Abstract
Arctic tundra and boreal forest soils have globally relevant functions that affect atmospheric chemistry and climate, yet the bacterial composition and diversity of these soils have received little study. Serial analysis of ribosomal sequence tags (SARST) and denaturing gradient gel electrophoresis (DGGE) were used to compare composite soil samples taken from boreal and arctic biomes. This study comprises an extensive comparison of geographically distant soil bacterial communities, involving the analysis of 12,850 ribosomal sequence tags from six composite soil samples. Bacterial diversity estimates were greater for undisturbed arctic tundra soil samples than for boreal forest soil samples, with the highest diversity associated with a sample from an extreme northern location (82oN). The lowest diversity estimate was obtained from an arctic soil sample that was disturbed by compaction and sampled from a greater depth. Since samples from the two biomes did not form distinct clusters on the basis of SARST data and DGGE fingerprints, factors other than latitude likely influenced the phylogenetic compositions of these communities. The high number of ribosomal sequences analyzed enabled the identification of possible cosmopolitan and endemic bacterial distributions in particular soils.
Arctic and boreal environments cover 22% of the terrestrial surface of the planet and are sensitive to climate change, and changes in their productivity have substantial impacts on the global climate (7). Considering the critical role that the microbial components of these soils play, it is surprising how little is known about their composition and distribution. In particular, arctic tundra soil is poorly studied, and its microbial communities are commonly assumed to be species poor (15, 17). In fact, the application of genomic research in polar biology is considered a “test bed” for extrapolation to more complex ecosystems (28). However, recent results have suggested that polar environments may contain substantial microbial diversity. Schadt et al. (32) used biomass measurements and fungal sequence libraries to describe unexpectedly high fungal diversity and activity in snow-covered tundra soils. Furthermore, DNA reassociation analysis from a variety of soils indicated that genetic diversity in high arctic tundra was similar to that in temperate soils (31). Previously, only one study investigated tundra bacterial diversity by examining a 16S rRNA gene clone library. Zhou et al. (39) screened 43 clones from a Siberian tundra by using restriction fragment length polymorphism. They demonstrated maximum possible diversity, because all clones had unique restriction fragment length polymorphism patterns. Similar high diversity was observed for Wisconsin agricultural soil (4) and a tropical forest soil (5). However, all these studies involved small clone libraries, which preclude relative comparisons of diversity. Prior to this study, there was no published evidence suggesting that bacterial diversity in arctic tundra was higher or lower than that in different geographical regions.
Cloning and sequencing of PCR-amplified 16S rRNA genes are commonly used methods for profiling microbial community composition (14). However, labor and cost limitations have precluded sample sizes of greater than a few hundred sequences. These sample sizes are too small to adequately describe and compare multiple microbial communities containing thousands of species (19), such as those found in pristine soil and sediment samples (21, 36). Toward overcoming these limitations, serial analysis of ribosomal sequence tags (SARST) was developed for amplification of a highly variable region of the 16S rRNA gene and ligation of these fragments into concatemers that are cloned and sequenced (22, 29, 30). The power of this method is that variable regions from many different organisms are obtained from each sequencing reaction.
In this study, SARST and denaturing gradient gel electrophoresis (DGGE) were used to examine the relative abundance and diversity of bacteria in composite soil samples from five undisturbed sites in the boreal forest and arctic tundra biomes. A sample from a disturbed arctic site was also characterized, in which the soil was compacted during construction of a pad supporting a fuel storage tank. Analysis of between 1,487 and 2,659 ribosomal sequence tags (RSTs) from each sample, with a total of 12,850 RSTs, provided the basis for robust estimates of phylotype richness and composition.
RST library analysis indicated a positive correlation between diversity and latitude, contrary to the latitudinal biodiversity gradient observed for most biodiversity on earth (38). Similarity analysis of SARST and DGGE data determined that samples did not form discrete biome-specific clusters, indicating that factors other than those represented by latitude governed the microbial community compositions of these geographically distant soils. The large collection of RSTs from each sample provided evidence for potentially endemic and cosmopolitan distributions of bacteria within these soil environments.
MATERIALS AND METHODS
Soil samples.
Similar sampling regimens were applied to all pristine soil sites. Surface mineral soil subsamples (3 to 10 samples; 0 to 10 cm, ∼100 to 200 g) were collected during the summer from two undisturbed arctic tundra sites and from three undisturbed boreal forest sites in Canada (Fig. 1A; Table 1). Multiple samples were taken from within an area of approximately 20 by 20 meters, with specific sample locations chosen as being representative of the particular boreal forest or arctic tundra sites. For the forest soils, samples were obtained from beneath the litter layer from within three different forest types (balsam fir, jack pine, and spruce-aspen mixed wood) to obtain soil samples that represented several boreal forest systems. In addition, a composite soil sample was taken from different depths within the top 100 cm of a soil pad constructed to support a fuel tank at a former Distant Early Warning Line station (DYE-MAIN), although the individual samples had petroleum hydrocarbon levels below detectable levels (data not shown). All soils, associated letter codes, and measured soil properties are indicated in Table 1. Subsamples were kept at 4°C for transport back to the laboratory and were used immediately or frozen at −80°C. For each soil, subsamples were sieved (5 mm) and an equal portion (by weight) was added from each to form a composite, which was thoroughly mixed. A portion of each composite was sent for physical and chemical analyses to Pacific Soil Analysis (Richmond, British Columbia, Canada).
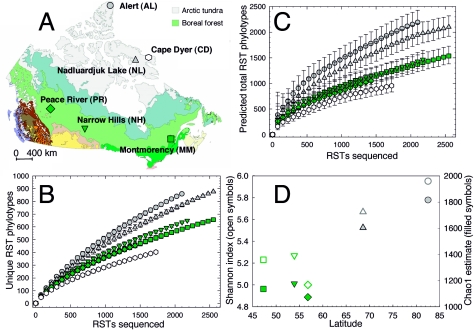
FIG. 1.
Source and diversity of soil RST libraries. All symbols correspond to sources of libraries as shown in panel A. (A) Geographical locations and biomes of sampling sites. This map was modified with permission from the Canadian Wildlife Service. (B) Rarefaction curves. Error bars are 95% confidence intervals from 100 randomizations of each library. (C) Chao1 richness estimates. Error bars are 95% confidence intervals from 100 randomizations of each library. (D) Relationship between latitude and diversity for undisturbed soil samples, shown as both Shannon-Weiner indices (richness and evenness; open symbols) and Chao1 estimates (richness; filled symbols) for equivalent-size RST library subsamples (1,487 RSTs).
TABLE 1.
Composite soil sample characteristics and associated information
| Soil sample | Site | Sample datea | Latitude | Longitude | Location | H2O (%) | Soil type | pH | C/N ratio | DNA (μg/g) | Organic (%) | RSTs sequenced |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alert | Tundra | 980804 | 82°30′N | 62°19′W | Nunavut | 19.0 | Silty loam | 7.6 | 31.4 | 30.0 | 9.2 | 2,117 |
| Nadluardjuk Lake | Tundra | 020815 | 68°37′N | 73°12′W | Nunavut | 2.5 | Sandy loam | 5.3 | 16.0 | 8.5 | 1.4 | 2,562 |
| Cape Dyer | Fuel pad | 021104 | 66°36′N | 61°34′W | Nunavut | 11.0 | Loamy sand | 6.0 | 8.7 | 0.4 | 0.3 | 1,785 |
| Montmorency | Balsam fir | 030909 | 47°19′N | 71°06′W | Quebec | 24.6 | Sandy loam | 4.7 | 25.3 | 26.0 | 6.1 | 2,659 |
| Narrow Hills | Jack pine | 030624 | 53°54′N | 104°41′W | Saskatchewan | 7.9 | Sand | 5.3 | 37.1 | 19.2 | 3.2 | 2,240 |
| Peace River | Mixed wood | 030613 | 56°46′N | 118°22′W | Alberta | 27.2 | Silty loam | 4.5 | 24.4 | 19.3 | 6.3 | 1,487 |
Year, month, day.
SARST.
DNA extraction, SARST, colony PCR, and sequencing of inserts were done as described by Neufeld and coworkers (29, 30). Briefly, DNA was extracted from triplicate 0.5-g subsamples from each composite soil sample by using the soil FastDNA SPIN kit in conjunction with a FastPrep Instrument (Qbiogene, Carlsbad, CA) with a repeated lysis step to maximize DNA yield. Combining all DNA solutions from the first and second lysis steps generated a DNA extract for SARST. A total of 18 25-cycle PCRs were conducted for each composite DNA extract, amplifying the V1 region from the 16S rRNA genes in the DNA extracts. For each library, all PCR products were pooled, and biotin-labeled primers were removed with simultaneous BpmI and BsgI digestion and subsequent purification with streptavidin beads. Linker oligonucleotides were ligated to each end of the RSTs, and RST-linker molecules were purified with polyacrylamide gel electrophoresis. Linkers were released with simultaneous SpeI and NheI digestion and subsequent streptavidin-bead purification. RSTs were ligated in the presence of SpeI and NheI for consistent 5′-to-3′ ligation. Polyacrylamide gel electrophoresis-purified concatemers of 300 to 500 bp served as inserts for generating clone libraries by using a SpeI-cut pZErO-2 vector (Invitrogen, Burlington, Ontario, Canada). Four 96-well plates were used for colony PCR of insert-containing colonies for each composite sample, and all inserts were sequenced regardless of size. DNA sequences were manually verified for base-calling accuracy using Chromas version 2.23 (Technelysium, Queensland, Australia), and RSTs were extracted from the resulting sequence text files using SARSTeditor (30).
All RSTs, site coordinates, and associated soil chemical properties were deposited in the Gene Expression Omnibus (GEO) database (10) of the National Center for Biotechnology Information. GEO was designed to hold gene expression data such as those generated by serial analysis of gene expression and microarray analysis, but it also accepts other forms of data such as those generated by SARST. GEO storage is helpful because most RST sequences are too short for GenBank submissions. Within GEO, SARST data are stored in platform GPL919, and the RST libraries from this study are entered in series GSE949. Corresponding sample accession numbers are Nadluardjuk Lake replicates (GSM14854 and GSM14855), Alert (GSM35149), Narrow Hills (GSM35162), Peace River (GSM35163), Montmorency (GSM35161) and Cape Dyer (GSM35159). RST data from the Nadluarjuk Lake site (FOX-B) were previously published as a duplicate library to confirm the reproducibility of SARST (30) and are included in this report for the purpose of comparison.
In order to make comparisons of RST library diversity and composition, RSTs from all libraries were clustered by similarity using SARSTgrouper (http://www.microbiology.ubc.ca/Mohn/SARST). By specifying a similarity threshold of 95% for clustering RSTs, the influence of PCR errors and variable intraspecific 16S rRNA gene operons on clustering was minimized (30). For identifying potential endemic and cosmopolitan RSTs, libraries were grouped together by exact matching using SARSTgrouper and then sorted to identify abundant RSTs (>10 total) found in one or more arctic soil libraries or in one or more boreal forest libraries or predominant (>20 total) in all of the libraries.
RST analysis.
Rarefaction curves, Chao1 richness estimates, and Shannon-Weiner diversity indices were calculated from clustered RST libraries by using EstimateS (version 5.0.1; R. Colwell, University of Connecticut [http://viceroy.eeb.uconn.edu/estimates]) as described previously (18, 19). Chao1 95% confidence intervals and Bray-Curtis similarity indices (6) were calculated using formulas formatted for compatibility with clone libraries (18). Because the RST libraries contain different numbers of RSTs (Table 1), 1,487 RSTs were randomly extracted from each library for generating Bray-Curtis similarity indices and Shannon-Weiner diversity indices because these measures are sensitive to sample size. Subtraction of the Bray-Curtis similarities from 100% provided a dissimilarity matrix for creating dendrograms (unweighted-pair group method using average linkages [UPGMA]) using the neighbor-joining program of the Phylip package (11). Divisions were assigned to individual RSTs based on the phylogenetic affiliation of the closest database hit in the Ribosomal Database Project (RDP-II) version 9.0 (8). Bray-Curtis similarity indices were calculated for division-level profiles, and UPGMA dendrograms were created as described above.
DGGE.
Using primers 63f-GC and 518r and 5 ng extracted soil DNA as the template in each reaction, PCR and DGGE were done as described previously (24) with minor modifications. PCR (25 cycles) amplified a ∼490-bp fragment and was carried out in a PTC-100 thermocycler (MJ Research, Waltham, MA). PCR products were quantified by comparison to a 1-kb ladder (Invitrogen, Burlington, Ontario, Canada) in a 1.5% agarose gel. DGGE electrophoresis was done using the Bio-Rad D-Code System (Bio-Rad, Hercules, CA) according to the manufacturer's directions. Gels had a denaturing gradient of 40 to 70% (100% denaturant contains 7.0 M urea and 40% deionized formamide) and were poured with an additional nondenaturing surface layer. Standard markers were generated with equal-volume mixtures of PCR products from 10 16S rRNA gene fragments cloned from cultured isolates or sample DGGE fingerprint bands. Electrophoresis was carried out for 14 h at 60°C and 85 V. Gels were stained with SYBR Green I nucleic acid gel stain (Molecular Probes, Eugene, OR) at a 1:10,000 dilution for 1 h. DGGE gels were scanned with a Typhoon 9400 imager (Amersham Biosciences, Piscataway, NJ). DGGE fingerprints were compared using Gel Compar II (Applied Maths, Belgium). Gel images were normalized using standards run in the outside and middle lanes. Fingerprint patterns were analyzed using Pearson's product moment correlations, providing pairwise percent similarity values for all fingerprint densitometric curves. A UPGMA dendrogram was created from this similarity matrix as described above.
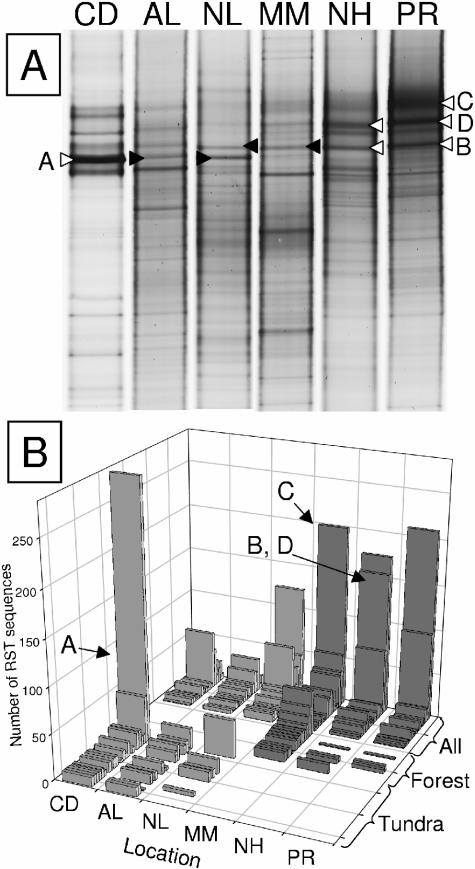
Bands of interest were excised from the gel with large-bore pipette tips. The gel fragments were incubated overnight at 4°C in 5 μl of Tris-EDTA buffer, and 1 μl of this mixture was then used as the template for PCR with the same primers and conditions used above but without a GC clamp. The PCR products were cleaned with Sephadex G-50 and sequenced as described previously (30). Bands providing high-quality sequence data without any ambiguous base calling were submitted to GenBank. Other bands yielded less clear sequence data but were still useful for confirming their similarity to bands in other lanes, and specifically for confirming an RST identical to other bands. Because one selected band yielded unclear sequence data (band C; see Fig. 4A), the PCR products were cloned using the TOPO-TA cloning kit (Invitrogen) and five inserts were sequenced to identify the insert that most closely resembled the data in the original sequencing reaction.
FIG. 4.
Comparison of abundant phylotypes with potential cosmopolitan and endemic distributions for each biome. (A) Soil DGGE fingerprints, with an indication of bands selected for sequencing. Open arrowheads indicate bands that provided excellent sequence data. Filled arrowheads generated mixed sequence data but were nonetheless sufficient for confirming the sequence as being identical to the corresponding high-quality sequences. (B) Abundant RST phylotypes associated with either biome or with all samples. Dark bars indicate boreal forest soil samples. Letter codes with arrows indicate RSTs that were present in all libraries (C, B, and D) or only in the tundra libraries (A) that matched sequenced DGGE bands. Sample locations are Alert (AL), Nadluardjuk Lake (NL), Cape Dyer (CD), Montmorency (MM), Narrow Hills (NH), and Peace River (PR).
Nucleotide sequence accession numbers.
Sequences for DGGE bands A, B, and C were cropped to remove primer sequences and deposited in GenBank with accession numbers AY823417 (band A, Cape Dyer), AY823416 (band B, Peace River), AY823415 (band C, Peace River), and AY847702 (band B, Narrow Hills). Fingerprint band D also provided clear sequence data and was stored in GenBank with accession numbers AY847703 and AY847704 for Narrow Hills and Peace River, respectively.
RESULTS AND DISCUSSION
Composite soil samples were taken from three arctic tundra sites and three boreal forest locations (Fig. 1A). From each sample, between 1,487 and 2,659 RSTs were obtained using SARST (Table 1), for a total of 12,850 RSTs. With approximately 400 inserts sequenced per sample, regardless of insert size or quality, SARST generated an average of over five RSTs per sequencing reaction. The largest concatemer yielded 18 RSTs in a single reaction. These are the largest collections of 16S rRNA gene sequences from individual environmental samples reported to date. Because the majority of RSTs are genus or species specific (29, 30), clustering of RSTs is comparable to clustering of corresponding longer 16S rRNA gene sequences. Further, one of the libraries (Nadluardjuk Lake) was a previously published duplicate soil library (30), and high correlation between the duplicates indicated that RST libraries were reproducible. High reproducibility of RST libraries justified the comparison of single RST libraries generated from each soil composite in this study. Therefore, trends evident in the RST libraries should reflect trends evident by using traditional 16S rRNA gene clone libraries and should provide representative descriptions of bacterial community composition.
Three diversity measures consistently indicated that the undisturbed arctic tundra soil libraries possessed greater bacterial diversity than the boreal forest soil libraries. Rarefaction analysis, which averages randomizations of the species-accumulation curve, indicated that the observed diversities of RST libraries from the two tundra soils were greater than those of the three boreal forest soils (Fig. 1B). The most diverse sequence library originated from an extremely high latitude, the northern tip of Ellesmere Island, Nunavut. The library from the disturbed soil at Cape Dyer exhibited the lowest richness. Rarefaction is arguably the best means to compare the libraries, but a disadvantage of using rarefaction is that curves may cross with further sampling (19). In addition, the confidence intervals around rarefaction curves reflect the error associated with reordering individual subsamples and do not reflect the precision of the observed richness. Nonparametric estimators, compared to rarefaction, provide more meaningful projections of the actual diversity within the sampled environment (19). Nonparametric Chao1 estimates, which predict the point at which an accumulation curve will reach an asymptote, also indicated that the richness of the undisturbed arctic tundra soil RST libraries was significantly greater than that of the boreal forest soil RST libraries (Fig. 1C and D) and was significantly lesser in the disturbed arctic soil than in all the other soil sample libraries. For equivalent subsamples from undisturbed soils, the Chao1 richness estimates were positively correlated with latitude (r = 0.94; P = 0.017 [n = 5]). The Shannon-Weiner diversity index (H′) reflects both phylotype richness and evenness and is thus a good overall measure of diversity. The H′ values for the undisturbed soils (Fig. 1D) indicated that with equivalent subsample size, the tundra soil RST libraries had greater bacterial diversity than the forest soil RST libraries, and this diversity measure was also positively correlated with latitude (r= 0.88; P = 0.046 [n = 5]). The RST library from the disturbed Cape Dyer soil had the least diversity, with a Shannon index of only 4.61.
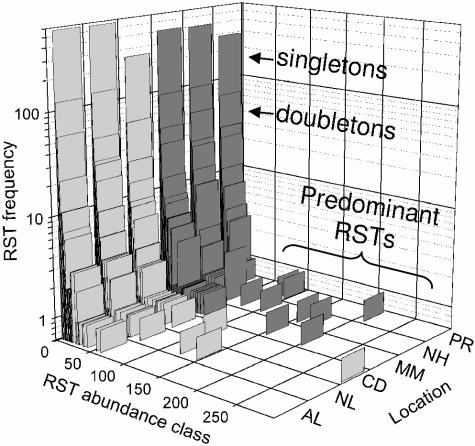
The distribution of unique RSTs in each library provided a visual explanation for the factors influencing the diversity estimates (Fig. 2). The three forest soils contained a higher proportion of predominant RSTs, and the disturbed arctic soil contained a clearly dominant RST. Strong predominance of individual RSTs indicated a lower evenness of RST distributions and affected the Shannon-Weiner diversity index, in particular. In addition, the undisturbed tundra libraries had a higher proportion of rare RST sequences than did the forest, or the Cape Dyer, libraries. Rare RSTs are those that occur once (singletons) or twice (doubletons) in each library. Because the Chao1 diversity estimate uses the relative proportions of singletons and doubletons for calculating estimated diversity, this abundance of rare sequences in tundra soils leads to higher estimates of richness.
FIG. 2.
RST frequency histogram for the six soil composites. RST frequency is plotted on a logarithmic scale against abundance class. Singletons, doubletons, and predominant RSTs are indicated within the graph area for convenience. Dark bars indicate boreal forest soil samples. Sample locations are Alert (AL), Nadluardjuk Lake (NL), Cape Dyer (CD), Montmorency (MM), Narrow Hills (NH), and Peace River (PR).
These results challenge a longstanding observation in ecology: that the taxonomic diversity of flora and fauna decreases as one samples closer to polar regions (38). Previous studies of microbial community diversity along latitudinal gradients are almost nonexistent. Staddon et al. (34) reported decreasing soil functional diversity moving northward along a latitudinal transect through Canadian boreal forest in parts of Saskatchewan and Manitoba. However, this transect was relatively short, and it is unclear how functional diversity relates to taxonomic diversity. Another study used clone libraries to measure the diversity of soil microbial eukaryotic organisms along a latitudinal transect proximal to the South Pole (23). While the most southerly sample possessed the lowest diversity, they discovered an unexpected increase in diversity with proximity to the South Pole within the maritime Antarctic (60 to 72oS). Notably, many exceptions to the latitudinal biodiversity gradient occur in studies that sample across relatively short latitudinal ranges of less than 20o (38), suggesting that local inversions of the gradient may not be uncommon.
The samples analyzed here were obtained from a relatively broad latitudinal range (47 to 82oN) and involved 16S rRNA gene libraries of sufficient size to enable the detection of statistically significant differences in diversity estimates for these samples (Fig. 1). Overlapping confidence intervals for diversity estimates are a result of insufficient sampling and are common for comparisons of rarefaction and Chao1 estimates in species-rich environments. Large confidence intervals have precluded detection of statistically significant differences in diversity estimates (3, 16, 33). Here, RST library subsets of a magnitude similar to that of traditional 16S rRNA gene clone libraries (∼100 to 300 clones) were insufficient to discriminate between any of the soils. Subsets of approximately 1,000 RST sequences were required to statistically discriminate between forest and tundra soil diversity estimates (Fig. 1B and C). However, the proportion of rare sequences in each library is high (Fig. 2), and the Chao1 estimates do not reach asymptotes (Fig. 1C). As with rarefaction curves, unstable Chao1 estimates might cross with further sampling. Therefore, even with thousands of RSTs sequenced, library sizes were inadequate for comprehensive coverage of these soil bacterial populations, and diversity estimates should be interpreted cautiously, as additional sequencing may affect these conclusions.
Additional caution should accompany these results because the data were obtained from relatively few composite samples due to the effort involved in collecting data from each location. Also, the impacts of DNA extraction (26), PCR amplification (37), and variable copy numbers of 16S rRNA gene operons (9) may have contributed further bias. As this is the first substantial investigation of arctic tundra soil microbial diversity, further research is needed to confirm the observations reported here. For example, measuring the diversity of additional tundra and boreal forest samples, as well as the diversity in lower-latitude samples from tropical regions and regions in the Southern Hemisphere would provide additional insight into this possible biodiversity trend. Methodologies such as DNA reassociation (35) would help provide confirmation of sequence-based results. Nucleic acid fingerprinting could enable rapid comparisons of replicate samples to assess within-site spatial variability.
High bacterial diversity measured in arctic tundra soils suggests that factors governing biodiversity in macrobiological communities may have different influences on microbiological communities. Relative to arctic tundra soils, boreal forest soils have higher carbon flux due to leaf decomposition and higher average temperatures leading to longer annual periods of high metabolic activity (7). These productivity factors might be selective pressures contributing to decreased bacterial diversity, although the inverse could also be argued (38). The lower pH of forest soils (Table 1) might favor fungal populations (2), leading to increased competition between fungal and bacterial populations. The influence of soil pH on bacterial diversity is unknown, but this may have been a factor contributing to lower bacterial diversity observed in the forest libraries. Another possibility is that the relatively great bacterial diversity of tundra soils may largely reflect allochthonous organisms having low metabolic activity and little functional significance in the soil systems, an example of which is viable mesophilic and thermophilic bacteria isolated from cold soil environments (25, 27). Such organisms may enter the soil via atmospheric transport, and low arctic temperatures may foster their persistence. If allochthonous populations, preserved by low temperatures, contributed substantially to the observed diversity of these RST libraries, these organisms might be represented by sequences of low relative abundance. However, by eliminating all singletons from each library and repeating the diversity analyses, the relative ranking of soil diversity was identical to that using the full sequence set (data not shown). This suggests that high bacterial diversity observed in these arctic tundra samples was not simply an artifact of cell preservation. Further investigations focusing on metabolically active bacteria (e.g., rRNA analysis) would help determine the effect of allochthonous organisms on microbial diversity in arctic soils and other environments and help in understanding the functional significance of microbial diversity.
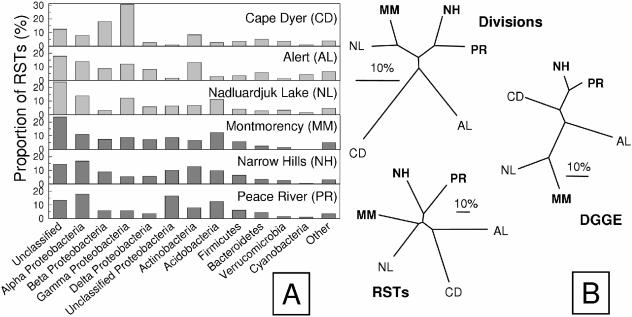
A closer examination of the structures of the RST libraries indicated that despite geographic isolation, as well as differences in soil chemistry and ground vegetation, the undisturbed tundra and forest soils had similar division-level representation (Fig. 3A). The taxonomic affiliations of RSTs demonstrated a dominance of Proteobacteria and substantial proportions of Actinobacteria, Acidobacteria, Firmicutes, Bacteroidetes, Verrucomicrobia, and Cyanobacteria. All libraries contained a large proportion of RSTs (∼10 to 25%) with close affiliations to 16S rRNA gene sequences of unknown phylogenetic affiliation. The Bray-Curtis index (6) was used for a similarity comparison of overall RST composition and relative abundance and also for comparing the division-level distribution for each of the soils (Fig. 3B). There was no strong clustering among the samples and, particularly, no separation of forest from tundra samples. The Bray-Curtis index indicated that the Narrow Hills and Peace River soils had the greatest similarity. Also, one of the arctic tundra samples (Nadluardjuk Lake) was more similar to one of the boreal forest soils (Montmorency) than to other samples. The DGGE fingerprints for each of these soil samples (Fig. 4A) were compared to one another, and the resulting DGGE fingerprint dendrogram (Fig. 3B) also clustered the Narrow Hills and Peace River samples together, as well as the Nadluardjuk Lake and Montmorency samples. Thus, similar topographical features of these dendrograms indicated agreement between the analyses. The lack of biome-specific clustering suggests that the overall structure of these soil microbial communities was governed by more factors than those related to latitude (annual temperature, insolation, and seasonality). If anything, undisturbed soils from the east and west clustered distinctly, and the RST composition for Alert was unique among the soil samples (Fig. 3B). Longitudinal clustering may be an initial indication that bacterial distribution by atmospheric vectors is an important determinant of soil community structure (12).
FIG. 3.
Comparison of RST library composition. (A) Relative abundance of phylogenetic divisions for each soil library in which RST sequences were assigned to the same taxonomic group as the closest relative in the RDP-II database. Dark bars indicate boreal forest samples. (B) UPGMA dendrograms showing Bray-Curtis similarity indices for phylogenetic divisions (Divisions), RST distributions (RSTs), and Pearson correlations of DGGE fingerprint patterns (DGGE). The scale bar indicates 10% dissimilarity between samples. Boldface sample codes indicate boreal forest soil samples.
The Cape Dyer soil sample is unique in its low carbon and DNA concentrations (Table 1), reduced RST library diversity (Fig. 1B and C), dominance of a single phylotype (Fig. 2), and having a different phylogenetic composition than the other soil samples (Fig. 3). This could be an example of the effect of disturbance on microbial community composition. Perturbation has been associated previously with reduced microbial biodiversity (1) and may be the cause of this sample's uniqueness. Alternatively, the uniqueness may be because the Cape Dyer composite was generated from samples taken from a greater depth (surface to 100 cm) than the other surface soil composites. Depth has been associated with lower microbial diversity in soil environments (40), which is attributable to higher water saturation. Comparison of pad samples with pristine soil samples obtained from surrounding Cape Dyer tundra would be required to further clarify the causes of this observation.
Several relatively abundant RSTs were associated solely with soils from one or the other biome, potentially representing populations endemic to either arctic tundra or boreal forest soils (Fig. 4; Table 2). Of the abundant RST groups (195 total), 25 were unique to one or more tundra libraries, and 17 were present in at least one of the three forest libraries but not in the tundra libraries. These phylotypes potentially represent populations endemic to their respective biomes. Of the very abundant RST groups (90 total), 18 were common to all libraries, potentially representing cosmopolitan populations. The RDP-II contained sequences identical to all of the ubiquitous RSTs (Table 2), indicating their frequent occurrence in clone libraries from other sources. In contrast, database sequences were identical to only 24% of the RSTs found solely in forest libraries and to only 60% of the RSTs found solely in tundra libraries. Most of the RSTs that were solely associated with either tundra or forest soils were collected from one particular soil (primarily Cape Dyer, Montmorency, and Alert) instead of being associated with multiple soils from a given biome.
TABLE 2.
Distributions of abundant RSTs associated with all samples (RST code C), boreal forest soils (RST code B), or arctic tundra soils (RST code A), corresponding taxonomic affiliations, and similarity score (S_ab) of the closest match in the RDP-II
| RST (DGGE band) | S_ab | Closest RDP affiliation | No. of RSTs ata:
|
RST Sequence | |||||
|---|---|---|---|---|---|---|---|---|---|
| CD | AL | NL | MM | NH | PR | ||||
| C1 (band C) | 1 | Unclassified Proteobacteria | 6 | 13 | 123 | 200 | 175 | 210 | AGCGGTAACAGGTGTAGCAATACATGCTGACGAGC |
| C2 (bands B and D) | 1 | Alpha Proteobacteria | 23 | 5 | 48 | 57 | 157 | 98 | AGCGGGCGTAGCAATACGTCAGC |
| C3 | 1 | Beta Proteobacteria | 16 | 36 | 10 | 39 | 72 | 15 | AACGGCAGCACGGGGGCAACCCTGGTGGCGAGT |
| C4 | 1 | Actinobacteria | 10 | 22 | 37 | 36 | 11 | 16 | AGCGGAAAGGCCCTTCGGGGTACTCGAGC |
| C5 | 1 | Beta Proteobacteria | 43 | 25 | 18 | 4 | 12 | 6 | AGCGGCAGCGCGGGGCAACCTGGCGGCGAGC |
| C6 | 1 | Beta Proteobacteria | 64 | 12 | 7 | 4 | 15 | 1 | AACGGCAGCACGGGAGCAATCCTGGTGGCGAGT |
| C7 | 1 | Gamma Proteobacteria | 5 | 1 | 66 | 23 | 4 | 2 | AGCGGTAACGCGGGAGCAATCCTGGCGACGAGC |
| C8 | 1 | Gamma Proteobacteria | 8 | 12 | 19 | 17 | 11 | 9 | AGCGGCAGCGCGGGGGCAACCCTGGCGGCGAGC |
| C9 | 1 | Unclassified | 4 | 2 | 15 | 28 | 5 | 1 | AACGCGAAAGTCCCGCAAGGGATCAGTAGAGT |
| C10 | 1 | Unclassified | 6 | 11 | 5 | 21 | 4 | 3 | AACGCGAAAGTCCCGCAAGGGATGAGTAGAGT |
| C11 | 1 | Acidobacteria | 1 | 5 | 9 | 24 | 4 | 2 | AACGAGAAAGGGGAGCAATCCCTGAGTAAAGT |
| C12 | 1 | Unclassified | 11 | 3 | 14 | 6 | 8 | 3 | AACGCGAAAGTCCCGCAAGGGATAAGTAGAGT |
| C13 | 1 | Alpha Proteobacteria | 2 | 2 | 7 | 2 | 20 | 8 | AACGCGTGTAGCAATACACGAGT |
| C14 | 1 | Gamma Proteobacteria | 3 | 1 | 5 | 8 | 9 | 13 | AACGGTAACAGGCCCGCAAGGGTGCTGACGAGT |
| C15 | 1 | Beta Proteobacteria | 2 | 7 | 5 | 3 | 13 | 2 | AACGGTAACGCGGGGGCAACCCTGGCGACGAGT |
| C16 | 1 | Unclassified Proteobacteria | 6 | 1 | 5 | 6 | 10 | 4 | AACGCGAAAGTCCCGCAAGGGATTAGTAGAGT |
| C17 | 1 | Alpha Proteobacteria | 6 | 1 | 11 | 3 | 3 | 3 | AACGATAAGCCACCTTCGGGTGGTGGACAGT |
| C18 | 1 | Alpha Proteobacteria | 1 | 4 | 9 | 3 | 3 | 1 | AACGCCCCGCAAGGGGAGT |
| B1 | 0.89 | Gamma Proteobacteria | 49 | AACGTGAAAGCGGGGCAACCCGCAAGTAGAGT | |||||
| B2 | 0.67 | Delta Proteobacteria | 33 | AGGGAGAAAGCCCGCAAGGGTTAGTAAACC | |||||
| B3 | 0.50 | Firmicutes | 24 | AACGAGCTGGTTTTCTTCGGAGAGCGAGCGAGT | |||||
| B4 | 0.50 | Alpha Proteobacteria | 16 | AACGCCCTTTGTCTCGCAAGAGAGGGAGGGAGT | |||||
| B5 | 1 | Unclassified | 12 | 1 | 1 | AACGCGCAGCAGTTTGTAGCAATACAGATTGTGGGCGCGT | |||
| B6 | 0.56 | Delta Proteobacteria | 14 | AACGCCCTTTGTCTCGCAAGAGAGGAAGGGAGT | |||||
| B7 | 0.75 | Aquificae | 14 | AACGGGTGCTAACTGCCCGCAAGGGT | |||||
| B8 | 0.68 | Unclassified | 13 | AACGCGCAGTTTCCTGTAGCAATACAGGGAATGGGCGCGT | |||||
| B9 | 0.60 | Actinobacteria | 12 | AACGGTGACCTCACTTCGGTGGGTGATCAGT | |||||
| B10 | 0.49 | Unclassified | 11 | AACGAGGATCATCGGGTTAGCAATAATTCGGTGGTCCTAGT | |||||
| B11 | 0.72 | Firmicutes | 5 | 6 | AGCGAATCTTTAGGAGCTTGCTCCTATTGGTTAGC | ||||
| B12 | 1 | Alpha Proteobacteria | 2 | 5 | 4 | AACGCGACCTTCGGGTCGAGT | |||
| B13 | 0.40 | Actinobacteria | 10 | AGCGTGGGCGTGCTGTCTCGCAAGAGATGGCACGTTCTAGC | |||||
| B14 | 1 | Unclassified | 10 | CACGAGAAACTCTGTAGCAATACGGGGCGGTAAAGT | |||||
| B15 | 0.52 | Firmicutes | 10 | AACGAGTTTGTCTTCTTCGGAAGATGAGCGAGT | |||||
| B16 | 1 | Nitrospira | 10 | AACGAGAAGGCGTAGCAATACGCTTGTAAAGT | |||||
| B17 | 0.96 | Delta Proteobacteria | 10 | AGCGAGAAAGGGGCAACCCCGGTAAAGC | |||||
| A1 (band A) | 1 | Gamma Proteobacteria | 284 | 23 | 42 | AACGGCAGCACAGAGGAGCTTGCTCCTTGGGTGGCGAGT | |||
| A2 | 1 | Beta Proteobacteria | 51 | 2 | AACGGTAGAGTAGCAATACTCGAGAGT | ||||
| A3 | 1 | Actinobacteria | 10 | 13 | AACGGTGAACCGGGCTTCGGCCCGGGGATCAGT | ||||
| A4 | 1 | Beta Proteobacteria | 10 | 10 | AACGGTAACGCGGGGCAACCTGGCGACGAGT | ||||
| A5 | 0.57 | Beta Proteobacteria | 19 | AACGGCAGCACGGGACTCAGGCAACTGAGCCCTGGTGGCGAGT | |||||
| A6 | 0.67 | Planctomycetes | 18 | AGCGAGAACCTAGGCTTCGGCTTAGGGGACAGC | |||||
| A7 | 0.78 | Gamma Proteobacteria | 17 | AACGGTAACAGACCCTTCGGGGTGCTGACGAGT | |||||
| A8 | 1 | Unclassified | 6 | 11 | AACGCGAAAGGGGCTTCGGCCCTGAGTAGAGT | ||||
| A9 | 0.46 | Cyanobacteria | 16 | TACGGGAAATATCCTAGTGGTGTTTCCAGT | |||||
| A10 | 1 | Delta Proteobacteria | 16 | AACGCGAAAGTCCTTCGGGATGAGTAAAGT | |||||
| A11 | 1 | Verrucomicrobia | 6 | 9 | AACGGGATTACTTTTGGTAGCAATACCGAAAGTGATTCAGT | ||||
| A12 | 1 | Actinobacteria | 11 | 3 | AGCGAGAACCAGGCCTTCGGGCCTGGGGACAGC | ||||
| A13 | 0.65 | Unclassified | 3 | 11 | AACGGGTGTTAGCTGCCCGCAAGGGT | ||||
| A14 | 0.89 | Gamma Proteobacteria | 12 | AACGGTAACGGGCCCTTCGGGGTGCTGACGAGT | |||||
| A15 | 0.49 | Verrucomicrobia | 11 | AACGGGAACTCTTTTGGTAGCAATACCGGGAGAGTTCTAGT | |||||
| A16 | 1 | Cyanobacteria | 11 | AACGGTAACAGGAAACAGCTTGCTGTTTCGCTGACGAGT | |||||
| A17 | 1 | Unclassified | 2 | 9 | AACGCGAAAGTCCCCTTCGGGGGGCGAGTAGAGT | ||||
| A18 | 1 | Actinobacteria | 11 | AGCGAGAACCGGACCTTCGGGTCCGGGGACAGC | |||||
| A19 | 0.59 | Alpha Proteobacteria | 11 | AACGGGGTCTTTCGGGATCTAGT | |||||
| A20 | 1 | Gamma Proteobacteria | 6 | 4 | AACGGCAGCACAGAGGAGCTTGCTCCTTGGGTGGCGAG | ||||
| A21 | 1 | Beta Proteobacteria | 10 | AGCGGACAGATGGGAGCTTGCTCCCTGATGTTAGC | |||||
| A22 | 0.89 | Unclassified | 9 | 1 | AACGCGAAAACCCCGCAAGGGGTTAGTAGAGT | ||||
| A23 | 0.64 | Firmicutes | 10 | AACGGATCTTTTCCTTCGGGGAAAGGTTAGT | |||||
| A24 | 1 | Actinobacteria | 10 | AACGAGAAAGCCCTTCGGGGTTAGTAAAGT | |||||
| A25 | 1 | Cyanobacteria | 10 | AACGGAATCTTTCGGGATTTAGT | |||||
CD, Cape Dyer; AL, Alert; NL, Nadluardjuk Lake; MM, Montmorency; NH, Narrow Hills; PR, Peace River.
The ubiquitous distribution of RSTs was not difficult to demonstrate, but true endemicity is not possible to confirm by sampling sequences from the environment, even when the sampled coverage of a population is high (23). However, complete database representation for ubiquitous RSTs and only partial representation of the uniquely distributed RSTs (Table 2) provided additional support for the ubiquitous and limited distributions of certain phylotypes observed in this study. To confirm the cosmopolitan or endemic nature of organisms and the underlying causes of their distributions, further physiological and phylogenetic information would be required. Culture-based approaches are thus critical to understanding bacterial distributions and are becoming possible even for previously uncultured organisms (20). Because RSTs can be used for designing phylotype-specific PCR primers (30), more phylogenetic information (a larger portion of the 16S rRNA gene sequence) can be obtained for selected RSTs.
DGGE analysis confirmed the most abundant RST distributions, because relatively intense bands in the fingerprints (Fig. 4A) were sequenced and found to correspond to predominant RSTs. The relative band intensities for each sample were similar to the relative abundances of the corresponding RST in the sequence libraries. For example, band A was visibly apparent only in arctic soil DGGE fingerprints. The sequence obtained from this band corresponds to an RST found only in the arctic soil RST libraries. The 417-bp sequence was identical to the corresponding sequences of clinical and environmental isolates of antibiotic- and siderophore-producing strains of Stenotrophomonas maltophilia, which have been isolated from a variety of environments. Isolates of this organism possess high genetic diversity despite low 16S rRNA gene heterogeneity (13). Thus, the ecological significance of this abundant sequence in arctic tundra soils is unknown. Bands B and C were apparent in many of the sample DGGE fingerprints, and corresponding RSTs for these bands were associated with all soil libraries. Band B had 100% identity to strains of Afipia broomeae, which are common soil inhabitants and closely related to Bradyrhizobium species. Band C differed by only one base from bacterial 16S rRNA gene clones from soil (unknown taxonomic affiliation) and was 93% similar to clones from cultured gamma Proteobacteria from Australian soil isolates. Band D was pronounced in the Narrow Hills and Peace River samples, and the sequences from these samples possessed a single nucleotide mismatch to band B across the ∼400-bp sequences. The RSTs in band B and D sequences were identical.
Based on the high number of soil-specific and ubiquitous sequences identified in RST libraries (Table 2), the ability of SARST to identify and compare potentially endemic and cosmopolitan populations of soil microorganisms surpasses that of DGGE or any other available method. This ability will help guide culture-based identification of ecologically important, endemic organisms. SARST provided an efficient approach for quantifying microbial diversity and distributions that potentially reflected the environmental conditions enabling phylotype growth and persistence in specific environments.
Despite the difficulty and great expense of accessing arctic study sites, organized research efforts are beginning to recognize the substantial ecological and industrial importance of investigating arctic tundra soils (28). Not only are cold-adapted organisms and enzymes likely abundant in arctic tundra environments, but this report demonstrates that the Arctic serves as an unrecognized reservoir of microbial diversity and thus of biochemical potential. An appreciation of the magnitude of arctic microbial biodiversity is a critical foundation for studies of its ecological and industrial significance and an important first step toward gauging the impact of climate change on this poorly studied biome.
Acknowledgments
This project was partly supported by a Postgraduate Fellowship to J.D.N. and a Discovery Grant, both from the Natural Sciences and Engineering Research Council (NSERC) of Canada. J.D.N. also acknowledges support from a Killam postgraduate scholarship (UBC).
We are grateful to David Paré, Cindy Prescott, Joe Bennett, Barb Zeeb, and Ken Van Rees for providing us with soil samples. We thank Benli Chai and Jim Cole of the RDP-II for help with batch sequence match analysis. We thank Paul Sue for contributing programming skills. Robert Hancock is thanked for providing access to DNA sequencing facilities. Zhongtang Yu, Klaus Nüsslein, Sue Grayston, Julian Davies, and Matthew Kane provided helpful suggestions on the manuscript.
REFERENCES
- 1.Atlas, R. M. 1984. Use of microbial diversity measurements to assess environmental stress, p. 540-545. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. American Society for Microbiology, Washington, D.C.
- 2.Bååth, E., and T.-H. Anderson. 2003. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35:955-963. [Google Scholar]
- 3.Bohannan, B., and J. Hughes. 2003. New approaches to analyzing microbial biodiversity data. Curr. Opin. Microbiol. 6:282-287. [DOI] [PubMed] [Google Scholar]
- 4.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray, J., and J. T. Curtis. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325-349. [Google Scholar]
- 7.Chapin, F., A. McGuire, J. Randerson, R. Pielke, D. Baldocchi, S. Hobbie, N. Roulet, W. Eugster, E. Kasischke, E. Rastetter, S. Zimov, and S. Running. 2000. Arctic and boreal ecosystems of western North America as components of the climate system. Global Change Biol. 6:211-223. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-802. [DOI] [PubMed] [Google Scholar]
- 10.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Griffin, D. W., C. A. Kellogg, V. H. Garrison, and E. A. Shinn. 2002. The global transport of dust. Am. Sci. 90:228. [Google Scholar]
- 13.Hauben, L., L. Vauterin, E. Moore, B. Hoste, and J. Swings. 1999. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 49: 1749-1760. [DOI] [PubMed] [Google Scholar]
- 14.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 15.Heal, O. W. 1999. Looking north: current issues in Arctic soil ecology. Appl. Soil Ecol. 11:107-109. [Google Scholar]
- 16.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 17.Hodkinson, I. D., and P. A. Wookey. 1999. Functional ecology of soil organisms in tundra ecosystems: towards the future. Appl. Soil Ecol. 11:111-126. [Google Scholar]
- 18.Hughes, J. B., and B. J. M. Bohannan. 2004. Application of ecological diversity statistics in microbial ecology, p. 1321-1344. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishing, London, United Kingdom.
- 19.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller, M., and K. Zengler. 2004. Tapping into microbial diversity. Nat. Rev. Microbiol. 2:141-150. [DOI] [PubMed] [Google Scholar]
- 22.Kysela, D. T., C. Palacios, and M. L. Sogin. 2005. Serial analysis of V6 ribosomal sequence tags (SARST-V6). Environ. Microbiol. 7:356-364. [DOI] [PubMed] [Google Scholar]
- 23.Lawley, B., S. Ripley, P. Bridge, and P. Convey. 2004. Molecular analysis of geographic patterns of eukaryotic diversity in antarctic soils. Appl. Environ. Microbiol. 70:5963-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leckie, S. E., C. E. Prescott, S. J. Grayston, J. D. Neufeld, and W. W. Mohn. 2004. Characterization of humus microbial communities in adjacent forest types that differ in nitrogen availability. Microb. Ecol. 48:29-40. [DOI] [PubMed] [Google Scholar]
- 25.Marchant, R., I. M. Banat, T. J. Rahman, and M. Berzano. 2002. The frequency and characteristics of highly thermophilic bacteria in cool soil environments. Environ. Microbiol. 4:595-602. [DOI] [PubMed] [Google Scholar]
- 26.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G.Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67: 2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita, R. Y. 1992. Low-temperature environments. Encyc. Microbiol. 2:625-637. [Google Scholar]
- 28.National Research Council. 2003. Frontiers in polar biology in the genomic era. National Research Council, Washington, D.C.
- 29.Neufeld, J. D., Z. Yu, W. Lam, and W. W. Mohn. 2004. SARST, serial analysis of ribosomal sequence tags, p. 543-568. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishing, London, United Kingdom.
- 30.Neufeld, J. D., Z. Yu, W. Lam, and W. W. Mohn. 2004. Serial analysis of ribosomal sequence tags (SARST): a high-throughput method for profiling complex microbial communities. Environ. Microbiol. 6:131-144. [DOI] [PubMed] [Google Scholar]
- 31.Øvreås, L. 2004. Personal communication.
- 32.Schadt, C. W., A. P. Martin, D. A. Lipson, and S. K. Schmidt. 2003. Seasonal dynamics of previously unknown fungal lineages in tundra soil. Science 301:1359-1361. [DOI] [PubMed] [Google Scholar]
- 33.Stach, J. E. M., L. A. Maldonado, D. G. Masson, A. C. Ward, M. Goodfellow, and A. T. Bull. 2003. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl. Environ. Microbiol. 69:6189-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staddon, W. J., J. T. Trevors, L. C. Duchesne, and C. A. Colombo. 1998. Soil microbial diversity and community structure across a climatic gradient in western Canada. Biodivers. Conserv. 7:1081-1092. [Google Scholar]
- 35.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torsvik, V., L. Øvreås, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 37.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 38.Willig, M. R., D. M. Kaufman, and R. D. Stevens. 2003. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol. Evol. Syst. 34:273-309. [Google Scholar]
- 39.Zhou, J., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, J., B. Xia, D. S. Treves, L. Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]