Abstract
In many industrialized countries, the incidence of campylobacteriosis exceeds that of salmonellosis. Campylobacter bacteria are transmitted to humans mainly in food, especially poultry meat products. Total prevention of Campylobacter colonization in broiler flocks is the best way to reduce (or eliminate) the contamination of poultry products. The aim of this study was to establish the sources and routes of contamination of broilers at the farm level. Molecular typing methods (DNA macrorestriction pulsed-field gel electrophoresis and analysis of gene polymorphism by PCR-restriction fragment length polymorphism) were used to characterize isolates collected from seven broiler farms. The relative genomic diversity of Campylobacter coli and Campylobacter jejuni was determined. Analysis of the similarity among 116 defined genotypes was used to determine clusters within the two species. Furthermore, evidence of recombination suggested that there were genomic rearrangements within the Campylobacter populations. Recovery of related clusters from different broiler farms showed that some Campylobacter strains might be specifically adapted to poultry. Analysis of the Campylobacter cluster distribution on three broiler farms showed that soil in the area around the poultry house was a potential source of Campylobacter contamination. The broilers were infected by Campylobacter spp. between days 15 and 36 during rearing, and the type of contamination changed during the rearing period. A study of the effect of sanitary barriers showed that the chickens stayed Campylobacter spp. free until they had access to the open area. They were then rapidly colonized by the Campylobacter strains isolated from the soil.
Thermophilic Campylobacter species (particularly Campylobacter jejuni and Campylobacter coli) have been recognized as major causes of acute diarrheal disease in humans (43) for the last 20 years. The incidence of Campylobacter infection is higher than that of salmonellosis in many western countries (49). Campylobacter rarely causes death or spectacular outbreaks of food poisoning (31), so these organisms do not trigger the same degree of concern as Escherichia coli O157:H7 or Salmonella. Nevertheless, C. jejuni is one of the most common causes of bacterial enteritis in humans (13, 26) and may lead to serious complications, such as Guillain Barré syndrome (51) or mucosa-associated lymphoid tissue (23). A recent retrospective Danish study indicated that the risk of death was significantly increased after infection with Campylobacter, especially in patients older than 55 years (20). The main source of Campylobacter infections highlighted in epidemiological studies is consumption of contaminated food, particularly raw or insufficiently cooked poultry products (11, 15, 18, 34).
The following factors contribute to the high correlation between poultry products and human infection: (i) chicken guts, particularly ceca, can be colonized at very high levels (about 109 organisms per g of cecal contents [5]) without symptoms; (ii) usually the entire flock is colonized once an infection becomes established in a poultry house (39), and thus, most flocks are contaminated on the day of slaughter (9, 22); and (iii) the Campylobacter spp. on the carcasses originate mainly from the guts of live birds, as shown by various studies in abattoirs (33, 44). Cross-contamination between different batches of broilers at the slaughterhouse is almost impossible to prevent due to the current slaughtering processes (38) and the high level of contamination of broilers. Thus, by reducing Campylobacter infection in broiler flocks, it should be possible to limit human campylobacteriosis (22). Total prevention of Campylobacter colonization of broilers at the farm level is the best way to prevent contamination of poultry products.
Despite various epidemiological studies, the origin of Campylobacter and its route of colonization in poultry are still incompletely understood. Some authors (2, 4, 22) reported that a poultry flock could be infected by one or a limited number of Campylobacter strains characterized by one or two typing methods. The prevalence of such strains could be explained by their greater ability to survive in a hostile environment and their better adaptation to colonization of the poultry gut. In these studies, the strains were defined by characterization of the Campylobacter isolates by biotyping and serotyping, which are not particularly discriminating.
Molecular methods, such as DNA macrorestriction analysis by pulsed-field gel electrophoresis (PFGE) or PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of different loci, are highly discriminatory (36). Several studies have demonstrated the ability of PFGE to discriminate subtypes within serotypes and to type strains that are untypeable with antisera (14, 41). In 1991, Yan et al. (50) demonstrated that DNA macrorestriction with the SmaI enzyme could be used to distinguish the two major species of Campylobacter and to define genotypes within these species. The discriminatory power of PFGE typing can be increased if two enzymes, e.g., SmaI and KpnI (14) or SacII (17), are used in combination. Recently, the recommendation (32) that a second enzyme should be used to determine relatedness between isolates has been emphasized (24).
In this study the role of biosecurity measures against Campylobacter was evaluated in epidemiological investigations of free-range broiler flocks reared for a minimum of 81 days. The poultry house density was lower than the density in conventional broiler flocks (11 chickens per m2), and, in addition, access to a free-range area (1 ha) was provided after 6 weeks. The facility had to be vacant for 3 weeks for reasons of sanitation. The purpose of this study was to establish the routes of Campylobacter infection of chickens in this kind of rearing system by determining the genomic diversity of thermophilic Campylobacter strains on seven free-range poultry farms using macrorestriction combined with PFGE and analysis of the restriction fragment length polymorphism (RFLP) of different genomic regions amplified by PCR. The biodiversity of Campylobacter strains isolated from free-range poultry production systems was compared with the biodiversity of strains having various origins.
MATERIALS AND METHODS
Strains.
The origins and dates of isolation of the C. coli and C. jejuni strains obtained from a laboratory collection are shown in Table 1.
TABLE 1.
Characterization by PFGE and PCR-RFLP of the different loci of the strains with different origins belonging to the laboratory collection
| Species | Strain | Animal | Year | Source | Geographic origin | PFGE profile
|
PCR-RFLP profile
|
Cluster-genotypeb | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SmaI | KpnI | hipO | flaA | pflA/gyrA or rRNA genea | |||||||
| C. jejuni | CWH260199 | Human | 1999 | Hospital | Western France | S78 | K78 | h25 | fl10 | m5 | J35 |
| BOF | Human | 1994 | Hospital | Western France | NDc | K79 | h6 | fl6 | m8 | J36 | |
| UA580 | Human | 1983 | D. E. Taylor | Canada | S79 | K80 | h12 | fl9 | m12 | J37 | |
| ATCC 33560 | Bovine | 1991 | Pasteur Institute | France | S80 | ND | h26 | fl22 | m18 | J38 | |
| 3J01 | Poultry | 1994 | AFSSAd | Brittany, France | S81 | K81 | h6 | fl9A | m15 | J39 | |
| 3J4.5 | Poultry | 1994 | AFSSA | Brittany, France | S82 | K82 | h19 | fl17 | m13 | 15-J40 | |
| 3J32.30 | Poultry | 1994 | AFSSA | Brittany, France | S83 | K83 | h22 | fl1 | m6 | J41 | |
| A800 | Poultry | 1996 | Aerial | Eastern France | S84 | K84 | h1 | fl1 | m1 | 1-J42 | |
| A805 | Poultry | 1996 | Aerial | Eastern France | S7 | K5 | h1 | fl1 | m1 | 1-J1 | |
| A728 | Poultry | 1996 | Aerial | Eastern France | S85 | K85 | h20 | fl18 | m3 | 5-J43 | |
| A922 | Poultry | 1996 | Aerial | Eastern France | S86 | K86 | h19 | fl17 | m3 | 15-J44 | |
| A940 | Poultry | 1996 | Aerial | Eastern France | S87 | K87 | h21 | fl10 | m3 | J45 | |
| A943 | Poultry | 1996 | Aerial | Eastern France | S88 | K88 | h2 | fl21 | m17 | J46 | |
| A1020 | Poultry | 1996 | Aerial | Eastern France | S89 | K89 | h24 | fl20 | m16 | J47 | |
| A1306 | Poultry | 1996 | Aerial | Eastern France | S90 | K90 | h23 | fl19 | m4 | J48 | |
| C. coli | CWH020399 | Human | 1999 | Hospital | Western France | S62 | K60 | fl31 | rc7 | C35 | |
| UA417 | Human | 1983 | D. E. Taylor | Canada | S63 | K61 | fl23 | rc4 | C36 | ||
| A9821 | Pork | 1996 | Aerial | Eastern France | S64 | K62 | fl30 | rc9 | 14-C37 | ||
| A992 | Pork | 1996 | Aerial | Eastern France | S64 | K63 | fl30 | rc9 | 14-C38 | ||
| A1538 | Pork | 1996 | Aerial | Eastern France | S65 | K64 | fl30 | rc9 | C39 | ||
| A1581 | Pork | 1996 | Aerial | Eastern France | S66 | K65 | fl9A | rc2 | C40 | ||
| A1642 | Pork | 1996 | Aerial | Eastern France | S67 | K66 | fl9A | rc2 | C41 | ||
| A1635 | Pork | 1996 | Aerial | Eastern France | S68 | K67 | fl9A | rc2 | C42 | ||
| A1649 | Pork | 1996 | Aerial | Eastern France | S69 | K68 | fl10 | rc2 | 15-C43 | ||
| A1578 | Pork | 1996 | Aerial | Eastern France | S70 | K69 | fl10 | rc2 | 15-C44 | ||
| A1575 | Pork | 1996 | Aerial | Eastern France | S71 | K70 | fl32 | rc2 | 15-C45 | ||
| A1552 | Pork | 1996 | Aerial | Eastern France | S70 | K71 | fl33 | rc2 | 15-C46 | ||
| ATCC 33559 | Pork | 1980 | Pasteur Institute | France | S72 | K72 | fl9A | rc8 | C47 | ||
| MJ4.3 | Poultry | 1994 | AFSSA | Brittany, France | S73 | K73 | NCe | NC | C48 | ||
| A849 | Poultry | 1996 | Aerial | Eastern France | S74 | K74 | fl25 | rc2 | C49 | ||
| A879 | Poultry | 1996 | Aerial | Eastern France | S75 | K75 | fl4 | rc2 | 16-C50 | ||
| A846 | Poultry | 1996 | Aerial | Eastern France | S76 | K76 | fl4 | rc2 | 16-C51 | ||
| A963 | Poultry | 1996 | Aerial | Eastern France | S77 | K77 | Fl34 | rc10 | C52 | ||
pflA/gyrA PCR-RFLP profile for C. jejuni strains and rRNA gene profile for C. coli strains.
The number before the hyphen indicates the cluster to which the genotype belongs (e.g., in 15-J40 15 indicates cluster 15-J and in 14-C37 14 indicates cluster 14-C), and the number after the hyphen indicates the genotype. When there is no hyphen, the number indicates the genotype, which may belong to any cluster.
ND, not digested. The isolate was refractory to digestion by the restriction enzyme.
AFSSA, Agence Française de Sécurité Sanitaire des Aliments.
NC, isolate was not characterized by the typing method used.
Farms.
This study was conducted from 1996 to 1999 and involved seven French broiler farms, designated farms A to G, belonging to different poultry companies. Only one of the separate broiler houses (houses 1 to 3) on each farm was chosen for the epidemiological study. The surface area of each poultry house was 400 m2, with an open area of 1 ha. A maximum of 4,600 chickens were reared in each building. The all-in all-out system was used on all farms, which meant that the broiler houses were depopulated, left empty for at least 3 weeks, and then restocked simultaneously. The broiler houses were cleaned and disinfected within 3 days of depopulation. The total rearing period was 81 days. When the chickens were 6 weeks old, they had access to an open space during the day. Straw was used for litter, and the broilers were provided with chlorinated tap water and were fed with a minimum of 75% cereals.
Sampling.
The buildings were investigated by sampling outside each house (near the entrance doors and soil in the closed pen) and inside either by taking soil samples or swabbing the walls and floors. When there were bovine feces on the ground of the open space or near the buildings, samples were taken. Animals were sampled by cloacal pressure (10 pools of 10 droppings). Feed and drinking water were also sampled. A set of samples was obtained almost every week on farms A, B, and C from just before arrival of the chicks until their departure for the slaughterhouse. Only three sets of samples were taken on the four other farms: on the day of arrival, on the day just before the chickens were freed (6weeks), and on the day of their departure for the slaughterhouse.
Isolation of Campylobacter spp.
Isolation of Campylobacter spp. was carried out on the day of sample collection.
Swabs were added to 150 ml of Campylobacter selective enrichment Preston broth. This medium consisted of NO2 nutritive broth (Oxoid, Dardilly, France), 5% lysed horse blood (AES Laboratory, Combourg, France), and Preston antibiotic supplements (AES Laboratory). Ten grams of soil, litter, food, or feces was added to 90 ml of Preston broth. Five hundred milliliters of drinking water was filtered through a 0.2-μm Millipore filter. The filter was then transferred to 20 ml of Preston broth. All samples were plated onto two Campylobacter selective agar media: Virion medium made with Mueller-Hinton agar base (Merck, Coger, Paris, France) and Bacto agar (Difco, Fisher Scientific, Elancourt, France) with 5% (vol/vol) defibrinated horse blood (AES Laboratory) and antibiotic supplements (cefoperazone, rifampin, colistin, and amphotericin; Sigma Aldrich Chimie, La Verpillière, France) and Karmali medium (AES Laboratory) with selective supplement CV (AES Laboratory). The plates were incubated at 42°C for 72 h under a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2).
After the enrichment step, which consisted of incubation at 42°C for 24 h in a microaerobic atmosphere, the samples were streaked onto the Virion and Karmali selective agar media. The plates were incubated microaerobically at 42°C for 48 h. Three suspected colonies of Campylobacter spp. were isolated from each plate, placed onto blood agar (Mueller-Hinton agar base [Merck] supplemented with 5% defibrinated horse blood [AES Laboratory]), and incubated under similar conditions. Characteristic colonies were examined with a phase-contrast microscope for typical spiral-shaped cells and rapid motility. Approximately four isolates were collected from each positive sample, which resulted in collection of 2,880 isolates, which were frozen at −80°C in glycerol peptone broth before nearly one-half of them were genotyped. One or two isolates per sample were typed by molecular typing methods.
DNA preparation for PFGE and PCR-RFLP analysis.
The bacterial lawn obtained from an overnight culture on blood agar was suspended by adding 2.5ml of a Tris-NaCl solution (0.01 mol liter−1 Tris-HCl, 1 mol liter−1 NaCl, pH7.6). Cells were harvested and washed twice with 2 ml of the Tris-NaCl solution.
For PCR, the pellet was resuspended in 200 μl of the same solution, and a rapid DNA extraction technique (Kit Fisher, Osi, France) was performed according to the manufacturer's instructions. The DNA was precipitated, pelleted, dried, and resuspended in 100 μl of TE buffer (0.01 mol liter−1 Tris-HCl, 0.001 mol liter−1 EDTA, pH 7.6) and then stored at 4°C. Dilutions were prepared from the resulting stock solutions and adjusted with TE buffer to an optical density at 260 nm of 0.1.
For PFGE, agarose plugs were prepared as described by Ragimbeau et al. (36). Each plug was then cut into four thin slices and stored in TE buffer at 4°C.
Restriction endonuclease digestion and PFGE conditions.
One-quarter of a plug was used for restriction endonuclease digestion in each separate reaction using 40 U of either SmaI or KpnI (Boehringer) under the conditions recommended by the manufacturer in a 100-μl (final volume) mixture with incubation for 5 h at the appropriate temperature. PFGE was done using the CHEF-DRIII system (Bio-Rad Laboratories, United States). An agarose gel (1%) prepared in 0.5× TBE (45 mmol liter−1 Tris, 45 mmol liter−1 boric acid, mmol liter−1 EDTA) was subjected to electrophoresis for 23 h at 220 V and 14°C with ramped pulse times from 2 to 25 s for KpnI. Fragments generated by SmaI digestion were separated by electrophoresis for 24 h at 200 V and 14°C with ramped pulse times from 15 to 45 s for the first 22 h and from 2 to 8 s for the last 2 h.
flaA PCR-RFLP conditions.
PCR was performed using the RAA19 and pg 50 primers (1) and generated a 1,448-bp amplified product. The following reagents were used for this PCR (50-μl mixture): 1× PCR buffer II (Perkin-Elmer), 1.5 mmol liter−1 MgCl2, 0.5 μmol liter−1 of each primer, 200 μmol liter−1 of deoxynucleoside triphosphates (Advantage ultrapure PCR deoxynucleoside mixure; Clontech, Ozyme, France), and 0.2 U liter−1 of AmpliTaq polymerase (Perkin-Elmer). The PCR was conducted with a Gene AMP 9600 system (Perkin-Elmer Instruments, Norwalk, CT) under the following conditions: 94°C for 1 min and then 30 cycles of 94°C for 15 s, 45°C for 30 s, and a 2-min ramp to 72°C for 30 s. The reaction was completed by a final extension of 10 min at 72°C.
Following PCR amplification, 5 μl of the reaction mixture was first checked for the presence of the amplicon on a 1% agarose gel (agarose standard; Eurobio). To study polymorphism of the flaA gene, 7.5 μl of PCR product was digested a 15-μl (total volume) mixture with 5 U of restriction enzyme DdeI (New England Biolabs, Ozyme, France). Digestion was performed with buffer 3 (New England Biolabs, Ozyme, France) at 37°C for 3 h according to the manufacturer's instructions.
rib rRNA gene PCR-RFLP conditions.
PCR was performed using the Rib5 (48) and Therm2 (12) primers, and this generated a 3,925-bp amplified product. The PCR (50-μl mixture) was carried out using an XL PCR kit (Perkin-Elmer) with 1× PCR buffer II, 1 mmol liter−1 magnesium acetate, 0.4 μmol liter−1 of each primer, 200 μmol liter−1 of deoxynucleoside triphosphates (Advantage ultrapure PCR deoxynucleoside mixture; Clontech, Ozyme, France), and 2 U liter−1 of rTth DNA polymerase XL (Perkin-Elmer). The PCR was conducted using the hot start technique and the following conditions: 95°C for 1 min and then 16 cycles of 94°C for 15 s, 52°C for 30 s, and a 4-min ramp to 68°C for 10 s, followed by 16 other cycles with a 5-min ramp to 68°C. The reaction was completed by a final extension of 10 min at 72°C.
Following PCR amplification, 5 μl of the reaction mixture was first checked for the presence of the amplicon on a 1% agarose gel (agarose standard; Eurobio). To study polymorphism of the rib rRNA gene, 7.5 μl of PCR product was digested in a 15-μl (total volume) mixture. For C. jejuni, the rib rRNA gene was digested with 8 U of AluI in buffer 1 (New England Biolabs, Ozyme, France) at 37°C for 3 h. For C. coli, the rib rRNA gene was digested with 10 U of HhaI and 5 U of BsiHKAI in the same tube using buffer 4 (New England Biolabs, Ozyme, France) for 2 h at 37°C and for 1 h at 65°C.
hipO PCR-RFLP conditions.
PCR was performed using the Hipu1 and Hipl3 primers (designed in our laboratory), and this generated various 2,800 to 5,000-bp amplified products. The PCR was carried out with the same reagents and conditions that were used for rib rRNA gene amplification; the cycling conditions were also the same except for the use of a ramp to 68°C of 4.5 min and 5.5 min.
Following PCR amplification, 5 μl of the reaction mixture was first checked for the presence of the amplicon on a 1% agarose gel (agarose standard; Eurobio). To study polymorphism of the hip gene, 8 μl of PCR product was digested in a 15-μl (total volume) mixture. A set of three enzymes (5 U of RsaI, 10 U of HhaI, and 6 U of MnlI) was used with buffer 2 at 37°C for 3 h.
pflA/gyrA PCR-RFLP conditions.
PCR was performed using the set of primers and the cycling conditions described by Ragimbeau et al. (36). Four enzymes were used (10 U of HhaI, 20 U of HindIII, 5 U of HinfI, and 5 U of DdeI) at the same time with the buffer, temperature, and time conditions described by Ragimbeau et al. (36).
Electrophoresis conditions.
For all PCR-RFLP analyses, the digests were analyzed by submarine gel electrophoresis. A 2.5% agarose gel (agarose standard; Eurobio) was used with 1× TBE (89 mmol liter−1 Tris, 89 mmol liter−1 boric acid, 2 mmol liter−1 EDTA, pH 8.3). Electrophoresis was performed at 3V cm−1 for 4 h.
Analysis of the patterns.
The agarose gels were stained with ethidium bromide, and the images were captured using UV illumination with a video system (Gel DOC 1000 system; Bio-Rad). The electrophoretic patterns were compared by Molecular Analyst software fingerprinting (Bio-Rad). Similarities between the profiles, based on band positions, were derived by using the Dice correlation coefficient with a maximum position tolerance of 1%. Dendrograms were constructed to reflect the similarities between the strains in the matrix. Strains were clustered by the unweighted pair group method using the arithmetic mean (40).
The discriminatory power of the typing methods was calculated by using Simpson's index (D) (21), determined as follows:
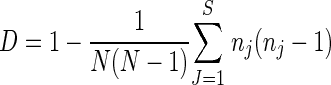 |
where N is the number of isolates tested that are not related, S is the number of different genotypes, and nj is the number of isolates belonging to type j.
RESULTS
A total of 2,880 Campylobacter isolates were recovered during the epidemiological investigations of the seven poultry farms, and 1,225 isolates were identified; 42.6% of the isolates (522 isolates) belonged to C. coli (Table 2), and 57.4% (703 isolates) belonged to C. jejuni (Table 3).
TABLE 2.
Characterization by PFGE and PCR-RFLP of different loci of the C. coli isolates collected on poultry farms
| Farm | Year | Total no. of typed isolates | PFGE
|
PCR-RFLP
|
Cluster-genotypea | ||||
|---|---|---|---|---|---|---|---|---|---|
| SmaI | KpnI | No. of isolates | flaA | rRNA gene | No. of isolates | ||||
| A | 1996 | 134 | S1 | K1 | 7 | fl23 | rc1 | 7 | 1-C1 |
| S2 | K1 | 1 | fl23 | rc1 | 1 | 1-C1A | |||
| S3 | K1 | 1 | fl23 | rc2 | 1 | 1-C2 | |||
| S4 | K2 | 1 | fl24 | rc2 | 1 | 2-C3 | |||
| S5 | K3 | 119 | fl25 | rc2 | 56 | 3-C4 | |||
| S6 | K4 | 5 | fl25 | rc2 | 1 | 3-C5 | |||
| B | 1997 | 224 | S11 | K10 | 124 | fl23 | rc2 | 33 | C6 |
| S12 | K11 | 3 | fl25 | rc2 | 3 | 4-C7 | |||
| S13 | K12 | 1 | fl25 | rc2 | 1 | 4-C8 | |||
| S28 | K18 | 1 | fl25 | rc2 | 1 | 5-C9 | |||
| S14 | K13 | 7 | fl25 | rc2 | 4 | 5-C10 | |||
| S15 | K14 | 57 | fl25 | rc2 | 36 | 5-C11 | |||
| S14 | K18 | 2 | fl25 | rc2 | 2 | 5-C15 | |||
| S16 | K15 | 18 | fl23 | rc3 | 14 | 6-C12 | |||
| fl24 | rc3 | 2 | 6-C12A | ||||||
| S17 | K16 | 8 | fl24 | rc2 | 5 | 7-C13 | |||
| S18 | K17 | 3 | fl26 | rc2 | 3 | 8-C14 | |||
| C | 1998 | 20 | S29 | K29 | 5 | fl24 | rc2 | 5 | 7-C16 |
| S31 | K31 | 4 | fl28 | rc2 | 4 | 7-C18 | |||
| S30 | K30 | 7 | fl23 | rc2 | 7 | 9-C17 | |||
| S32 | K32 | 4 | fl27 | rc2 | 4 | 9-C19 | |||
| D | 1998 | 16 | S38 | K41 | 5 | fl23 | rc4 | 1 | C20 |
| S39 | K42 | 11 | fl23 | rc5 | 2 | C21 | |||
| E | 1999 | 46 | S40 | NDb | 1 | fl29 | rc3 | 1 | C22 |
| S41 | K43 | 17 | fl25 | rc2 | 5 | 2-C23 | |||
| S42 | K44 | 20 | fl23 | rc3 | 5 | 10-C24 | |||
| S45 | K47 | 4 | fl23 | rc3 | 2 | 10-C27 | |||
| S43 | K45 | 2 | fl28 | rc2 | 2 | 11-C25 | |||
| S44 | K46 | 1 | fl28 | rc2 | 1 | 11-C26 | |||
| S46 | K48 | 1 | fl23 | rc6 | 1 | C28 | |||
| F | 1999 | 37 | S47 | K49 | 11 | fl28 | rc2 | 3 | 12-C29 |
| S47 | ND | 1 | fl28 | rc2 | 1 | 12-C29A | |||
| S51 | ND | 2 | fl28 | rc2 | 2 | 12-C33 | |||
| S49 | ND | 1 | fl28 | rc2 | 1 | C31 | |||
| S50 | K50 | 2 | fl25 | rc2 | 2 | 3-C32 | |||
| S18 | K17 | 2 | fl24 | rc2 | 2 | 8-C14A | |||
| S48 | ND | 14 | fl23 | rc2 | 6 | 13-C30 | |||
| S40 | ND | 4 | fl23 | rc2 | 4 | 13-C22A | |||
| G | 1999 | 45 | S40 | ND | 11 | fl23 | rc2 | 11 | 13-C22A |
| S18 | K17 | 9 | fl24 | rc2 | 9 | 8-C14A | |||
| S52 | ND | 25 | fl23 | rc2 | 4 | C34 | |||
See Table 1, footnote b.
ND, not digested. The isolate was refractory to digestion by the restriction enzyme.
TABLE 3.
Characterization by PFGE and PCR-RFLP of different loci of the C. jejuni isolates collected on the poultry farms
| Farm | Year | Total no. of typed isolates | PFGE
|
PCR-RFLP
|
Cluster-genotypea | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| SmaI | KpnI | No. of isolates | hipO | flaA | pflA/gyrA | No. of isolates | ||||
| A | 1996 | 67 | S7 | K5 | 17 | h1 | fl1 | m1 | 14 | 1-J1 |
| S8 | K6 | 13 | h2 | fl2 | m2 | 11 | J2 | |||
| S9 | K7 | 31 | h3 | fl3 | m3 | 20 | J3 | |||
| S7 | K8 | 4 | h1 | fl1 | m1 | 2 | 1-J4 | |||
| S10 | K9 | 2 | h4 | fl4 | m4 | 2 | J5 | |||
| B | 1997 | 358 | S19 | K19 | 174 | h4 | fl4 | m4 | 50 | 2-J6 |
| S19 | K20 | 26 | h4 | fl4 | m4 | 3 | 2-J7 | |||
| NDb | K21 | 7 | h6 | fl6 | m8 | 6 | 3-J8 | |||
| h6 | fl4 | m8 | 1 | 3-J8A | ||||||
| S20 | K22 | 101 | h5 | fl7 | m5 | 48 | 4-J9 | |||
| h5 | fl8 | m5 | 14 | 4-J9A | ||||||
| h5 | fl6 | m5 | 1 | 4-J9B | ||||||
| S21 | K23 | 1 | h1 | fl10 | m7 | 1 | 5-J10 | |||
| S22 | K24 | 12 | h9 | fl11 | m3 | 3 | J11 | |||
| S23 | K25 | 21 | h7 | fl9 | m9 | 16 | 6-J12 | |||
| h7 | fl7 | m9 | 1 | 6-J12A | ||||||
| S24 | K26 | 5 | h1 | fl1 | m6 | 5 | 7-J13 | |||
| S24 | K27 | 1 | h1 | fl1 | m6 | 1 | 7-J14 | |||
| S25 | K28 | 6 | h8 | fl1 | m10 | 3 | 8-J15 | |||
| h8 | fl9 | m10 | 2 | 8-J15A | ||||||
| S26 | ND | 3 | h9 | fl10 | m11 | 2 | 9-J16 | |||
| S27 | ND | 1 | h8 | fl1 | m10 | 1 | 9-J17 | |||
| C | 1998 | 225 | S33 | K33 | 1 | h10 | fl12 | m13 | 1 | J18 |
| S34 | K34 | 154 | h11 | fl9 | m10 | 73 | 10-J19 | |||
| h12 | fl9 | m12 | 4 | 10-J19A | ||||||
| S35 | K35 | 26 | h12 | fl9A | m12 | 9 | 11-J20 | |||
| h11 | fl9 | m10 | 5 | 11-J20A | ||||||
| h13 | fl7 | m7 | 2 | 11-J20B | ||||||
| S36 | K36 | 23 | h13 | fl7 | m7 | 20 | 12-J21 | |||
| h11 | fl9 | m10 | 1 | 12-J21A | ||||||
| S37 | K37 | 8 | h14 | fl9A | m6 | 8 | 13-J22 | |||
| S37 | K38 | 3 | h14 | fl9A | m6 | 3 | 13-J23 | |||
| S37 | K39 | 7 | h14 | fl9A | m6 | 5 | 13-J24 | |||
| h12 | fl9A | m10 | 1 | 13-J24A | ||||||
| h13 | fl7 | m7 | 1 | 13-J24B | ||||||
| S37 | K40 | 2 | h14 | fl9A | m6 | 2 | 13-J25 | |||
| S9 | K7 | 1 | h3 | fl3 | m3 | 1 | J3 | |||
| D | 1998 | 19 | S53 | K51 | 6 | h15 | fl13 | m4 | 1 | J26 |
| S54 | K52 | 5 | h4 | fl4 | m1 | 2 | J27 | |||
| S55 | K53 | 1 | h1 | fl1 | m1 | 1 | 1-J28 | |||
| S56 | K54 | 4 | h16 | fl14 | m14 | 2 | J29 | |||
| S57 | K55 | 1 | h6 | fl7 | m13 | 1 | 14-J30 | |||
| S58 | K56 | 2 | h17 | fl15 | m3 | 2 | J31 | |||
| F | 1999 | 11 | S59 | K57 | 8 | h18 | fl9A | m1 | 3 | J32 |
| S60 | K58 | 3 | h6 | fl16 | m13 | 3 | 14-J33 | |||
| G | 1999 | S61 | K59 | 23 | h1 | fl10 | m3 | 3 | J34 | |
See Table 1, footnote b.
ND, not digested. The isolate was refractory to digestion by the restriction enzyme.
PFGE analysis.
Digestion of the genomic DNAs of the 1,225 Campylobacter isolates and the 33 collection strains using restriction enzyme SmaI, followed by PFGE analysis (SmaI-PFGE), yielded 90 profiles, which were designated S1 to S90 (Tables 1, 2, and 3; see Fig. SA1 in the supplemental material). Seven C. jejuni isolates recovered from poultry farm B and C. jejuni strain BOF (Tables 1 and 2) were refractory to SmaI. Each SmaI profile contained 4 to 14 bands that ranged in size from approximately 45 kbp to 545 kbp (see Fig. SA1 in the supplemental material). The genetic similarities between patterns were analyzed using the Dice coefficient and the unweighted pair group method using the arithmetic mean for cluster analysis, and a dendrogram was constructed (see Fig. SA1 in the supplemental material). Two major groups could be distinguished in this dendrogram; the first consisted of the 50 patterns of the C. coli isolates, and the second consisted of the 40 patterns corresponding to the C. jejuni isolates (see Fig. SA1 in the supplemental material). Some patterns in these two main groups showed high levels of similarity (80 to 99%) (see Fig. SA1 in the supplemental material) and were closely related.
Macrorestriction by the KpnI enzyme (KpnI-PFGE) of the 1,258 Campylobacter isolates distinguished 90 different patterns, which were desiganted K1 to K90 (Tables 1, 2, and 3; see Fig. SA2 in the supplemental material). Four C. jejuni isolates recovered from poultry farm B and C. jejuni strain ATCC 33560, as well as 59 C. coli isolates (1 isolate from poultry farm E, 22 isolates from farm F, and 36 isolates from farm G) were refractory to KpnI (Tables 1, 2, and 3). The numbers of bands in these KpnI-PFGE patterns ranged from 8 to 17, and the sizes ranged from 40 kbp to 445 kbp. Analysis of the similarity of the different KpnI-PFGE patterns did not reveal any well-defined clusters related to the two species of Campylobacter (C. coli and C. jejuni) (see Fig. SA2 in the supplemental material). Nevertheless, some profiles of the two species showed high degrees of genetic similarity (see Fig. SA2 in the supplemental material).
Combining the two restriction enzyme PFGE analyses permitted determination of 102 PFGE genotypes, 54 genotypes corresponding to C. coli (Tables 1 and 2) and 48 genotypes corresponding to C. jejuni (Tables 1 and 3). The similarity analysis showed that there were two main clusters, one cluster corresponding to the C. coli PFGE genotypes and one cluster corresponding to the C. jejuni PFGE genotypes (Fig. 1) (genotypes composed of only one macrorestriction were not included). Within these two clusters, some PFGE genotypes were close to each other (e.g., C. coli genotypes S1K1, S2K1, and S3K1 and C. jejuni genotypes S7K5 and S55K53 or S7K8 and S84K84 [Fig. 1]).
FIG. 1.
Dendrogram showing the relatedness among the PFGE genotypes.
PCR-RFLP analysis.
Of the 1,225 Campylobacter farm isolates, 242 C. coli and 360 C. jejuni isolates were chosen on the basis of PFGE genotype and sample origin.
The C. coli isolates and 17 collection strains were characterized by two PCR-RFLP typing methods, flaA and rib rRNA gene typing (Tables 1 and 2). Fifteen and ten different patterns were discriminated by flaA and rib rRNA gene typing, respectively (Tables 1 and 2; see Fig. SA3 in the supplemental material). By combining the results of the two PCR-RFLP typing methods, we distinguished 22 PCR-RFLP types for the 259 C. coli isolates.
For the 15 flaA types, two major profiles (fl23 and fl25) represented 78.8% of the C. coli isolates tested (Table 2). In addition, profiles fl24 and fl26 exhibited more than 85% similarity with profile fl25 (see Fig. SA3 in the supplemental material). Similarly, most (216) of the 259 C. coli isolates studied by rib rRNA gene typing had the same rc2 profile, and all the profiles were very similar, demonstrating the genetic homogeneity of this region in C. coli.
Three PCR-RFLP typing methods (flaA, hipO, and gyrA/pflA typing) were used to characterize the 360 C. jejuni isolates and the 15 collection strains (Tables 1 and 3). The C. jejuni isolates characterized by flaA, hipO, and gyrA/pflA typing gave 22, 26, and 18 profiles, respectively. The three typing methods did not classify the C. jejuni isolates in the same way (Tables 1 and 3), and 43 combined PCR-RFLP types were distinguished. Some identical PCR-RFLP combinations (h1fl1m1, h4fl4m4, and h6fl6m8 [Tables 1 and 3]) were found for isolates having different origins.
Some profiles obtained with the three PCR-RFLP methods differed by only a few DNA bands and showed high degrees of similarity (more than 80%). Thus, for the 22 flaA types, profiles fl9A-fl15-fl9, fl21-fl10, and fl7-fl8 were very similar (see Fig. SA3 in the supplemental material). Some hipO types were also closely related (h5-h9-h25, h1-h21-h22-h24, and h3-h7) (see Fig. SA3 in the supplemental material). The patterns obtained by gyrA/pflA typing also exhibited many similarities (m7-m4-m2, m13-m3, m6-m1-m16, and m17-m5-m12-m18) (see Fig. SA3 in the supplemental material).
Evidence of clusters obtained by analysis of all genotyping methods.
A combination of all molecular typing methods led to definition of 116 genotypes for the two Campylobacter species, 57 genotypes corresponding to the C. coli isolates (genotypes C1 to C52) (Tables 1 and 2) and 59 genotypes corresponding to the C. jejuni isolates (genotypes J1 to J48) (Tables 1 and 3).
Some individual PFGE genotypes were divided slightly by the PCR-RFLP methods, and some similarities observed between similar PFGE genotypes were strengthened. This led to identification of clusters within the two species; 43 of the 57 C. coli genotypes could be grouped into 16 clusters (designated clusters 1-C to 16-C) (Tables 1 and 2), and 38 of the 59 C. jejuni genotypes could be grouped into 15 clusters (designated clusters 1-J to 15-J) (Tables 1 and 3).
Based on the identical PFGE genotypes and slight differences determined by PCR-RFLP analysis, eight clusters (clusters 3-J, 4-J, 6-J, 8-J, 10-J, 11-J, 12-J, and 13-J) (Table 3) were then defined for the C. jejuni isolates, and three clusters (clusters 6-C, 8-C, and 13-C) (Table 2) were defined for the C. coli isolates.
On the other hand, some similarities revealed by PFGE were strengthened by PCR-RFLP typing, which also led to identification of clusters for the two Campylobacter species. Thus, 14 and 8 additional clusters were described for C. coli and C. jejuni, respectively (Tables 1, 2, and 3); e.g., within the C. coli isolates, genotypes C1, C1A, and C2 were considered members of the same cluster (cluster 1-C) (Table 2) as they produced similar SmaI profiles (S1, S2, and S3) (Fig. 1) and identical KpnI, flaA, and rib rRNA gene profiles. Similarly, within the C. jejuni isolates, those having genotypes J1, J4, J28, and J42 were placed in the same cluster (cluster 1-J) (Table 3). Although the isolates with genotypes J1 and J4 produced a significantly different KpnI profile, they produced the same SmaI, hipO-, flaA, and pflA/gyrA profiles. Isolates with the J28 and J42 genotypes also produced the same hipO, flaA, and pflA/gyrA profiles, and their PFGE genotypes (S55K53 and S84K84) were similar to PFGE genotypes J1 and J4 (S7K5 and S7K8, respectively) (Fig. 1 and Tables 1 and 3).
Some genotypes, such as C6 or J2 (Tables 2 and 3), were not included in any cluster.
Biodiversity of Campylobacter: comparison with the laboratory collection strains.
Molecular characterization revealed 116 Campylobacter genotypes among the 1,225 isolates from the seven poultry farms and the 33 strains from the laboratory collection. As the frequencies of investigation were different for different farms (weekly for farms A, B, and C but only three times for farms D, E, F, and G), the numbers of isolates characterized from the farms differed considerably.
The two species of Campylobacter (C. coli and C. jejuni) were recovered at the same ratio throughout the study, and there was a slight preponderance of C. jejuni (57.4% versus 42.6%). However, large variations between different farms were apparent; e.g., 100% of the isolates from farm E belonged to C. coli, compared with only 8.2% of the isolates from farm C.
With the exception of farm E, all broiler flocks were contaminated by both Campylobacter species, and several genotypes within these species were described (Tables 2 and 3). On farm B, for example, 61.5% of the 582 isolates characterized belonged to C. jejuni, and 38.5% belonged to C. coli; 28 distinct genotypes were described, some of which could be further grouped into 13 clusters (Tables 2 and 3). In each species, some genotypes were predominant; 55.4% of the C. coli isolates collected on this farm were genotype C6 isolates (Table 2), and cluster 5-C accounted for 29.9% of the C. coli isolates studied (Table 2). The remaining C. coli isolates (14.7%) were grouped into six genotypes (genotypes C7 and C8 in cluster 4-C, genotypes C12 and C12A in cluster 6-C, and genotypes C13 and C14). On this farm, 55.9% and 28.2% of the C. jejuni isolates belonged to clusters 2-J and 4-J, respectively (Table 3). The remaining isolates (15.9%) grouped into five distinct clusters (clusters 3-J, 6-J, 7-J, 8-J, and 9-J) (Table 3) and two distinct genotypes (genotypes J10 and J11) (Table 3).
It was also apparent from the genotype distribution on the seven poultry farms that some isolates from different farms had identical or similar genotypes. A few similar or identical patterns were also observed in the collection strains. For C. coli, isolates belonging to cluster 8-C were collected in 1997 from farm B and in 1999 from farms F and G (Table 2). Isolates belonging to cluster 13-C were found on both farms F and G, and isolates belonging to cluster 3-C were collected from farm A during 1996 and from farm F in 1999 (Table 2). Similarly, some cluster 2-C isolates were collected from farm A in 1996 and from farm E in 1999, and some cluster 7-C isolates were found on farms B and C in 1997 and 1998, respectively (Table 2). Some strains in the laboratory collection were members of the same cluster. Thus, strains A9821 and A992 collected from pork during 1996 in eastern France belonged to cluster 14-C (Table 1). In the same way, strains A1649, A1578, A1575, and A1552, also collected from pork in eastern France in 1996, had similar profiles and seemed to belong to the same cluster (cluster 15-C) (Table 1). Strains A879 and A846 collected from poultry in eastern France in 1996 belonged to cluster 16-C (Table 1).
Within the C. jejuni isolates, genotype J3 was isolated from farm A in 1996 and from farm C in 1998 (Table 3). Genotype J1 was isolated from farm A and, during the same year, from a poultry farm located in eastern France (Tables 1 and 3). Moreover, this organism belonged to the same cluster as strains having genotypes J4, J28, and J42 that were collected from farms A and D in 1998 and from a poultry farm in eastern France in 1996, respectively. Some isolates belonging to cluster 14-J were collected from farms D and F in 1998 and 1999, respectively (Table 3). One isolate collected from farm B in 1997 belonged to cluster 5-J, the same cluster as strain A940 isolated from a poultry farm in eastern France in 1996 (Tables 1 and 3). Strains 3J4.5 and A922, which were isolated from geographically distant poultry farms (in Brittany and Alsace) and were studied in different years (1994 and 1996), belonged to the same cluster, cluster 15-J (Table 1).
Tracing Campylobacter spp. on the broiler farms.
No Campylobacter strains were detected in the farm buildings on any of the seven poultry farms before arrival of the chickens (Table 4). The 1-day-old chicks and the transport cases were also free of Campylobacter spp. No Campylobacter was detected in any feed or drinking water samples. On farms A, B, and C, the chickens were contaminated by Campylobacter during the rearing period inside the farm buildings (Table 4). No Campylobacter was detected in the broiler droppings on farms D, E, F, and G before the chickens went outside into the open rearing space (Table 4). Nevertheless, all seven poultry flocks were contaminated by Campylobacter spp. before their departure for the slaughterhouse.
TABLE 4.
Tracing of Campylobacter spp. on seven broiler farms
| Farm | Chicken age (days) | Outside sampling | Bovine feces | Entrance hall | Inside environment | Chicken |
|---|---|---|---|---|---|---|
| A | 1 | 1-C1, 1-C1A, 1-C2, 1-J1a | −b | − | − | − |
| 8 | C3 | − | − | − | − | |
| 15 | − | − | − | − | − | |
| 22 | − | − | − | − | − | |
| 29 | − | J2 | − | − | − | |
| 36 | 1-J1, J2 | J2 | − | C4 | 3-C4 | |
| 43c | 3-C4 | J2 | 3-C4 | C4, C5 | 3-C4 | |
| 50 | 3-C4, 3-C5 | − | 3-C4, 1-J1 | C4 | 3-C4 | |
| 78 | 3-C4, J3, 1-J4 | J5 | NDd | ND | 3-C4, 1-J1, J3, 1-J4 | |
| B | 1 | − | ND | − | − | |
| 8 | − | ND | − | − | ||
| 15 | − | ND | − | − | ||
| 22 | − | ND | C6, 2-J6 | C6, 2-J6, 3-J8, 4-J9 | ||
| 29 | 5-C11, 2-J6, 4-J9 | ND | C6, 2-J6, 4-J9 | C6, 2-J6, 2-J7, 4-J9 | C6, 5-C11, 2-J6, 2-J7, 4-J9 | |
| 36c | − | ND | − | − | − | |
| 48 | 4-C7, 5-C10, 5-C11, 2-J6, 2-J7, 4-J9, J11, 6-J12, 7-J13 | ND | 5-C10, 4-J9, 6-J12, 7-J13, 7-J14 | J6, J9 | C6, 5-C11, 2-J6, 3-J8, 4-J9, 6-J12, 7-J13, 8-J15 | |
| 87 | ND | ND | 6-C12, 7-C13, 4-J9, 5-J10 | ND | 4-C7, 4-C8, 5-C10, 5-C11, 6-C12, 7-C13, 8-C14, 5-C15, 4-J9, J11, 6-J12, 7-J13, 8-J15, 9-J16, 9-J17 | |
| C | 1 | − | ND | − | − | − |
| 12 | J18 | ND | − | − | − | |
| 27 | 10-J19, 11-J20, 12-J21 | ND | 10-J19 | − | 10-J19, 11-J20 | |
| 33 | 10-J19 | ND | − | 10-J19 | 10-J19, 11-J20, 12-J21 | |
| 40c | 10-J19 | ND | 10-J19 | − | 10-J19, 12-J21, 13-J24 | |
| 48 | 10-J19, 12-J21 | ND | − | − | 10-J19, 11-J20, 12-J21, 13-J23, 13-J24, 13-J25 | |
| 68 | 10-J19 | ND | − | − | 10-J19, 12-J21, 13-J22, 13-J23, 13-J24, 7-C16, 9-C17 | |
| 83 | ND | ND | ND | ND | 10-J19, 11-J20, J3, 7-C16, 9-C17, 7-C18, 9-C19 | |
| D | 1 | C20 | J26 | − | − | − |
| 36c | − | ND | − | − | − | |
| 86 | ND | ND | − | − | C21, J27, 1-J28, J29, 14-J30, J31 | |
| E | 1 | C22, 2-C23, 10-C24, 11-C25, 11-C26 | ND | − | − | − |
| 41c | − | ND | ND | ND | − | |
| 81 | ND | ND | ND | ND | 2-C23, 10-C24, 10-C27, C28 | |
| F | 1 | 8-C14, 12-C29, 13-C22A, 13-C30, C31 | ND | − | − | − |
| 41c | − | ND | ND | ND | − | |
| 81 | ND | ND | ND | ND | 13-C22A, 13-C30, 3-C32, J32, J33, 12-C29, 12-C29A, 12-C33 | |
| G | 1 | 8-C14, 13-C22A | ND | − | − | − |
| 41c | − | ND | ND | ND | − | |
| 81 | ND | ND | ND | ND | 13-C34, J34 |
Boldface type indicates genotypes and clusters that were isolated from the soil at the beginning of the rearing period and from animals before their departure for the slaughterhouse. See Table 1, footnote b.
−, negative samples.
After this date the chickens could go outside.
ND, sampling not done.
On farm A (Table 4), some Campylobacter strains (C. coli genotypes C1, C1A, and C2 and C. jejuni genotype J1) were detected in soil samples collected in the open rearing space on the first sampling day. Other strains of C. coli (genotype C3) and C. jejuni (genotype J2) were detected in soil samples and in bovine feces, respectively, in the second set of samples. C. jejuni genotype J5 was also detected in bovine feces during the epidemiological study of this farm. Broilers were contaminated between days 29 and 36 by C. coli genotype C4. This was the only genotype detected in the poultry droppings during the next two sampling times (Table 4). Nevertheless, just before their departure for the slaughterhouse, the chickens were found to carry several Campylobacter strains (C. coli genotype C4 and C. jejuni genotypes J1, J3, and J4). It should be noted that the birds were contaminated by the same C. jejuni strain (genotype J1) isolated from the soil on the first day of the rearing period and also detected in the entrance hall during sampling on day 50 of rearing.
No Campylobacter was detected in the environment outside the broiler building on farm B (Table 4) before the birds became contaminated. Some strains of C. coli (genotypes C6 and C11) and C. jejuni (genotypes J6, J7, J8, and J9) were isolated from birds on days 22 and 29. Before departure of the chickens for the slaughterhouse, eight distinct genotypes of C. coli and seven genotypes of C. jejuni were isolated from the 100 chickens sampled (10 samples of 10 individual droppings) (Table 4).
One C. jejuni strain on farm C (Table 4) was isolated from a soil sample in the second set of samples. This strain (genotype J18) was never recovered during the rearing period. Birds carried C. jejuni (genotypes J19 and J20) from day 27. During the same sampling, genotype J21 was also isolated from soil in front of the entrance door. This organism was isolated from the chicken droppings in the next set of samples (Table 4). C. jejuni genotype J19 was the genotype that was most frequently isolated from chicken droppings during most of the rearing period. On day 68, 2 C. coli isolates (genotypes C16 and C17) were detected among 18 isolates obtained from droppings; the 16 other isolates belonged to C. jejuni and were genotypes J19, J21, J22, J23, J24, and J25. However, just before their departure for the slaughterhouse, the chickens seemed to carry a majority of C. coli. Sixteen of the 20 isolates from droppings belonged to C. coli (genotypes C16, C17, C18, and C19).
On farm D (Table 4), C. coli genotype C20 and C. jejuni genotype J26 were isolated from soil and bovine feces, respectively, on the first day of the rearing period. At the end of this period, the chickens were contaminated by one C. coli strain (genotype C21) and five different C. jejuni strains (genotypes J27, J28, J29, J30, and J31).
On farm E (Table 4), several soil samples were Campylobacter positive on the first day of the rearing period. Twenty-three isolates were identified and characterized. They belonged to five C. coli genotypes, genotypes C22, C23, C24, C25, and C26. At the end of the rearing period, the chickens were contaminated by four distinct C. coli genotypes (genotypes C23, C24, C27, and C28). It is important to note that some isolates with identical genotypes (genotypes C23 and C24) were found in the soil at the beginning of the rearing period and in droppings before transport of the chickens to the slaughterhouse. Moreover, the genotype C27 isolates recovered from the droppings belonged to the same cluster as the genotype C24 isolates (cluster 10-C) (Table 2).
On farm F (Table 4), five distinct C. coli genotypes were isolated from soil samples on the first day of the rearing period (genotypes C14, C29, C22A, C30, and C31). Genotypes C22A and C30 belonged to the same cluster (cluster 13-C) (Table 2). Before their departure for the slaughterhouse, the chickens were contaminated by six C. coli genotypes (genotypes C22A, C30, C32, C29, C29A, and C33) and two C. jejuni genotypes (genotypes J32 and J33) (Table 4). As on farm E, the same or closely related genotypes (genotypes C29, C29A, C22A, and C30) were isolated in the soil on the first day and in the chicken droppings at the end of the rearing period.
On farm G (Table 4), two C. coli genotypes (genotypes C22A and C14) were isolated from the soil on the first day of sampling. The chickens were contaminated by C. coli genotype C34 isolates and C. jejuni genotype J34 isolates before their departure for the slaughterhouse (Table 4). It is important to note that the genotype C22A and C34 isolates belonged to the same cluster (cluster 13-C) (Table 2).
DISCUSSION
The best way to evaluate the genomic diversity of C. coli and C. jejuni is DNA sequencing. However, this technique is both time-consuming and expensive, so a number of subtyping methods have been developed to differentiate bacterial isolates beyond the species level. In this study, PFGE and PCR-RFLP methods were used to analyze the whole genome and specific sequences, respectively.
Thus, Campylobacter isolates were first characterized by PFGE using two restriction enzymes, SmaI and KpnI. Gibson et al. (14) showed that the divergence between some strains varied significantly according to the restriction endonuclease used and that matches between PFGE profiles obtained with at least two enzymes were required to prevent misinterpretation of strain affinities. In the same way, Lindmark et al. (24) showed that 19 isolates with identical SmaI profiles displayed 15 different profiles after digestion with KpnI. This clearly underlines the need to use a second enzyme when the relatedness between isolates is determined.
Characterization of selected Campylobacter isolates was completed by studying the polymorphism of three loci (hipO, flaA, and gyrA/pflA) for the C. jejuni isolates and two loci for C.coli (flaA and rib rRNA gene). Molecular characterization by PCR-RFLP typing is less discriminatory than macrorestriction using two restriction enzymes (102 PFGE genotypes versus 65 PCR-RFLP genotypes). Simpson's index of discrimination (21) for the different techniques was calculated using the poultry farm results (Table 5). Thus, KpnI-PFGE analysis gave better discrimination than the other techniques used in this study (SmaI-PFGE analysis, in particular) (27). Nevertheless, as previously described (50), SmaI-PFGE analysis was able to discriminate between C. coli and C. jejuni.
TABLE 5.
Simpson's index of discrimination calculated using poultry farm data shown in 2 and 3
| Typing method | Enzyme or amplified gene | Simpson's index
|
|
|---|---|---|---|
| Single method | Combined methods | ||
| Macrorestriction | SmaI | 0.9237 | 0.9313 |
| KpnI | 0.9317 | ||
| PCR-RFLP (C. jejuni) | hipO | 0.8852 | 0.8943 |
| flaA | 0.8468 | ||
| gyrA/pflA | 0.8741 | ||
| PCR-RFLP (C. coli) | flaA | 0.7717 | 0.7972 |
| rRNA gene | 0.2957 | ||
A total of 116 genotypes for the 1,258 Campylobacter isolates were defined by combined PFGE and PCR-RFLP typing. With minor changes in profiles, PCR-RFLP typing permitted further discrimination of the Campylobacter isolates characterized by PFGE typing. Thus, three PFGE genotypes for the C. coli isolates (S16K15, S18K17, and S40ND) displayed different PCR-RFLP genotypes. Eight PFGE genotypes for the C. jejuni isolates (NDK21, S20K22, S23K25, S25K28, S34K34, S35K35, S36K36, and S37K39) were slightly divided by PCR-RFLP typing. These differences could have been due to intra- and interstrain recombination in the different amplified sequences, as was shown previously for the flagellin gene (19). On the other hand, the PCR-RFLP typing methods strengthened some of the similarities observed between similar PFGE genotypes. Thus, by combining the different molecular typing methods some clusters could be defined within the two species; 43 of the 57 C. coli genotypes were grouped into 16 clusters (designated clusters 1-C to 16-C) (Tables 1 and 2), and 38 of the 59 C. jejuni genotypes were grouped into 15 clusters (clusters 1-J to 15-J) (Tables 1 and 3).
This study demonstrated the relative genetic diversity of Campylobacter isolates from poultry (84 genotypes for 1,225 isolates). In this study, six of the seven poultry flocks investigated were contaminated by both species of Campylobacter, while only C. coli was isolated from farm E. To our knowledge, no previous study has revealed multiple Campylobacter types in broiler flocks. In most previous studies, flocks were contaminated by only one species (generally C. jejuni), and when typing was used, the contamination was generally due to a single serotype or genotype of Campylobacter (24, 28, 30). Thus, during a 1-year epidemiological study of 287 poultry flocks, Berndston et al. (4) showed that 75 of 77 contaminated flocks were contaminated by C. jejuni and only 2 flocks were contaminated by C. coli. Only 11 distinct serotypes were detected for the Campylobacter isolates collected from these 77 poultry flocks. Moreover, most of the flocks were contaminated by a single Campylobacter serotype, and only four flocks carried more than one serotype. Perko-Mäkelä et al. (35) also found that chicken flocks were contaminated by a single Campylobacter species, generally C. jejuni (31 of 33 flocks). In another study, 24 flocks reared on the same farm were contaminated by a single species (C. jejuni) with the same serotype (serotype HS2) (3).
The greater diversity found in our study could have been due to the use of powerful discriminating techniques, to the characterization of a larger number of Campylobacter isolates, and to the kinds of rearing systems studied. Thus, on farm B, 11 C.coli genotypes and 17 C. jejuni genotypes were identified among the 582 poultry isolates tested. However, dominant genotypes were found for each species, and several genotypes showed relatedness; almost 85% of the isolates tested could be placed into two C. coli groups (genotype C6 and cluster 5-C) and two C. jejuni groups (clusters 2-J and 4-J). Similarly, on farm A, although five C. jejuni and six C. coli genotypes were found among the 201 isolates, 87.5% of these isolates could be grouped into two C. jejuni clusters (clusters 1-J and J3) and one C. coli cluster (cluster 3-C). On farm C, 62.8% of the isolates belonged to cluster 10-J. C. coli was much less frequent on this farm (8.2% of the isolates), and the isolates could be divided into two clusters (clusters 7-C and 9-C). Although fewer Campylobacter isolates were found on the four other farms, several genotypes were still apparent. Thus, on farm D, two distinct genotypes were found for the 16 C. coli isolates studied, and six genotypes were found for the 19 C. jejuni isolates. No relatedness was observed for these various isolates, but 73.3% of the isolates were members of two genotypes of C. jejuni (genotypes J26 and J27) and one genotype of C. coli (genotype C21). The genotypic heterogeneity observed on all seven poultry farms was probably due to the existence of different sources of Campylobacter in the environment.
The well-known genetic instability of Campylobacter species (46) could also explain our results, which included finding 16 clusters for the 57 C. coli genotypes and 15 clusters for the 59 C. jejuni genotypes. The Campylobacter species were naturally transformable and had the capacity to acquire exogenous DNA that could be integrated into the chromosome by illegitimate recombination (37, 45). Hanninen et al. (16) showed that C. jejuni could undergo genetic recombination (insertions, deletions, acquisition of foreign DNA, inversions, crossovers, etc.) during chicken intestine colonization. Similarly, Boer et al. (6) demonstrated that interstrain genetic exchange and intragenomic alterations occurred in vivo during C. jejuni infection, which might explain the genome plasticity observed for this pathogen. Our results corroborate these findings and also demonstrate the genetic instability of Campylobacter within a single poultry flock. Thus, on farm B, genotypes C9, C10, and C15 of C. coli probably originated from genotype C11. All four of these genotypes had identical flaA and rib rRNA gene profiles, whereas minor modifications were observed with the SmaI and KpnI profiles. Genotype C11, which was found more frequently throughout the rearing period, was considered the major genotype. Thus, genotypes C9, C10, and C15 could have been derived from genotype C11 by genomic rearrangements that could have occurred in vivo or in vitro. Indeed, they may have occurred in vitro during treatment of the samples. Recombination in genotypes J6 and J7 (cluster 2-J), collected during the same rearing period, could have occurred in vivo, because very small amounts of genotype J7 of C. jejuni were found at several times throughout the rearing period. This genetic instability can be explained by the fact that Campylobacter and other bacteria with small genomes (Helicobacter, for example) need to undergo genetic rearrangements in order to increase their potential and adapt to the environment (25, 29, 47).
Another important result of this study was that identical strains or strains belonging to the same cluster were observed on different farms and at different times. In certain cases, the farms belonged to the same poultry company and therefore received chickens from the same hatchery and food from the same factory. This was the case for strain C14, found on farms B, F, and G, for strain C22A found on farms F and G, and for strain J3 found on farms A and C. These results highlight the problems associated with a common source of chicks and a common source of feed. However, some strains that were identical or were members of a given cluster were also found on farms without any such connection (Table 4). These results suggest that some Campylobacter strains could be adapted to poultry; the relative genetic stability in space and time could have been the result of adaptation to environmental pressure (7, 25). Thus, Broman et al. (7) showed that some C. jejuni subtypes could be associated with different biotopes. Moreover, this hypothesis was supported by a multilocus sequence typing investigation, in which it was shown that certain clonal complexes could be found only among isolates from certain sources (e.g., sand samples) (10). However, in our study, the presence of a particular strain may have been entirely stochastic or may have been the result of pressures outside the present niche that influenced the migration.
Campylobacter spp. could be traced throughout the rearing period on all seven poultry farms in this study. On farms A, B, and C, on which no particular biosecurity measures were used by the farmers, chickens were contaminated by Campylobacter from the second week of rearing, as has been described in other studies (3, 22). The farmers on the four other farms (farms D, E, F, and G) had been informed of the need for biosecurity measures, such as systematic changing of boots before the rearing house is entered. The chickens on these farms were free from Campylobacter during the first 6 weeks of rearing inside the building. Nevertheless, all seven poultry flocks were contaminated by Campylobacter before their departure for the slaughterhouse.
The molecular typing methods used in this study revealed that the soil around the farm building was a source of Campylobacter contamination. On farm A, C. jejuni strain J1 was isolated from the soil on the first day, from the entrance hall on day 50, and from the poultry feces at the end of the rearing period. Similarly, the same strains were isolated on farms E, F, and G from the soil on the first day of sampling and from the birds before their departure for the slaughterhouse.
In this study, the soil was clearly identified as a potential source of Campylobacter contamination, but we also demonstrated that there were multiple sources of contamination as several Campylobacter strains were isolated from each poultry flock. This study also showed that the carriage of Campylobacter by the birds may change during the rearing period. Thus, broilers on farm C were primarily contaminated by C. jejuni strains at the beginning of contamination (day 27) until day 48. For the last two sets of samples (days 68 and 83), C. coli was isolated from the majority of the samples before departure of the chickens for the slaughterhouse. Several hypotheses could be advanced to explain these changes in chicken colonization by Campylobacter. First, the broilers were exposed to several sources of Campylobacter during the rearing period, and certain Campylobacter strains colonized the chickens. Then a physiological alteration (food change, additives, immune status, etc.) might have resulted in the establishment of another type of strains. Moreover, the method used to recover the Campylobacter isolates (direct plating, prior enrichment, choice of medium, etc.) may influence the subtypes of strains (30).
No Campylobacter was isolated from the soil in the second set of samples obtained before the end of the in-house period on farms D, E, F, and G, even when Campylobacter strains had been isolated from the soil in the first set of samples. One possible explanation for this is that the number of samples was not sufficient for the real soil contamination to be evaluated. This is unlikely, however, as some Campylobacter strains were isolated from the soil in the open area during the first sampling in the same number of samples on all four farms. The other explanation is that the Campylobacter-contaminated chickens from the previous batch were the source of soil contamination. On the first day of the rearing period, the open rearing area had been vacant for 3 weeks (the conventional sanitary procedure), and the Campylobacter strains excreted by chickens from the previous batch might still have been present. On the other hand, by the time of the second sampling, no chicken would have had access to the open rearing space for 9 weeks (3 weeks empty plus 6 weeks with the poultry in the house), by which time the Campylobacter strains previously present could have died and the number of soil samples taken would have been insufficient to detect the presence of the bacterium. Moreover, it is possible that, given the conditions of stress encountered in the soil, the Campylobacter strains might have transformed into viable but noncultivable forms and might have become cultivable after passage in the intestinal tract of chickens, as was observed in laboratory conditions by Cappellier (8). On these farms, the Campylobacter strains isolated from the soil and from the chickens at the end of the rearing period belonged to C. coli. It is known that survival is different from strain to strain (42) and that injuries can be repaired through intestinal passage. It would be interesting to compare the survival of the different strains and their capacities to recover the ability to grow.
This study is the first study to reveal relatively significant biodiversity within Campylobacter strains collected from broiler flocks. This biodiversity could have resulted from the fact that the flocks studied were free-range broilers exposed to multiple sources of contamination. The use of several methods to characterize the Campylobacter isolates might also explain this biodiversity. We also showed that a broiler flock colonized by one Campylobacter strain could be colonized by one or more other strains. In this study, we also observed that Campylobacter exhibited different related profiles that could be due to genomic rearrangements which could have occurred in vitro or in vivo throughout the rearing period. This work also demonstrated that the soil around the rearing facilities was a potential source of contamination and that biosecurity measures were useful for preventing or minimizing Campylobacter colonization of broilers.
Supplementary Material
Acknowledgments
This work was supported by SYNALAF (Syndicat National des Labels Avicoles de France) and OFIVAL (Office National Interprofessionnel des Viandes de l'Elevage et de l'Aviculture).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alm, R. A., P. Guerry, and T. J. Trust. 1993. Distribution and polymorphism of the flagellin genes from isolates of Campylobacter coli and Campylobacter jejuni. J. Bacteriol. 175:3051-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayling, R. D., S. Woodward, S. J. Evans, and D. G. Newell. 1996. Restriction fragment length polymorphism of polymerase chain reaction products applied to the differentiation of poultry campylobacters for epidemiological investigations. Res. Vet. Sci. 60:168-172. [DOI] [PubMed] [Google Scholar]
- 3.Berndston, E., M. L. Danielsson-Tham, and A. Engvall. 1996. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int. J. Food Microbiol. 32:35-47. [DOI] [PubMed] [Google Scholar]
- 4.Berndston, E., U. Emanuelson, A. Engvall, and M. L. Danielsson-Tham. 1996. A 1-year epidemiological study of campylobacters in 18 Swedish chickens farms. Prev. Vet. Med. 26:167-185. [Google Scholar]
- 5.Berndston, E., M. Tivemo, and A. Engvall. 1992. Distribution and numbers of Campylobacter in newly slaughtered broiler chickens and hens. Int. J. Food Microbiol. 15:45-50. [DOI] [PubMed] [Google Scholar]
- 6.Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 7.Broman, T., J. Waldenstrom, D. Dahlgren, I. Carlsson, I. Eliasson, and B.Olsen. 2004. Diversities and similarities in PFGE profiles of Campylobacter jejuni isolated from migrating birds and humans. J. Appl. Microbiol. 96:834-843. [DOI] [PubMed] [Google Scholar]
- 8.Cappelier, J. 1997. L'état viable non cultivable chez une bactérie responsable de toxi-infections alimentaires: Campylobacter jejuni. Ph.D. thesis. Université de Nantes, Nantes, France.
- 9.Denis, M., J. Refrigier-Petton, M. J. Laisney, G. Ermel, and G. Salvat. 2001. Campylobacter contamination in French chicken production from farm to consumers. Use of a PCR assay for detection and identification of Campylobacter jejuni and Camp. coli. J. Appl. Microbiol. 91:255-267. [DOI] [PubMed] [Google Scholar]
- 10.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiological investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, M. R., W. Lane, J. A. Frost, and G. Nylen. 1998. A campylobacter outbreak associated with stir-fried food. Epidemiol. Infect. 121:275-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyers, M., S. Chapelle, G. Van Camp, H. Goossens, and R. De Wachter. 1993. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J. Clin. Microbiol. 31:3340-3343. (Erratum, 32: 1623.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, C., S. Reddy, M. Samuel, R. Marcus, J. Bender, S. Desai, B.Shiferaw, D. Helfrick, M. Carter, B. Anderson, and M. Hoekstra. 2000. Risk factors for sporadic Campylobacter infections in United States: a case-control study on FoodNet sites. Centers for Disease Control and Prevention, Atlanta, Ga. [DOI] [PubMed]
- 14.Gibson, J., E. Lorenz, and R. J. Owen. 1997. Lineages within Campylobacter jejuni defined by numerical analysis of pulsed-field gel electrophoretic DNA profiles. J. Med. Microbiol. 46:157-163. [DOI] [PubMed] [Google Scholar]
- 15.Graves, T., K. Bradley, and J. Crutcher. 1998. Outbreak of Campylobacter enteritis associated with cross-contamination of food. Morb. Mortal. Wkly. Rep. 47:129-131. [PubMed] [Google Scholar]
- 16.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanninen, M. L., M. Niskanen, and L. Korhonen. 1998. Water as a reservoir for Campylobacter jejuni infection in cows studied by serotyping and pulsed-field gel electrophoresis (PFGE). Zentbl. Vetmed. Reihe B 45:37-42. [DOI] [PubMed] [Google Scholar]
- 18.Hanninen, M. L., P. Perkomakela, A. Pitkala, and H. Rautelin. 2000. A three-year study of Campylobacter jejuni genotypes in humans with domestically acquired infections and in chicken samples from the Helsinki area. J. Clin. Microbiol. 38:1998-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrington, C. S., F. M. Thomsoncarter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helms, M., P. Wastrup, P. Gerner-Smidt, and K. Molbak. 2003. Short and long term mortality associated with foodborne bacterial gastrointestinal infections: registry based study. Br. Med. J. 326:357-361. [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter, P. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 23:163-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs-Reitsma, W. F., A. W. van de Giessen, N. M. Bolder, and R. W. Muller. 1995. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol. Infect. 114:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecuit, M., E. Abachin, A. Martin, C. Poyart, P. Pochart, F. Suarez, D. Bengoufa, J. Feuillard, A. Lavergne, J. I. Gordon, P. Berche, L. Guillevin, and O. Lortholary. 2004. Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N. Engl. J. Med. 350:239-248. [DOI] [PubMed] [Google Scholar]
- 24.Lindmark, H., B. Harbom, L. Thebo, L. Andersson, G. Hedin, B. Osterman, T. Lindberg, Y. Andersson, A. Westoo, and E. O. Engvall. 2004. Genetic characterization and antibiotic resistance of Campylobacter jejuni isolated from meats, water, and humans in Sweden. J. Clin. Microbiol. 42:700-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning, G., B. Duim, T. Wassenaar, J. A. Wagenaar, A. Ridley, and D. G. Newell. 2001. Evidence for a genetically stable strain of Campylobacter jejuni. Appl. Environ. Microbiol. 67:1185-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresse, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaud, S., R. D. Arbeit, and C. Gaudreau. 2001. Molecular strain typing of Campylobacter jejuni by pulsed-field gel electrophoresis in a single day. Can. J. Microbiol. 47:667-669. [PubMed] [Google Scholar]
- 28.Nadeau, E., S. Messier, and S. Quessy. 2002. Prevalence and comparison of genetic profiles of Campylobacter strains isolated from poultry and sporadic cases of campylobacteriosis in humans. J. Food Prot. 65:73-78. [DOI] [PubMed] [Google Scholar]
- 29.Newell, D. G., A. M. Ridley, M. J. Toszeghy, and T. M. Wassenaar. 2003. Genetic instability as a mechanism for Campylobacter jejuni for environmental survival and enhanced colonisation potential. Abstr. 1st Meet. WG4 COST920: Behaviour of Zoonotic Bacteria along the Food Chain, Budapest, Hungary, 7 to 8 October 2003. [Online.] http://www.cost920.com.
- 30.Newell, D. G., J. E. Shreeve, M. Toszeghy, G. Domingue, S. Bull, T. Humphrey, and G. Mead. 2001. Changes in the carriage of Campylobacter strains by poultry carcasses during processing in abattoirs. Appl. Environ. Microbiol. 67:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notermans, S., and A. Hoogenboom-Verdegaal. 1992. Existing and emerging foodborne disease. Int. J. Food Microbiol. 15:197-205. [DOI] [PubMed] [Google Scholar]
- 32.On, S. L. W., E. M. Nielsen, J. Engberg, and M. Madsen. 1998. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol. Infect. 120:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono, K., and K. Yamamoto. 1999. Contamination of meat with Campylobacter jejuni in Saitama, Japan. Int. J. Food Microbiol. 47:211-219. [DOI] [PubMed] [Google Scholar]
- 34.Pearson, A. D., M. H. Greenwood, J. Donaldson, T. D. Healing, D. M. Jones, M. Shahamat, R. K. A. Feltham, and R. R. Colwell. 2000. Continuous source outbreaks of campylobacteriosis traced to chicken. J. Food Prot. 63:309-314. [DOI] [PubMed] [Google Scholar]
- 35.Perko-Mäkelä, P., M. Hakkinen, T. Honkanen-Buzalski, and M. L. Hänninen. 2002. Prevalence of campylobacters in chicken flocks during the summer of 1999 in Finland. Epidemiol. Infect. 129:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragimbeau, C., G. Salvat, P. Colin, and G. Ermel. 1998. Development of a multiplex PCR gene fingerprinting method using gyrA and pflA polymorphisms to identify genotypic relatedness within Campylobacter jejuni specie. J. Appl. Microbiol. 85:829-833. [DOI] [PubMed] [Google Scholar]
- 37.Richardson, P. T., and S. F. Park. 1997. Integration of heterologous plasmid DNA into multiple sites on the genome of Campylobacter coli following natural transformation. J. Bacteriol. 179:1809-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivoal, K., M. Denis, G. Salvat, P. Colin, and G. Ermel. 1999. Molecular characterization of the diversity of Campylobacter spp. isolates collected from a poultry slaughterhouse: analysis of cross-contamination. Lett. Appl. Microbiol. 29:370-374. [DOI] [PubMed] [Google Scholar]
- 39.Shanker, S., A. Lee, and T. C. Sorell. 1990. Horizontal transmission of Campylobacter jejuni amongst broiler chicks: experimental studies. Epidemiol. Infect. 104:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. San Francisco, Calif.
- 41.Suzuki, Y., M. Ishihara, M. Saito, N. Ishikawa, and T. Yokochi. 1994. Discrimination by means of pulsed-field electrophoresis between strains of Campylobacter jejuni Lior type 4 derived from sporadic cases and from outbreaks of infection. J. Infect. 29:183-187. [DOI] [PubMed] [Google Scholar]
- 42.Talibart, R., M. Denis, A. Castillo, J. M. Cappelier, and G. Ermel. 2000. Survival and recovery of viable but noncultivable forms of Campylobacter in aqueous microcosm. Int. J. Food Microbiol. 55:263-267. [DOI] [PubMed] [Google Scholar]
- 43.Tauxe, R. V. 1992. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 9-19. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 44.Uyttendaele, M., P. De Troy, and J. Debevere. 1999. Incidence of Salmonella, Campylobacter jejuni, Campylobacter coli, and Listeria monocytogenes in poultry carcasses and different types of poultry products for sale on the Belgian retail market. J. Food Prot. 62:735-740. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weisburg, W. G., J. G. Tully, D. L. Rose, P. J. P., H. Oyaizu, D. Yang, L. Mandelco, J. Sechrest, T. G. Lawrence, and J. Van Etten. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells, J. M., and M. H. J. Bennik. 2003. Genomics of food-borne bacterial pathogens. Nutr. Res. Rev. 16:21-35. [DOI] [PubMed] [Google Scholar]
- 50.Yan, W., W. Chang, and D. E. Taylor. 1991. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiological application. J. Infect. Dis. 163:1068-1072. [DOI] [PubMed] [Google Scholar]
- 51.Yuki, N. 1998. Anti-ganglioside antibody and neuropathy: review of our research. J. Periph. Nerv. Syst. 3:3-18. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



