinary pathogen. Contaminated silage commonly leads to disease in livestock, but the pervasive nature of the bacterium can make it difficult to identify the source of infection. An investigation of bovine listeriosis that occurred on a Pacific Northwest dairy farm (“farm A”) revealed that the clinical strain was closely related to fecal strains from asymptomatic cows, and that farm environment was heavily contaminated with a diversity of L. monocytogenes strains. In addition, the farm A clinical strain was closely related to clinical and environmental strains obtained 1 year prior from a second Northwest dairy farm (“farm B”). To investigate the source(s) of contamination on farm A, environmental samples were collected from farm A at two time points. Pulsed-field gel electrophoresis characterization of 538 isolates obtained from that farm identified 57 different AscI pulsovars. Fecal isolates obtained from individual cows were the most genetically diverse, with up to 94% of fecal samples containing more than one pulsovar. The maximum numbers of pulsovars and serotypes isolated from a fecal sample of one cow were 6 and 4, respectively. Serotype 1/2a was isolated most frequently at both time points. Microarray genotyping of bovine listeriosis, fecal, and silage strains from both farms identified four probes that differentiated listeriosis strains from environmental strains; however, no probe was common to both bovine listeriosis strains.
Listeria monocytogenes is a gram-positive, facultative intracellular bacterial pathogen capable of causing serious disease in animals and humans. L. monocytogenes infection of cattle and sheep can lead to abortion, central nervous system disease, and death. Although the feeding of poor-quality silage is thought to be the main source of on-farm infection, the ubiquitous distribution of L. monocytogenes makes identification of the source(s) of infection difficult.
Human listeriosis accounts for approximately 2,500 cases and 500 deaths each year (12). Although relatively rare compared to other food-borne diseases, listeriosis frequently presents with serious complications, including meningitis, septicemia, endocarditis, nonmeningitic central nervous system infection, febrile gastroenteritis, or abortion in people with predisposing conditions. Neonates, elderly, and immunocompromised patients are particularly at risk.
Ready-to-eat meats, milk, and milk-related products have been implicated in several outbreaks of listeriosis (17). The organism is excreted in the feces of healthy cows for months to several years (11, 20), and it has been hypothesized that fecal and mammary contamination serve as sources of contamination of raw milk at the dairy farm (24). Raw milk has been suggested as a source of L. monocytogenes in the dairy processing environment (8). However, because L. monocytogenes is widely distributed in the environment, the source(s) of L. monocytogenes in food-borne illness and the role of domestic animals in the contamination of dairy products both pre- and postprocessing are not clearly understood.
We describe here the use of molecular epidemiological methods to investigate two bovine listeriosis cases that occurred on a Pacific Northwest dairy farm (“farm A”). Sampling and testing of the farm A environment revealed that the entire environment was heavily contaminated with L. monocytogenes. The silage was improperly stored, resulting in pockets of high pH and fungal contamination, and numerous L. monocytogenes isolates were obtained from silage samples and many other environmental samples. A second visit to farm A 9 months later revealed that although the storage conditions of the silage had improved, L. monocytogenes contamination persisted throughout the farm environment. However, despite the high levels of contamination, no new listeriosis cases had been reported. In order to identify the source(s) of the listeriosis cases and to investigate the ecology and epidemiology of various L. monocytogenes subtypes on farm A, 538 isolates were subtyped by using serotyping and pulsed-field gel electrophoresis (PFGE). Comparison of the pulsovars present on farm A with regional subtypes (4) revealed that the farm A bovine clinical isolate had a pulsovar that was closely related to that of another bovine listeriosis isolate collected from a different dairy farm in the region (“farm B”). To identify genetic differences between farms A and B clinical strains and the closely related environmental strains from farm A, these strains were further subtyped by using DNA microarray analysis (2, 5, 7).
MATERIALS AND METHODS
Farm A.
On 3 and 4 May 2002, two Holstein cows in a herd of 800 showed signs typical of listerial encephalitis. Samples from one of the cows were submitted by the attending veterinarian to the Washington Animal Disease Diagnostic Laboratory (Pullman, WA), where listerial encephalitis was confirmed by histochemistry and isolation of L. monocytogenes from the brain tissue. We were contacted regarding a possible bovine listeriosis outbreak and collected 103 environmental samples (silage, commodity feeds, feces, trough water, milk filters, bedding, etc.) from the farm on 21 and 30 May 2002. Due to the large number of samples testing positive for L. monocytogenes and the diversity of PFGE types isolated from these visits (time point 1 [TP 1]), we returned to the farm on 20 February 2003 (TP 2) and collected 103 additional samples. In particular, we made an effort to collect fecal samples from cows that had positive fecal samples in the previous collections. To ensure that particular isolates could be associated with an individual cow, fecal samples were collected per rectum using a separate disposable glove for each sample.
Farm B.
In March of 2001 a Holstein cow in a herd of 115 showed signs typical of listerial encephalitis. Tissue samples were submitted by the attending veterinarian to the Washington Animal Disease Diagnostic Laboratory, where listerial encephalitis was confirmed by histochemistry and isolation of L. monocytogenes from the brain tissue. Environmental samples collected from the farm included silage (n = 31, collected over a 1-month period), fecal drag swabs (n = 2), grain (n = 1), milk filters (n = 2), and mouse and bird droppings (n = 2). Farm B was located within 50 miles of farm A.
L. monocytogenes environmental strain isolation and identification.
The method detailed in USDA-FSIS-Office of Public Health and Science document MLG 8.03 (http://www.fsis.usda.gov/OPHS/microlab/mlgbook.htm) was used to identify and isolate L. monocytogenes strains from farm environmental samples. Samples were stored at 4°C prior to processing, and a two-staged enrichment was used for strain isolation. A total of 25 g/25 ml of sample was added to 225 ml of University of Vermont Modified Listeria Enrichment Broth (UVM; Remel, Lenexa, KS), mixed well, and incubated at 30°C for 24 h. After mixing, 100 μl of the broth was transferred to 10 ml of Fraser Broth with supplement (Becton Dickinson, Sparks, MD), followed by incubation for a maximum of 48 h at 30°C. At 24 h and again at 48 h, 50 μl was removed from positive Fraser Broth tubes, plated on Modified Oxford Agar Base with supplement (Remel), and incubated for 24 to 48 h at 30°C. At least four colonies phenotypically characteristic of L. monocytogenes for each sample tested were spotted onto ALOA plates (Microbiology Int., Frederick, MD) and incubated at 30 to 37°C for 48 h to test for phospholipase activity. Phospholipase-positive isolates were then CAMP tested with Staphylococcus aureus using tryptic soy agar plates with 5% sheep blood (USDA-FSIS-MLG8.6.3) and tested for motility in brain heart infusion broth containing 0.4% agar (30°C for 24 h). PCR was used to verify the presence of the listeriolysin gene (1) for any isolate that was positive for all of the tests.
Serotyping.
Denka Seiken Listeria antisera (Tokyo, Japan) were obtained from Accurate Scientific (Westbury, NY). Serotyping was performed according to the manufacturer's recommendations or by using these reagents in an enzyme-linked immunosorbent assay format (15).
PFGE.
All L. monocytogenes isolates were subtyped by using the 30 h PFGE protocol (9). Two reference strains (H2446 and F2365) were used to ensure uniform DNA preparation between experiments. Lambda size standard (Bio-Rad, Hercules, CA) was used as a molecular weight marker. BioNumerics (Applied Maths, Kortrijk, Belgium) was used for band detection and construction of dendrograms by using Dice binary coefficients and UPGMA (unweighted pair-group method with arithmetic averages). Visual inspection of band identification was performed following BioNumerics' band assignment. A 1.5% tolerance was applied to band matching. Reference strains (H2446 and F2365) were included in each gel to minimize gel to gel variability. Gels in which the standard isolates did not cluster within 95% similarity in the dendrogram were rejected from the analysis. Restriction enzyme digestion profiles (REDP) that had >95% similarity according BioNumerics were visually inspected to ensure similarity.
Microarray analysis.
A mixed genome microarray from a previous study (5) was used to further analyze selected strains of L. monocytogenes. The strains analyzed included the two bovine listeriosis strains (LMB 468 and 113), six farm A environmental strains (LMB 630, 679, 680, 789, 1073, and 1154), and ten strains collected from other farms. These ten additional strains were included in the microarray analysis to allow microarray groups (phylogenetic divisions and subclusters) to be defined accurately.
The microarray was assembled from randomly selected, cloned fragments of genomic DNA from multiple strains and serovars of L. monocytogenes. All clones were sequenced before the array was constructed, and redundant clones were not used as probes for the array. Target labeling and hybridization and signal analysis were performed as described previously (5). All probes (n = 606), excluding misprinted probes, were used for data analysis. Misprinted probes were identified by annealing two L. monocytogenes strains representing both major phylogenetic divisions to the microarray at a low stringency (hybridization temperature of 40°C versus 55°C) to allow all probes present to be visualized. For dendrogram construction, probes with normalized intensity readings of <0.4 were assigned a score of “1,” probes with normalized intensity readings greater than 0.4 but less than 0.7 were scored as “2” (treated as ambiguous data in the phylogenetic analysis), and probes with normalized intensity readings of >0.7 were scored as “3.” A matrix was constructed and processed with PAUP (ver. 4.0b8a; Sinauer Associates, Inc., Sunderland, MA). Bootstrap confidence values (n = 100 iterations) were calculated for the nodes in the dendrogram, and the UPGMA and Treeview (14) were used to construct a dendrogram that summarized genetic relationships between samples. Probes of interest were identified by using discriminant function analysis (NCSS Statistical Software, Kaysville, UT) and visual examination of the normalized microarray data.
Statistical analysis.
GraphPad InStat version 3.05 (GraphPad Software, San Diego CA) and NCSS 2000 (NCSS Statistical Software) were used to analyze data. The Fisher exact test was used to analyze binomial data, and an unpaired t test was used to analyze parametric data.
RESULTS
One hundred and eight samples were collected from the first two visits to farm A in May 2002 (TP 1), and 247 isolates were characterized by PFGE. One hundred and three samples were collected from a visit to farm A in February 2003 (TP 2), and 291 isolates were subtyped by using PFGE.
Ninety-seven percent (32 of 33) of farm A fecal samples were positive at TP 2 compared to 56% (32 of 57) positive fecal samples at TP 1. The significant difference between time points may be due to the fact that cows that had previously tested positive for L. monocytogenes were specifically retested at TP 2. Therefore, to determine whether the percentage of positive fecal samples differed between time points, data from the fecal samples of cows tested only at TP 2 (n = 9; all positive) were compared to TP 1 data, and the difference was significant (P = 0.01; Fisher exact test).
A total of 46% (11 of 24) of farm A silage samples were positive at TP 1, and 29% (8 of 28) TP 2 silage samples were positive. A total of 33% (3 of 9) of commodity samples were positive at TP 1, and 45% (5 of 11) were positive at TP 2.
The pH was recorded for a subset of TP 1 silage samples (n = 20). Thirteen samples were classified as low pH (pH < 4.5), and seven samples were classified as high pH (pH > 4.5). Fifteen of the silage samples tested negative for L. monocytogenes, and five samples were positive. The average pH of the positive samples was 6.1 compared to an average pH of 4.4 for the L. monocytogenes-negative samples (18). The difference between the average pH of negative and positive samples was statistically significant (P < 0.05).
Reoccurrence of PFGE subtypes in asymptomatically shedding cows.
Individual fecal samples from 20 cows from farm A were tested at both time points. Of the 20 samples, 12 had different AscI pulsovars present at the different time points. The remaining eight cows had one to two of the same pulsovars present in their feces at both time points; however, fecal samples from five of these eight cows also contained other pulsovars that were not present at both time points.
Serotype diversity.
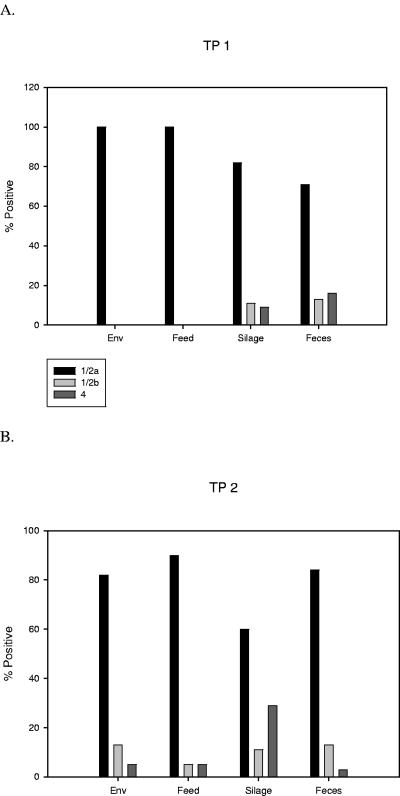
Serotype 1/2a was the most prevalent serotype on farm A for both time points, with 77 and 83% of all samples belonging to serotype 1/2a at TP 1 and TP 2, respectively (Fig. 1). A total of 9% of the samples were serotype 1/2b at TP 1, and this increased slightly to 11% at TP 2. We found that 14% of the samples were serovar 4 (4b, 4ab, or 4e) at TP 1, and this decreased to 6% at TP 2; the difference between the time points was statistically significant (P < 0.01).
FIG. 1.
Percentage of isolates from each sample source according to serotype. Environmental strains not obtained from commodity feeds, silage, or individual fecal samples are listed as “Env.”
PFGE subtype diversity.
PFGE characterization of farm A isolates by using AscI digestion identified 57 different pulsovars present on the farm and 58 subtypes (one pulsovar included two different serotypes [sample ID 0836, Fig. 2]). Twelve subtypes were unique to TP 1, and 27 subtypes were unique to TP 2. Simpson's index of diversity (10) was calculated for both time points to estimate the genetic diversity present in the populations sampled at the two time points. The genetic diversity was calculated to be 0.874 and 0.912 for TP 1 and TP 2, respectively.
FIG. 2.
Dendrogram showing PFGE subtype (AscI REDP) diversity of L. monocytogenes samples from farm A at two collection time points (TP 1 = 2002 collection, TP 2 = 2003 collection). Numbers listed in the first column on the left are the LMB (Listeria monocytogenes Bank) isolate identification numbers. Unique pulsovars are designated using the LMB number of the first strain to be identified with that pulsovar. Source codes are as follows: F (feed), S (silage), C (cow fecal), W (water tank), T (tractor blade), MS (milk sock/filter), B (bovine brain), CYD (commodity yard dust), and O (other). The serotypes associated with each pulsovar are listed in the third column; however, pulsovar/sample ID 0836 includes strains of two serotypes (1/2a and 1/2b). Numbers listed in the two columns on the right indicate the number of samples positive for a particular PFGE type at each time point. The UPGMA dendrogram was constructed from Dice coefficients with 1.5% band matching tolerance.
Of the 58 subtypes detected, 32 (55%) were serotype 1/2a, 9 (16%) were serotype 1/2b, 13 (22%) were serotype 4b, 3 (5%) were serotype 4e, and 1 (2%) was serotype 4ab. Twenty-two patterns were unique to feces, seven were unique to silage, three were unique to water tanks, one was unique to milk socks, one was found only in starling (Sturnus vulgaris) feces, and one was found only in commodity yard dust (Fig. 2).
We found that 53% of the positive fecal samples from TP 1 contained more than one pulsovar (average of two), whereas 94% of positive fecal samples from TP 2 contained more than one pulsovar (average of three); the difference was statistically significant (P < 0.001). The maximum number of AscI pulsovars isolated from a fecal sample from one cow was six. Multiple serotypes were isolated from individual fecal samples from six cows, with a maximum of four different serotypes isolated from one fecal sample (Fig. 3).
FIG. 3.
PFGE subtypes and serotypes of L. monocytogenes isolates from the cow 153 fecal sample. (A) AscI REDP; (B) ApaI REDP.
PFGE subtyping of environmental and bovine listeriosis strains.
As part of a separate project (4), regional farm-associated L. monocytogenes strains were compared to other Pacific Northwest human listeriosis strains by using PFGE (AscI). Interestingly, the farm A listeriosis isolate (LMB 468) was found to be closely related to a bovine listeriosis isolate from farm B (LMB 113). Further PFGE analysis (ApaI) showed that although LMB 113 had AscI and ApaI PFGE patterns that were distinct from farm B environmental isolates, it was identical to two fecal isolates obtained from two farm A asymptomatic cows (Fig. 4).
FIG. 4.
PFGE REDP of bovine clinical isolates (LMB 468 and 113) and closely related fecal strains present on farm A. The upper REDP cluster shows ApaI REDP, and the lower cluster shows AscI REDP. The numbers in the left column are LMB strain numbers. The letters in the center column indicate the source (FA, farm A; FB, farm B; CDC, PFGE reference strain obtained from the Centers for Disease Control and Prevention). The sample sources are listed in the right column (C, cow feces; B, bovine brain).
Microarray analysis.
A subset of strains including the bovine listeriosis strains (LMB 468 and 113), farm A environmental strains of identical, similar, or different PFGE subtypes, and farm-associated strains not associated with farm A or B were hybridized to a mixed genome DNA L. monocytogenes microarray (5) for genetic comparison and subtyping (Fig. 5). Strains were grouped according to phylogenetic division (6, 16, 21) and PFGE subtype, and these groupings were supported by high bootstrap values. Strains from farm A that were not similar by PFGE grouped separately. However, stepwise discriminant analysis (NCSS Statistical Software) failed to identify any probe unique to both bovine listeriosis strains (LMB468 and LMB113).
FIG. 5.
UPGMA representation of genetic relationships between 18 L. monocytogenes isolates based on hybridization data derived from 606 microarray probes. Bootstrap values are indicated next to the appropriate nodes (only values >60 are indicated), and phylogenetic divisions are indicated as Div I (Division I), Div II, and Div III. Strains are coded as follows: letter(s) to the left of the LMB number indicate the strain serotype (F, 4b; A, 1/2a; B, 1/2b; FED, 4e/d), and letters immediately following the three- to four-digit LMB number indicate the farm where the isolate was obtained from (FA, farm A; FB, farm B). If no letters are present the isolate was obtained from a farm source unassociated with farm A or farm B. Letter(s) after the period indicate the sample type from which the isolate was obtained from (S, silage; C, cow feces; B, bovine brain; BM, bulk milk; G, goat).
DISCUSSION
As part of a previous study, PFGE restriction enzyme digestion profiles obtained from farm A strains were compared to those of other regional human and farm L. monocytogenes strains (4). Interestingly, the PFGE subtype of the farm A bovine listeriosis isolate was closely related to an isolate obtained from a bovine listeriosis case that had occurred a year earlier on a different dairy farm (i.e., farm B). In addition, both bovine clinical isolates had PFGE subtypes that were identical or closely related to fecal strains present on farm A (Fig. 4). The farms were separated by a distance of approximately 50 miles, and no epidemiological link was identified (such as shared source of cattle, feed, silage, water, equipment, common veterinarian, or other personnel) based on interviews with the farm owners.
To better understand the ecology and epidemiology of farm A, samples were collected at two time points over a period of 9 months. Although silage storage conditions had improved by TP 2, the diversity of strains on the dairy farm increased. In particular, although fewer silage samples were positive at TP 2, a significantly greater percentage of cows were asymptomatically shedding L. monocytogenes in their feces. Studies have shown that up to 52% of asymptomatic cattle shed L. monocytogenes in their feces (13, 22). A recent study by Nightingale et al. (13) found that cattle farms with at least one recent listeriosis case had L. monocytogenes in 32% of fecal samples, whereas control farms with no listeriosis cases had a lower prevalence of L. monocytogenes in fecal samples (22%). Therefore, the rate of fecal shedding at farm A was exceptionally high at both time points.
The diversity of serotypes and PFGE subtypes in individual fecal samples also increased between time points. These data suggest that numerous subtypes were established on farm A, likely initiating a cycle of fecal spread and oral infection of cattle. It is likely that fecal shedding also led to contamination of water tanks and milk socks. In addition, the manure produced by the cattle was used to fertilize the corn fields that were later used for silage production.
The percentage of isolates belonging to serotypes 1/2a and 1/2b did not change significantly between time points; however, a significantly greater percentage of samples belonged to serotype 4 in TP 1 compared to TP 2. The percentage of each serotype also varied with sample type (Fig. 1); however, serotype 1/2a was the dominant serotype for all sample types at both time points. Although the reason for the dominance of serotype 1/2a is unclear, a previous study demonstrated that strains belonging to phylogenetic division I are generally more capable of biofilm formation compared to division II strains (3).
A greater percentage of L. monocytogenes-positive silage samples had serotype 4 at TP 2 compared to TP 1, whereas the percentage of L. monocytogenes-positive fecal samples contaminated with serotype 4 decreased at TP 2 compared to TP 1. The observed increase in serotype 4 isolation in silage samples by TP 2 may be a result of fertilization of TP 2 silage with feces from infected cows from TP 1.
Reoccurrence of PFGE subtypes in asymptomatic cows.
Individual fecal samples were collected from 20 cows at both time points, and all cows that tested positive for L. monocytogenes contamination at TP 1 were also positive at TP 2. PFGE subtyping of the fecal isolates showed that 12 of the 20 cows infected at both time points shed different AscI pulsovars at different time points. The remaining eight cows had one to two of the same pulsovars present in their feces at both time points; however, five of these eight cows had additional pulsovars present in their fecal samples not isolated at both time points. A single pulsovar (LMB 1044) accounted for six of the nine pulsovars isolated at both time points. Because this pulsovar was commonly isolated from environmental samples at both time points (including silage and feed; Fig. 1), it is likely that these cows were reinfected by feed, silage, and/or water. Alternatively, reoccurring strains may able to colonize the gastrointestinal tract and cause chronic shedding.
Microarray analysis.
The presence of strains with similar or identical PFGE subtypes in the brain tissue of cows suffering from listeriosis and in the feces of asymptomatic cows may be due to genetic differences not detected by PFGE subtyping contributing to strain virulence. Therefore, bovine listeriosis strains LMB 468 and 113 and genetically similar fecal strains LMB 630, 679, 680, and 789 were subtyped by using DNA microarray analysis to identify genetic differences that may account for differential virulence. As with previous studies (2, 5, 7), microarray subtyping grouped identical PFGE subtypes together and distinct from other PFGE subtypes present on the farm (Fig. 5). None of the ten strains collected from other farms or bulk milk tanks grouped with LMB 468 or 113. Four probes were identified that differentiated each clinical isolate from the environmental strains that clustered within the same microarray group (Fig. 5). Interestingly, although these four probes distinguished each clinical isolate from the environmental strains that grouped within the same microarray cluster, the signal intensities of the four probes did not correspond for the two clinical isolates (for example, the signal intensity was high for one clinical strain but low for the other clinical strain). All four of these probes had highest sequence similarity to phage proteins, indicating that the phage sequence was present in one clinical isolate but not the other.
Although human listeriosis is primarily a food-borne illness, bovine listerial encephalitis may be caused by factors other than invasion of the gastrointestinal tract. In particular, direct infection of the trigeminal nerve may occur during changes in dentition, leading to bovine listerial encephalitis (23). Therefore, a variety of risk factors, including strain virulence, route of entry, dose, host immune response, and physical stresses (calving, concurrent disease, etc.), may contribute to the occurrence of bovine listeriosis (19). Therefore, the inability of microarray analysis to identify probes characteristic of both listeriosis isolates may be due to assay format (not all open reading frames of the genome are represented on this array) or the influence of other risk factors mentioned previously that contribute to the clinical outcome of infection.
In conclusion, subtyping of 538 isolates from farm A revealed that the farm was contaminated with a diversity of L. monocytogenes subtypes. Silage contamination and fecal shedding were the major sources of strains, and although silage contamination decreased over time, fecal shedding increased. Numerous fecal samples collected directly from individual cattle yielded multiple serotypes and pulsovars. Serotype 1/2a was the most prevalent serotype isolated at both time points. Bovine listeriosis strains were identical or closely related to strains isolated from the feces of asymptomatic cows by PFGE and microarray subtyping.
Acknowledgments
We gratefully acknowledge the excellent technical assistance provided by Edith Orozco, Edward Kuhn, Tricia Trunek, Blanca Lopez, and James Murnighan.
Funding was provided by USDA-Agricultural Research Service grant CWU 5348-32000-017-00D and the Agricultural Animal Health Program (College of Veterinary Medicine, Washington State University).
REFERENCES
- 1.Border, P. M., J. J. Howard, G. S. Plastow, and K. W. Siggens. 1990. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Lett. Appl. Microbiol. 11:158-162. [DOI] [PubMed] [Google Scholar]
- 2.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 3.Borucki, M. K., J. D. Peppin, D. White, F. Loge, and D. R. Call. 2003. Variation in biofilm formation among strains of Listeria monocytogenes.Appl. Environ. Microbiol. 69:7336-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borucki, M. K., J. Reynolds, C. C. Gay, K. L. McElwain, S. H. Kim, D. P. Knowles, and J. Hu. 2004. Dairy farm reservoir of Listeria monocytogenes sporadic and epidemic strains. J. Food Prot. 67:2496-2499. [DOI] [PubMed] [Google Scholar]
- 5.Borucki, M. K., S. H. Kim, D. R. Call, S. C. Smole, and F. Pagotto. 2004. Selective discrimination of Listeria monocytogenes epidemic strains by a mixed-genome DNA microarray compared to discrimination by pulsed-field gel electrophoresis, ribotyping, and multilocus sequence typing. J. Clin. Microbiol. 42:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes.J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes.J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravani, R. 1999. Listeria in food-processing facilities, p. 657-709. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety. Marcel Dekker, New York, N.Y.
- 9.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovett, J., D. Francis, and J. Hunt. 1987. Listeria monocytogenes in raw milk: detection, incidence and pathogenicity. J. Food Prot. 50:188-192. [DOI] [PubMed] [Google Scholar]
- 12.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nightingale, K. K., Y. H. Schukken, C. R. Nightingale, E. D. Fortes, A. J. Ho, Z. Her, Y. T. Grohn, P. L. McDonough, and M. Wiedmann. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page, R. D. M. 1996. Treeview: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 15.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryser, E. T., S. M. Arimi, and C. W. Donnelly. 1997. Effects of pH on distribution of Listeria ribotypes in corn, hay, and grass silage. Appl. Environ. Microbiol. 63:3695-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryser, E. T. 1999. Foodborne listeriosis, p. 299-358. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety. Marcel Dekker, Inc., New York, N.Y.
- 19.Schukken, Y. H., Y. T. Grohn, and M. Wiedmann. 2003. Listeria monocytogenes, p. 257-266. In M. E. Torrence and R. E. Isaacson (ed.), Microbial food safety in animal agriculture. Iowa State Press, Ames.
- 20.Skovgaard, N., and C. A. Morgen. 1988. Detection of Listeria spp. in faeces from animals, in feeds, and in raw foods of animal origin. Int. J. Food Microbiol. 6:229-242. [DOI] [PubMed] [Google Scholar]
- 21.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes.J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wesley, I. 1999. Listeriosis in animals, p. 39-73. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety. Marcel Decker, Inc., New York, N.Y.
- 23.Wesley, I., M. Borucki, D. R. Call, D. Larson, and L. Schroeder-Tucker. 2003. Detection and diagnosis of Listeria monocytogenes and listeriosis in animals, p. 233-242. In M. E. Torrence and R. E. Isaacson (ed.), Microbial food safety in animal agriculture. Iowa State Press, Ames.
- 24.Yoshida, T., Y. Kato, M. Sato, and K. Hirai. 1998. Sources and routes of contamination of raw milk with Listeria monocytogenes and its control. J. Vet. Med. Sci. 60:1165-1168. [DOI] [PubMed] [Google Scholar]