Abstract
Repetitive extragenic palindromic PCR fingerprinting of Escherichia coli is one microbial source tracking approach for identifying the host source origin of fecal pollution in aquatic systems. The construction of robust known-source libraries is expensive and requires an informed sampling strategy. In many types of farming systems, waste is stored for several months before being released into the environment. In this study we analyzed, by means of repetitive extragenic palindromic PCR using the enterobacterial repetitive intergenic consensus primers and comparative analysis using the Bionumerics software, collections of E. coli obtained from a dairy farm and from a swine farm, both of which stored their waste as a slurry in holding tanks. In all fecal samples, obtained from either barns or holding tanks, the diversity of the E. coli populations was underrepresented by collections of 500 isolates. In both the dairy and the swine farms, the diversity of the E. coli community was greater in the manure holding tank than in the barn, when they were sampled on the same date. In both farms, a comparison of stored manure samples collected several months apart suggested that the community composition changed substantially in terms of the detected number, absolute identity, and relative abundance of genotypes. Comparison of E. coli populations obtained from 10 different locations in either holding tank suggested that spatial variability in the E. coli community should be accounted for when sampling. Overall, the diversity in E. coli populations in manure slurry storage facilities is significant and likely is problematic with respect to library construction for microbial source tracking applications.
Access to clean drinking water is a key factor underpinning public health (5). Even in areas of the world where significant investments are made to protect water quality, fecal contamination of surface water represents a threat to human and environmental health.
This is particularly true when freshwater resources are in proximity to land subject to increasing agricultural activity and burgeoning human populations, increasing the risk to adjacent waters from agricultural runoff, sewage effluent, leaking rural septic systems, and stormwater discharge. The microbiological quality of surface and drinking water has traditionally been evaluated by quantifying fecal indicator bacteria, notably Escherichia coli, using standard microbiological methods. The presence of this organism is implicit evidence for fecal contamination and indicates a possible risk of contamination with viral, bacterial, or parasitic pathogens of enteric origin. On this basis, many jurisdictions mandate compliance with drinking and recreational water standards (4, 5).
A watershed management approach has been proposed for protecting surface water quality (12, 28). A key requirement of this strategy is to identify and then eliminate or abate sources of significant fecal contamination. In watersheds with mixed urban, agricultural, and industrial activities, the identification of pollution sources can pose a significant challenge. There has thus been significant interest in identifying attributes of fecal indicator bacteria that distinguish the host source. Genotypic and phenotypic approaches that have been investigated for this purpose include repetitive extragenic palindromic-type PCR fingerprinting, ribotyping, AFLP, pulsed-field gel electrophoresis, detection of source-specific marker genes, antibiotic resistance profiles, and carbon utilization profiles (6, 8, 13, 15, 16, 19-21, 23, 27). Repetitive extragenic palindromic PCR methods are a method of choice because of the relatively low cost, operational ease, and success at correctly classifying the host source (3, 20). Typically with this approach, environmental isolates are compared with reference collections of bacteria obtained from potential sources of fecal pollution in the area and on the basis of similarity are ascribed a probable host source. For any given study, the likelihood of correct classification will depend on several factors, including the size and representability of the reference collection, geographic size of the study area, choice of methods for image analysis and pattern recognition, and statistical methods for comparison of environmental isolates with the reference collection (1, 8, 13, 19-22, 27). The construction of robust fingerprint libraries is expensive and therefore requires an informed sampling strategy.
In many types of farming systems, animals or poultry are raised confined in barns, and their manure is stored for several months prior to release into the environment. Swine, dairy, and egg-laying poultry operations notably store excreta as a liquid or slurry in sometimes extremely large manure holding tanks. In Canada, for example, about 85% of swine and 43% of dairy cattle are produced on farms that use liquid manure storage systems (25). These manure holding tanks therefore represent a key sampling point for construction of reference collections in areas with confined production systems. Furthermore, the strain composition of enteric bacterial communities shed by farm animals could be altered significantly during manure storage, biasing the genetic composition of populations released into the broader environment. In this context, we (i) compared the structure and diversity of E. coli collections obtained from manure holding tanks with those of concurrent collections obtained from fresh manure shed by the herds on a dairy farm and a swine farm, (ii) examined the E. coli populations in swine and dairy manure holding tanks at different collection times to assess potential for population differences over time, and (iii) evaluated the spatial variability in the E. coli community composition within large manure holding tanks.
MATERIALS AND METHODS
Farms and manure collection.
The dairy farm used in this study housed 90 Holstein animals, including 45 cows and all female offspring. The adult animals received a mixed feed consisting of alfalfa hay and haylage, corn silage, high-moisture shelled corn, and protein supplement, including ground roasted soybeans, corn gluten, and minerals. The young cattle were fed hay and a grain mix. The animals receive no antibiotics or growth promoters. The average residence time for animals is 4.3 years (the average age at calving is 24 months, and the average number of 365-day lactations is 2.3). Manure from the tie stall barn is stored as liquid manure in an open outdoor concrete manure tank with a capacity of about 2,000,000 liters. The tank is emptied once a year in the fall. The swine farm used in this study is a farrow-to-finish operation consisting of approximately 2,000 animals (240 sows, 2 boars, 600 nursery pigs, and 1,100 finishers). The approximate residence time for animals in this operation is 3 years for sows and boars and 175 days for hogs (birth to market). The animals receive a feed mix consisting of corn and soybean meal. Nursery pigs received a growth promotion level of Linco-Spectrin (lincomycin and spectinomycin), and finishing pigs received a dose of 40 g/metric ton of Tylan (tylosin phosphate). Manure from the barn is stored in an open concrete manure tank with a capacity of about 800,000 liters. The tank is emptied in the spring and in the fall. The holding tank is constantly aerated during its residence time by means of an electrically driven impeller supported at the center of the tank by a flotation device. The impeller created little visible agitation.
Approximately 500 g of freshly excreted fecal material were obtained from each animal in the dairy barn during morning feeding. Swine in-barn samples were taken by collecting a composite sample of approximately 500 g of freshly excreted fecal material from pens representing the various age groups in the barn. Sampling of individuals in the swine barn was not possible due to the number of individuals per pen and the type of pens used in the barn. Holding tank samples were collected in 1-liter sterile bottles (Systems Plus, Woodstock, Ontario, Canada) at a discrete depth of either 2.5 or 0.5 m below the surface, using a Sludge Judge Ultra sampler (NASCO Canada, Aurora, Ontario, Canada). Ten samples were taken around the circumference of the holding tank, five of which were taken at 2-foot depth and five of which were taken at 9-foot depth.
In-barn samples were pooled by thoroughly mixing each individual sample and transferring 100 g into a clean Ziploc bag. This composite sample was then mixed thoroughly prior to any subsampling for microbiological purposes.
E. coli isolation and identification.
Manure samples were kept at 4°C and processed within 24 h. Samples were serially diluted in sodium metaphosphate buffer (2 g/liter; Fisher, Mississauga, Ontario, Canada) and mixed thoroughly. Dilutions were spread plated onto mFC basal medium (Difco, Toronto, Ontario, Canada) supplemented with 100 mg/liter of 3-bromo-4-chloro-5-indolyl-β-d-glucopyranoside (BCIG) (Med-Ox Diagnostics, Ottawa, Ontario, Canada) and incubated overnight at 44.5°C. After overnight growth, single blue colonies were picked and streaked onto LB agar (Difco, Toronto, Ontario, Canada) (four isolates/plate) and grown at 37°C overnight. The isolates were purified by restreaking twice on LB agar. The purified colonies were inoculated into sterile 96-well microtiter plates containing 100 μl fresh LB broth (Difco, Toronto, Ontario, Canada) per well and grown statically overnight at 37°C. For confirmation, the isolate cultures were replica plated (10 μl per well) into sterile 96-well microtiter plates containing 100 μl of lactose broth (containing, per liter, 10 g Proteose Peptone no. 3, 3 g yeast extract, 5 g NaCl, 10 g lactose, and 20 mg bromcresol purple) or 100 μl of tryptone broth (containing, per liter, 10 g Bacto tryptone and 5 g NaCl) and were incubated overnight at 37°C. Positive confirmation was indicated by a color change from purple to yellow in lactose broth (lactose fermentation) and by the formation of a red-pink color upon addition of 40 μl of Kovac's reagent to the tryptone broth wells, indicating indole production (24). Isolates were considered to be Escherichia coli if they grew at 44.5°C, had a positive reaction for β-glucuronidase (blue color on mFC-BCIG agar), fermented lactose, and produced indole. Confirmed isolates were inoculated into sterile 96-well microplates containing 100 μl/well of LB broth and incubated overnight at 37°C. Sterile glycerol (Sigma, Mississauga, Ontario, Canada) was then added to each well at a final concentration of 15% (vol/vol), and the plates were stored at −70°C.
Template preparation and ERIC PCR.
Cell suspensions of E. coli were prepared by inoculating 100 μl of fresh LB broth per well in a sterile 96-well microtiter plate with frozen stock cultures. Cells were grown statically at 37°C overnight to an A600 of about 1 and centrifuged at 710 × g for 25 min (Centra CL3 microplate centrifuge; Thermo IEC, Needham Heights, MA). The pelleted cells were resuspended in 100 μl of sterile Milli-Q H2O (100 μl) and shaken at 1,000 rpm with a microplate shaker (Sarstedt, Montreal, Quebec, Canada) for 5 min. The resuspended cells were used directly as template for the PCR or frozen at −20°C until required.
Primers used for enterobacterial repetitive intergenic consensus (ERIC) PCR were the same as described by Versalovic et al. (29). The final reaction mix (25μl) consisted of 1× PCR buffer (Promega, Madison, WI), 3 mM MgCl2, 0.1 mg/ml gelatin, 200 μM of each deoxynucleoside triphosphate (Invitrogen, Burlington, Ontario, Canada), 2 μM each of forward and reverse primers ERIC-1 and ERIC, 1 U of Taq polymerase (Promega), and 2 μl of E. coli suspended cells as template. Amplification was performed in a Hybaid OmniGene thermocycler (InterSciences Inc., Markham, Ontario, Canada) as follows: after an initial denaturation at 95°C for 10 min, 34 cycles of denaturation (94°C, 3 seconds), (92°C, 30 seconds), annealing (50°C, 1 min), and extension (65°C, 1 min) were performed, followed by a final extension (65°C, 8 min).
PCR products were resolved by horizontal gel electrophoresis in a 25-cm by 50-cm gel (Gator A3-1; Owl Separations, Portsmouth, NH) prepared with 1.5% (wt/vol) agarose (Invitrogen, Mississauga, Ontario, Canada) and 1× Tris-borate-EDTA buffer. Six microliters of loading dye was added to 25 μl of PCR product, and 7 μl of this mixture was loaded into wells prepared with an 8-mm by 1-mm comb tooth size. Every eighth well received the MassRuler DNA ladder (Fermentas, Burlington, Ontario, Canada). Gels were subjected to 4 V/cm for 2.5 h in 1× Tris-borate-EDTA. The gel was stained with 1 μg/ml ethidium bromide solution for 10 min and destained in Milli-Q water for 10 min. Gel images were captured as 8-bit TIFF images, using Quantity One gel documentation software (Bio-Rad, Mississauga, Ontario, Canada) with a CCD gel documentation system (Bio-Rad, Mississauga, Ontario, Canada).
Computer-assisted image and data analysis.
Normalization of gel images and assignment of fingerprints to isolates were done with Bionumerics (version 3.5; Applied Maths, Kortrijk, Belgium). Positions of fingerprints on gels were normalized using the MassRuler DNA ladder as the external standard in the range of 300 bp to 3,000 bp. Similarity coefficients were generated using the curve-based cosine correlation coefficient. Similarity trees were generated using the unweighted-pair group method using average linkage, and a similarity cutoff of 80% was used in order to determine related fingerprint types. Fingerprint types and numbers of isolates per fingerprint type were tabulated in Microsoft Excel. The diversity captured in the E. coli collections was estimated by rarefaction analysis using the analytical approximation algorithm of Hurlbert (11) and 95% confidence intervals estimated as described by Heck et al. (9). Calculations were performed with the freeware program Analytical Rarefaction 1.3, available at http://www.uga.edu/∼strata/software/. Curves were plotted using SigmaPlot (version 8.02; SPSS Inc., Chicago, IL). The asymptotes of the rarefaction curves were estimated using the Michaelis-Menten equation, which is available in SigmaPlot as the one-site saturation ligand model (10). The asymptote is a measure of richness at sampling saturation and was used to estimate the fraction of total community diversity captured within our E. coli collections. The SigmaPlot curve fitter uses the Marquardt-Levenberg algorithm to find the coefficients that give the best fit between the equation and the data (17).
Evaluation of spatial distribution variability in manure holding tanks.
Five sampling sites were located horizontally at about 70o angles from each other and about 1 m from the holding tank edge. At each site 1-liter samples were taken from depths of 0.5 m and 2.5 m. A Classification and Regression Trees (CART)-based classification tree approach was used to classify the dominant fingerprints on the basis of sampling depth and lateral sampling location (2, 26). CART is a widely accepted automated, binary recursive partitioning technique that selects predictor variables (independent variables) and their interactions that optimally predict a dependent measure. Dominant fingerprints in both the dairy and swine data sets were defined as being those with ≥30 observations. The classification trees were produced using a Gini splitting criterion. Each fingerprint class was also treated as if it was uniformly distributed in the population regardless of the observed sample proportions, essentially treating each fingerprint class as equally important for classification accuracy purposes. Misclassification rates were determined using a CART-based cross-validation procedure. In this study, a 10-fold approach was employed.
RESULTS
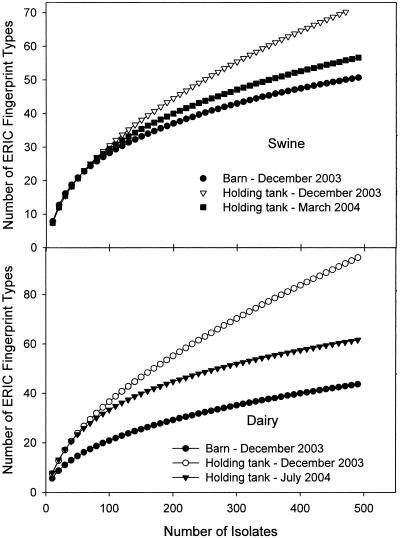
Manure was sampled from the barns and the manure holding tanks of a dairy farm and a swine farm in December 2003. The abundance of genotypes for the number of individual strains obtained from fresh and stored manure were estimated with rarefaction curves (Fig. 1). None of the rarefaction curves reached a horizontal asymptote, indicating that further acquisition of isolates would be required to capture all of the diversity within these communities. The rarefaction data were fitted with the Michaelis-Menten equation and used to estimate the asymptote (saturation of richness) and the number of isolates required to capture half of the predicted diversity (Table 1). The Michaelis-Menten fit with the experimental data was excellent (r2 of >0.98). The smallest number of genotypes detected in the six collections was 44 (dairy barn, December 2003), and the largest was 95 (dairy holding tank, December 2003). The predicted asymptotes for richness at sampling saturation ranged widely, from 57 to 158 genotypes. By comparing the measured to the predicted genotype numbers, the collections are estimated to have captured 60% to 85% of the predicted richness. For both the swine and the dairy collection sources, for sampling that was conducted on the same day, the diversity among strains obtained from the manure holding tanks was consistently greater than the diversity derived from the corresponding fresh manure found in the barn.
FIG. 1.
Rarefaction curves indicating the relative richness of E. coli collections obtained from fresh manure and the manure holding tank from a swine farm and a dairy farm (n = 500 in each case). All of the collections obtained in December 2003 were obtained on the same day. In order to evaluate the temporal stability of the populations in the stored manure, the holding tanks were subsequently resampled.
TABLE 1.
Estimated total richness in E. coli populations
| Source of collection | r2a | Genotypesb
|
No. of isolates required to capture 50% of predicted genotypes (mean ± SD)d | ||
|---|---|---|---|---|---|
| Predicted no. (mean ± SD)c | Detected
|
||||
| No. | % of predicted | ||||
| Dairy barn, December 2003 | 0.9854 | 57 ± 1 | 44 | 77 | 178 ± 9 |
| Dairy holding tank, December 2003 | 0.9908 | 158 ± 4 | 95 | 60 | 352 ± 19 |
| Dairy holding tank, July 2004 | 0.9910 | 75 ± 1 | 61 | 81 | 127 ± 4 |
| Swine barn, December 2003 | 0.9820 | 60 ± 1 | 51 | 85 | 111 ± 5 |
| Swine holding tank, December 2003 | 0.9878 | 104 ± 3 | 70 | 67 | 250 ± 14 |
| Swine holding tank, March 2004 | 0.9886 | 70 ± 1 | 57 | 81 | 141 ± 6 |
Coefficient of determination of the goodness of fit to the Michaelis-Menten equation.
The number of genotypes detected in the rarefaction analyses of Figure 1 is used to estimate the percentage of total community diversity that was captured in the collection.
The Vmax parameter in the Michaelis-Menten equation.
The Km parameter in the Michaelis-Menten equation.
The manure holding tanks were resampled several months later to evaluate the stability over time of the richness of the E. coli communities (Fig. 1). In both cases, the diversity was significantly lower than that measured previously (P < 0.05). The E. coli collection obtained from dairy manure contained 61 genotypes, approximately 81% of the 75 predicted by Michaelis-Menten modeling of the asymptote; while the swine manure yielded 57 genotypes, 81% of the 70 predicted via Michaelis-Menten modeling (Table 1). The number of isolates required to capture 50% of the predicted genotype diversity ranged from 111 to 352 isolates (Table 1).
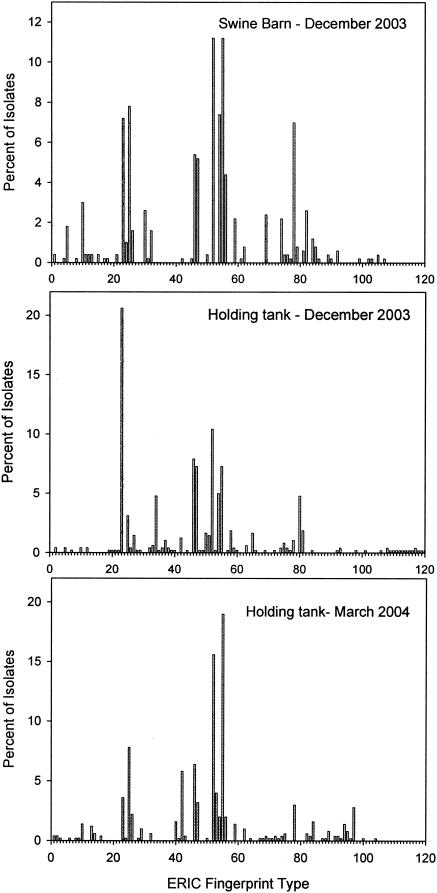
The distributions of genotypes in the swine collection sources were compared (Fig. 2). The 2003 swine barn, 2003 holding tank, and 2004 holding tank collections had 19, 39, and 22 unique genotypes (i.e., represented by only one isolate), respectively. In general, the genotypes that were most well represented in the barn were also dominant in the holding tank at both sampling times. Taking into account the relative abundance of each distinct genotype, 63% of the isolates from the December 2003 holding tank collection were represented in the barn collection. Eighty-four percent of isolates from the March 2004 holding tank collection were represented in the barn collection. Overall, the majority of the isolates obtained from the holding tank were also detected in the barn, but each collection had a significant number of unique genotypes.
FIG. 2.
Frequency of occurrence of genotypes (ERIC fingerprint types) of E. coli obtained from the barn and the manure holding tank of a swine farm (n = 500 in each case). The holding tank was sampled on two different dates.
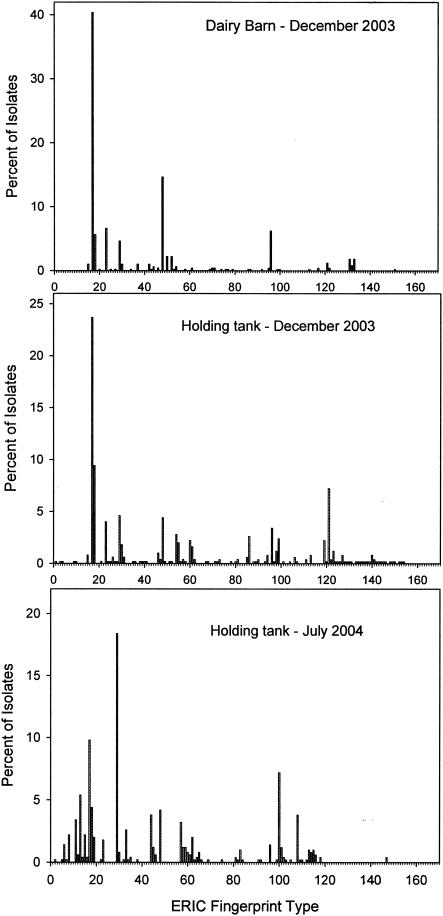
The distributions of genotypes in the 2003 dairy barn and dairy manure holding tank collections were compared (Fig. 3). There were 16, 66, and 37 unique genotypes found in the 2003 barn, 2003 manure holding tank, and 2004 holding tank collections, respectively. Sixteen genotypes detected in the barn were absent in the holding tank, whereas 66 genotypes found in the December 2003 holding tank sample were not otherwise detected, and 37 genotypes found in the March 2004 holding tank sample were unique. Taking into account the relative abundance of each distinct genotype, 72% of the isolates found in the December 2003 holding tank collection were also found in the barn collection, and 58% of the March 2004 holding tank collection isolates were found in the barn collection.
FIG. 3.
Frequency of occurrence of genotypes (ERIC fingerprint types) of E. coli obtained from the barn and the manure holding tank of a dairy farm (n = 500 in each case).
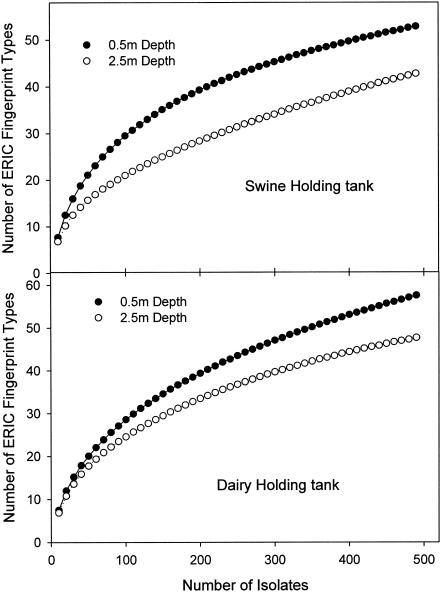
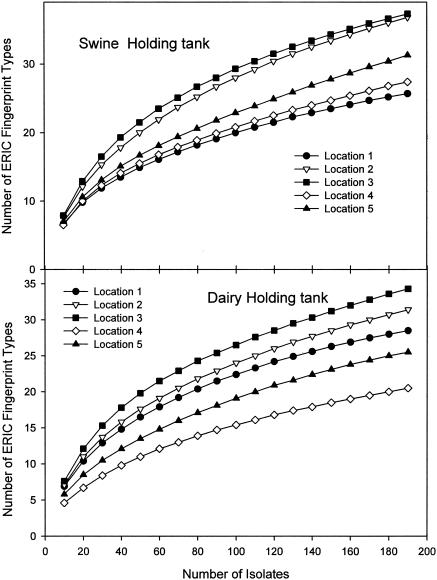
Differences in E. coli community composition between sampling locations within the swine and the dairy manure holding tanks were evaluated. One hundred E. coli isolates were obtained from each of the 10 spatially distributed sampling points taken from each holding tank. When considered together, the 500-member collections isolated from the 0.5-m-depth samples was more diverse than the collections obtained from the 2.5-m depth (Fig. 4). Samples obtained from the five different locations varied in their richness, as indicated in the rarefaction data (Fig. 5). In the dairy manure holding tank, as many as 34 genotypes and as few as 19 genotypes were detected at any one location.
FIG. 4.
Rarefaction curves for manure collections obtained from the manure holding tanks at a depth of 0.5 m or 2.5 m. Samples taken from these depths at five different lateral locations were pooled (n = 500).
FIG. 5.
Rarefaction curves for manure collections obtained from the manure holding tanks at five different lateral locations. Samples from depths of 0.5 and 2.5 m at each lateral location were pooled (n= 200).
The CART-based classification tree analysis was used to quantify the capacity for the vertical and horizontal sample locations, as independent variable criteria, to delineate dominant fingerprint genotypes for 2003 swine and 2003 dairy collections. For the dairy manure holding tank data, there were a total of 703 observations. The classification tree with the best predictive accuracy was achieved via stratification of all data on the basis of the five lateral and two sampling depth locations (10 discrete spatial locations). Output consisted of learning and testing sample results; the former was generated from the application of the input data to the selected tree, and the test sample information corresponds to the best estimate of the results that would occur if the tree was applied to new data.
There was excellent classification (percentage classified correct) on the basis of spatial location in the holding tank for genotypes designated 17, 66, and 67. Approximately 87% (for both testing and learning sets) of genotype 67 observations were located at one 0.5-m-depth location in the tank, while 97% (for both testing and learning sets) of genotype 66 observations were found at one 2.5-m-depth location in the tank. However, for genotype 17, where there was excellent classification prediction (93% for learning and 88% for testing), the fingerprint was found at three distinct lateral locations (two at 0.5-m depth and one at 2.5-m depth). Variable importance as defined on the basis of the primary splitting variables in the classification indicated that lateral location and depth were essentially of equal relative importance in discriminating the genotype data (2).
For the swine manure holding tank, as with the dairy tank, the optimal classification tree consisted of genotype classes representing each of the lateral and depth locales (total of 10 sites). There were a total of 739 observations. However, there was much poorer classification accuracy on the basis of the independent variables, relative to the dairy data set. The greatest prediction success occurred for the genotype designated 19, at only 50% correct classification for both the learning and test sample sets. The variable importance indicated dramatic differences between lateral location and depth as classification tree predictor variables (lateral location was scaled at 100% importance, relative to 19% relative importance for depth).
DISCUSSION
It has been suggested that inhospitable conditions outside of the digestive tract will alter the genetic composition of E. coli populations once shed by the host (7, 30). We had anticipated that the diversity of the holding tank communities would be significantly lower than that of the bacteria obtained from the barn, reasoning that conditions outside of the primary host habitat would eliminate less fit individuals. This would be advantageous from the microbial source tracking (MST) perspective, reducing the complexity of bacterial populations prior to their release into the broader environment. On the contrary, when sampled on the same date, the holding tank populations were more diverse than the barn populations (Fig. 1, 2, and 3). There are a number of factors that could be contributing to the higher diversity in the holding tank relative to the barn. The manure samples obtained from the barn yielded a collection of E. coli strains that were representative of the community at the time of sampling. In contrast, the manure holding tanks are continuously or periodically inoculated with fresher manure from the barn throughout the year. Thus, at any given sampling time, the community composition will integrate the diversity of E. coli shed by the herd since the previous occasion of emptying. The genetic composition of E. coli populations shed by livestock changes during the lifetime of the animal, varies from individual to individual, and is influenced by the feed composition (14, 22). Thus, the composition of E. coli populations entering the holding tank is likely to be temporally variable. Furthermore, the stored manure slurry is heterogeneous in many respects and will be presenting shed E. coli populations with a variety of environmental conditions. For example, in the absence of vigorous agitation, the solid materials settle, creating a vertical gradient of particulate material. The surface of the stored manure is exposed to the atmosphere, whereas the bulk of the material is highly reduced. Fresh manure enters at one point in the tank, from which a plume of fresh material diffuses. The concentration and temperature of the slurry are seasonably variable, increasing with evaporation during warm dry weather and decreasing with precipitation during wet cool weather. The design features of the holding tank (for example, whether it is covered, above ground, or below the barn) and management of the manure (whether it is agitated or receives odor-controlling amendments) will affect the chemical and physical compositions of the manure. Finally, there may be an opportunity for genetic rearrangements or exchange between organisms within the holding tank. Overall, it can be expected that the E. coli population in a holding tank containing many thousands or millions of liters will be diverse, seasonally dynamic, and subject to significant farm-to-farm variability.
Our sample size captured from 60% (dairy holding tank, December 2003) to 85% (swine barn, December 2003) of the predicted genotypes in the manures (Table 1). The number of isolates required to capture 50% of the predicted numbers of genotypes in the manures ranged from 111 to 353 isolates. Clearly, on a watershed that has many confined livestock production farms, the number of isolates that will be required to build representative sample libraries for this potential source will be very large. This finding is in agreement with the large E. coli population diversity now having been characterized in other potential sources of fecal pollution (7, 8, 13, 18). In any MST study, the choice of source reference library size will be determined by the size and complexity of the study area, the accuracy of source identity required by the investigators, and the financial resources available (13, 19, 21, 27).
In both farms studied here, the diversity in the E. coli community was higher in the December samples than those of the following March (swine) or July (dairy) (Fig. 1). We did not simultaneously sample the barns in the later samplings and therefore do not know if the holding tank communities are reflective of changes in the community shed by the herd at those times. In southern Ontario, Canada, the stored manure is typically at a temperature of 0 to 5°C in December and in the mid-20oCs in the summer (data not shown). It is highly likely that the E. coli population is more dynamic at warmer temperatures. A cursory examination of the genotype distributions (Fig. 2 and 3) suggests that libraries constructed from the herd would miss a number of the genotypes found at low representation in the holding tank. In the swine farm, the dominant genotypes found in the barn in December were also well represented in the holding tank in both December and March. In the dairy farm, the December barn and holding tank communities were heavily dominated by one genotype, whereas the July holding tank community was heavily dominated by genotypes that were relatively underrepresented in the December samples. Overall, these data suggest that the composition of E. coli populations released into the environment following manure storage will be variable, subject to seasonal effects that will vary according to the local climate, and that libraries constructed from that farm at any time may misrepresent the composition of E. coli populations subsequently released by that farm into the environment.
We detected clear differences in dominant genotype spatial structure between the swine and dairy manure tank data. For dairy, there are distinct locations in the tanks that harbor specific genotypes, and a sampling strategy should therefore ensure lateral and vertical sampling components. However, for swine, the results here suggested that a bulk sample from any location would likely capture the dominant fingerprints. The more homogeneous population distribution in the swine manure holding tank may be due to the aeration that it received and whatever agitation this provided. Nevertheless, from a conservative perspective, it is likely best that a holding tank sampling scheme include lateral and vertical sampling components.
An intrinsic dilemma that plagues many environmentally based sampling programs is the development of a sampling design that captures spatial and temporal variability while maintaining logistical feasibility. This dilemma is underscored for watershed-scale MST studies, where it is critical that as many representative fecal sources are sampled as feasible and that the numbers of bacteria obtained be sufficiently large to be representative of what will be released into the broader environment. This study indicated that even at the scale of a single source point, diversity and genotype representation can vary with respect to spatial location in manure storage facilities as well as with respect to the time the sample was taken. Overall, the size of reference libraries required to capture the genetic diversity of E. coli from potential fecal sources on a watershed scale is one of a number of operational constraints that limit the applicability and likely accuracy of MST methods that require host source reference libraries (8, 13, 19, 27).
Acknowledgments
This research was supported in part by funding from Ontario Pork and Health Canada through the National Water Quality Surveillance Program.
We sincerely thank K. and K. Nagelschmitz and C. and M. Bontja for access to their farms. L. Sabourin and P. Bastedo assisted with farm sampling. A. Lachance provided valuable advice.
REFERENCES
- 1.Albert, J. M., J. Munakata-Marr, L. Tenorio, and R. L. Siegrist. 2003. Statistical evaluation of bacterial source tracking data obtained by rep-PCR DNA fingerprinting of Escherichia coli. Environ. Sci. Technol. 15:4554-4560. [DOI] [PubMed] [Google Scholar]
- 2.Breiman, L., J. Friedman, R. Olshen, and C. Stone. 1984. Classification and regression trees. Wadsworth, Pacific Grove, Calif.
- 3.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequencing and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edberg, S. C., E. W. Rice, R. J. Karlin, and M. J. Allen. 2000. Escherichia coli: the best biological drinking water indicator for public health protection. J. Appl. Microbiol. 88:106S-116S. [DOI] [PubMed] [Google Scholar]
- 5.Fewtrell, L., and J. Bartram. 2001. Water quality—guidelines, standards and health: assessment of risk and risk management for water-related infectious disease. World Health Organization, London, United Kingdom.
- 6.Field, K. G., E. C. Chern, L. K. Dick, J. Fuhrman, J. Griffith, P. A. Holden, M. G. LaMontagne, J. Le, B. Olson, and M. T. Simonich. 2003. A comparative study of culture-independent, library-independent genotypic methods of fecal source tracking. J. Water Health 1:181-194. [PubMed] [Google Scholar]
- 7.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 8.Harwood, V. J., B. Wiggins, C. Hagedorn, R. D. Ellender, J. Gooch, J. Kern, M. Samadpour, A. C. H. Chapman, B. J. Robinson, and B. C. Thompson. 2003. Phenotypic library-based microbial source tracking methods: efficacy in the California collaborative study. J. Water Health 1:153-166. [PubMed] [Google Scholar]
- 9.Heck, K. L., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 10.Hughes, J. B., J. J. Hellman, T. H. Ricketts, and B. J. M. Bohanan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 12.International Joint Commission. 2004. 12th Biennial report on Great Lakes water quality. [Online.] http://www.ijc.org.
- 13.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katouli, M., A. Lund, P. Wallgren, I. Kühn, O. Söderlind, and R. Möllby. 1995. Phenotypic characterization of intestinal Escherichia coli of pigs during suckling, postweaning, and fattening periods. Appl. Environ. Microbiol. 61:778-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khatib, L. A., Y. L. Tsai, and B. H. Olson. 2003. A biomarker for the identification of swine fecal pollution in water, using the STII toxin gene from enterotoxigenic Escherichia coli. Appl. Microbiol. Biotechnol. 63: 231-238. [DOI] [PubMed] [Google Scholar]
- 16.Leung, K. T., R. Mackereth, Y.-C. Tien, and E. Topp. 2004. A comparison of AFLP and ERIC-PCR analyses for discriminating Escherichia coli from cattle, pig, and human sources. FEMS Microbiol. Ecol. 47:111-119. [DOI] [PubMed] [Google Scholar]
- 17.Marquardt, D. W. 1963. An algorithm for least squares estimation of parameters. J. Soc. Ind. Appl. Math. 11:431-441. [Google Scholar]
- 18.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myoda, S. P., C. A. Carson, J. J. Fuhrmann, B. K. Hahm, P. G. Hartel, I. H. Yampara, L. Johnson, R. L. Kuntz, C. H. Nakatsu, M. J. Sadowsky, and M. Samadpour. 2003. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 1:167-180. [PubMed] [Google Scholar]
- 21.Ritter, K. J., E. Carruthers, C. A. Carson, R. D. Ellender, V. J. Harwood, K. Kingsley, C. Nakatsu, M. J. Sadowsky, B. Shear, B. West, J. E. Whitlock, B. A. Wiggins, and J. D. Wilbur. 2003. Assessment of statistical methods used in library-based approaches to microbial source tracking. J. Water Health 1:209-223. [PubMed] [Google Scholar]
- 22.Russell, J. B., G. F. Diez, and G. N. Jarvis. 2000. Effects of diet shifts on Escherichia coli in cattle. J. Dairy Sci. 83:863-873. [DOI] [PubMed] [Google Scholar]
- 23.Simpson, J. M., J. W. Santodomingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 24.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular microbiology. American Society for Microbiology, Washington D.C.
- 25.Statistics Canada. 2002. Farm environmental management survey, 2001. Collection registration: STC/AGR-450-75054. Statistics Canada, Ottawa, Ontario, Canada.
- 26.Steinberg, D., and P. L. Colla. 1995. CART: tree-structured non-parametric data analysis. Salford Systems, San Diego, Calif.
- 27.Stoeckel, D. M., M. V. Mathes, K. E. Hyer, C. Hagedorn, H. Kator, J. Lukasik, T. L. O'Brien, T. W. Fenger, M. Samadpour, K. M. Strickler, and B. A. Wiggins. 2004. Comparison of seven protocols to identify fecal contamination sources using Escherichia coli. Environ. Sci. Technol. 38:6109-6117. [DOI] [PubMed] [Google Scholar]
- 28.U.S. Environmental Protection Agency. 2001. Protocol for developing pathogen TMDLs. EPA-R-00-002. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 29.Versalovic, J., M. Schneider, F. J. De-Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 30.Whittam, T. S. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Leeuwenhoek 55:23-32. [DOI] [PubMed] [Google Scholar]