Abstract
The role of a mitogen-activated protein kinase (MAPK) TmkA in inducing systemic resistance in cucumber against the bacterial pathogen Pseudomonas syringae pv. lacrymans was investigated by using tmkA loss-of-function mutants of Trichoderma virens. In an assay where Trichoderma spores were germinated in proximity to cucumber roots, the mutants were able to colonize the plant roots as effectively as the wild-type strain but failed to induce full systemic resistance against the leaf pathogen. Interactions with the plant roots enhanced the level of tmkA transcript in T. virens and its homologue in Trichoderma asperellum. At the protein level, we could detect the activation of two forms reacting to the phospho-p44/42 MAPK antibody. Biocontrol experiments demonstrated that the tmkA mutants retain their biocontrol potential against Rhizoctonia solani in soil but are not effective against Sclerotium rolfsii in reducing disease incidence. Our results show that, unlike in many plant-pathogen interactions, Trichoderma TmkA MAPK is not involved in limited root colonization. Trichoderma, however, needs MAPK signaling in order to induce full systemic resistance in the plant.
The perception of inductive signals from plant surfaces by pathogenic fungi has received considerable attention. As a result of these studies, cyclic AMP (cAMP)- and mitogen-activated protein kinase (MAPK)-mediated signaling pathways have been directly implicated in regulating infection-related development in diverse plant pathogenic fungi (21), including Magnaporthe grisea (23), Fusarium oxysporum (6), Botrytris cinerea (27), and Ustilago maydis (16). MAPK cascades can transduce multiple signals sensing different stimuli from the environment, thus regulating several different processes. In filamentous fungi, MAPKs, apart from being involved in pathogenicity, have been reported to be involved in sporulation, hyphal growth, and development (3, 4, 5, 13, 19).
Plant fungal pathogens deleted in members of the yeast and fungal extracellular signal-related kinase 1 (YERK1) class of MAPKs lose virulence against the host (24). The primary reason for this in pathogens that require appressoria for penetration is likely to be their inability to form these structures (14, 23). Pathogens that do not require appressoria, however, still depend on YERK1 signaling for virulence. The YERK1 family member in F. oxysporum, fmk1, was found to be indispensable for root colonization (6). Other mechanisms are likely to be important, for example, host-specific gene expression patterns that may be related to invasive growth (12, 15).
Several strains of Trichoderma are effective biological control agents. It has recently been recognized that the basis for antagonism is not only the direct mycoparasitic interaction with the pathogen but also a systemic defense response induced in the plant by root colonization (11). Some Trichoderma species are able to colonize the intercellular spaces of plant roots up to the cortex layer (25). Though there are plenty of reports on the role of signaling pathways in plant-pathogen interactions, no literature is available on the role of these events in Trichoderma-plant interactions. An understanding of the signaling events leading to induced systemic resistance would help in the genetic manipulation of Trichoderma strains for improving biocontrol potential. In our previous study (18), the tmkA gene, whose product is a MAPK homologue belonging to the YERK1 class, was isolated from Trichoderma virens and loss-of-function mutants were constructed. These mutants sporulated in the dark and had attenuated mycoparasitism on the sclerotia of Rhizoctonia solani and Sclerotium rolfsii. Mendoza-Mendoza and coworkers (17), however, reported that deletion of the homologous gene tvk1 in a different strain of T. virens improves the disease control potential.
The objective of the present work was to study the role of Trichoderma MAPK in root colonization, plant-induced resistance, and in vivo disease control ability using TmKA loss-of-function mutants. We used two plant model systems. For the induced systemic resistance assay, we used cucumber seedlings and a leaf bacterial pathogen (26), and for the in vivo disease control mycoparasitism assay in roots, we used bean seedlings and two soilborne fungi.
MATERIALS AND METHODS
Plant material and axenic growth system.
Seeds of cucumber (Cucumis sativus L. cv. Kfir) from Gedera Seeds Co. (Gedera, Israel) were used in this experiment. Surface-sterilized seeds (25 per box) were grown in an axenic hydroponic growth system (25). Plants were grown in a controlled environment as follows: 26°C, 80% relative humidity, light at 300 μE/m2/s, and a daily cycle of 14 h of light and 10 h of darkness. The growth chambers enabled the development of cucumber seedlings for up to 3 weeks.
Fungal material.
All Trichoderma strains were grown on potato dextrose agar (Difco) or synthetic medium (SM) (22). T. virens IMI 304061 wild type (WT) and tmkA mutants were obtained from our earlier study (18).
Trichoderma plant inoculation.
Trichoderma inoculum was added under aseptic conditions to the plant growth medium of 7-day-old seedlings to a final concentration of approximately 105 germinated conidia/ml (25). Control plants were treated with sterile distilled water. To observe the extent of root colonization by Trichoderma, roots were detached 48 h postinoculation and extensively washed in water. After sterilization in 1% NaOCl for 2 min, the roots of five plants per replicate were washed with sterile distilled water, weighed, and homogenized using an ULTRA-TURRAX apparatus (Janke & Kunkel, Staufen, Germany) in 20 ml water for 1 min. Serial dilutions were assayed for CFU on Trichoderma-selective medium (8) at 30°C. For each isolate, three separate experiments were set for a total of 60 plants. Data were subjected to one-way analysis of variance data analysis at the P = 0.05 level.
Bacterial inoculum.
Pseudomonas syringae pv. lachrymans was grown in tryptic soy broth overnight at 30°C. Bacterial cells were pelleted at 5,000 rpm and resuspended in sterile saline-phosphate buffer (5 mM, pH 7.2) containing 0.01% (vol/vol) surfactant (Tween 20). The bacterial suspension (20 μl; optical density at 600 nm = 0.5) was applied to the surface of the cotyledons of Trichoderma 48 h postinoculation and gently smeared with a sterile tip. Multiplication of P. syringae pv. lachrymans in the cotyledon was assessed 48 h after inoculation. For each treatment, 15 randomly selected leaves were rinsed thoroughly in sterile water and homogenized in a sterile solution of 10 mM phosphate-saline buffer. Dilutions were plated onto Pseudomonas-selective King's B agar supplemented with 1 ml/liter of 9 mg/ml basic fuchsin, 200 mg/ml cycloheximide, 10 mg/ml nitrofurantoin, and 23 mg/ml nalidixic acid. After incubation at 28°C for 2 days, the number of P. syringae pv. lachrymans CFU per gram of infected tissue was determined.
RNA isolation.
For RNA analysis, roots and leaves were harvested at a set of time points after the inoculation and stored at −70°C until required. Total RNA was extracted using the EZ-RNA total RNA isolation kit (Biological Industries Co., Beit-Haemek, Israel). Total RNA was extracted from Trichoderma mycelium (22) colonizing roots of cucumber seedlings grown in hydroponic medium or from mycelium grown for 24 to 48 h in hydroponic medium supplemented with 0.05% glucose in the presence of inert nylon fibers (Nilit, Migdal-Haemek, Israel) to mimic roots. The mycelium was removed from the roots or nylon fibers by a direct gentle water jet and recovered by centrifugation. RNA was treated with RNase-free DNase I (Roche) in 40 mM Tris-HCl (pH 7.9)-10 mM NaCl-6 mM MgCl2-10 mM CaCl2 for 30 min at 37°C. This was followed by a phenol-chloroform and chloroform extraction and a subsequent ethanol precipitation.
Reverse transcription (RT) and RT-PCR.
Total RNA (4 μg) was used as a template for first-strand synthesis using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's procedure with oligo(dT) as a primer. As a negative control, the same reactions were performed in the absence of the enzyme. One-tenth of the reaction mixture was used for PCR amplification in the linear range, determined by calibration for each gene. PCR products were separated on 1.0% agarose gel containing ethidium bromide (10 μg ml−1) and visualized under UV light. PCR products that appeared in the captured images were quantified by the National Institutes of Health Image processing and analysis program. Housekeeping genes coding for Trichoderma β-tubulin and C. sativus actin were used as references for semiquantitative analysis. Primers for β-tubulin (AY390326) were 5′ CGAGCCTTACAACGCCACCCT 3′ and 5′ AGTTCTTGTTCTGGACGGTGC 3′ (22 cycles at 54°C). Primers for the amplification of actin from C. sativus (AB010922) were 5′ TGACGCAGATAATGTTTGAGA 3′ and 5′ AGAGATGGCTGGAATAGAACT 3′. Primers for the amplification of tmkA as described by Mukherjee et al. (18) and for task1 from Trichoderma asperellum were 5′ TCAGCGAGCAGTATG 3′ and 5′ ACCGCATAATCTCTTGG 3′. Primers for the amplification of hpl and pal1 were as described in Yedidia et al. (24). The amplification conditions were 95°C for 1 min, followed by 22 cycles of 95°C for 45 s, 54°C for 45 s, and 72°C for 45 s. A final extension step of 72°C for 4 min was employed.
Isolation of tmkA homologue from T. asperellum (T203).
Primers were designed according to the conserved nucleotide sequences (5′ TCAGCGAGCAGTATG 3′ and 5′ ACCGCATAATCTCTTGG 3′) of tmkA and tvk1 from T. virens IMI 304061 and Gv29-8, respectively (17, 18). The full genomic sequence of the gene, named task1 (AY757955), was obtained using the Universal GenomeWalker kit (Clontech) as described by Viterbo et al. (22) with a gene-specific primer (5′ TTCATCTCTCGCAGGGTT 3′) and the adaptor primer provided in the kit.
Western blot analysis.
Proteins were extracted from frozen mycelia as described by Mendoza-Mendoza et al. (17). Equivalent amounts of protein (30 μg) were loaded on a sodium dodecyl sulfate-12% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane and probed according to the instructions included in the Phospho-Plus p42/p44 MAP kinase (Thr202/Tyr204) antibody kit (Cell Signaling Technology, Beverly, MA).
Phenolics extraction and microbial bioassay.
Phenolics extraction and bioassay were performed as described by Yedidia et al. (26).
Greenhouse biocontrol experiments.
Bean seeds were planted in sterile sand soil. Ten seeds were planted in each pot, and six pots were used for each treatment. Trichoderma spores were added together with the pathogen inoculum at a concentration of 107 spores/ml/kg sand. For R. solani, the inoculum was 1 g of fresh ground mycelia/kg sand, and for S. rolfsii, sclerotia were used at a concentration of 150 mg/kg sand. Observations were taken after 10 days on the number of plants infected (percentage of plants showing disease symptoms).
RESULTS
T. virens tmkA mutants retain root penetration ability but cannot induce full systemic resistance against P. syringae pv. lachrymans.
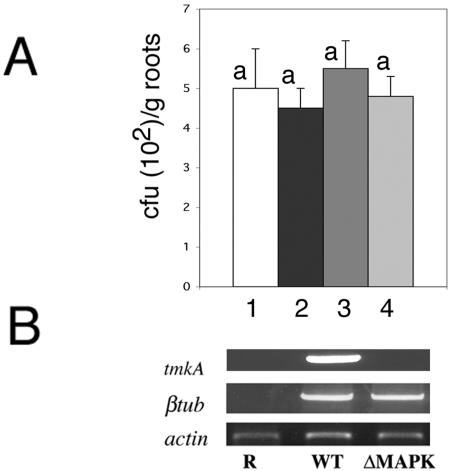
Microscopic analysis did not reveal any differences in the morphology of root-interacting hyphae between the wild type and mutants. Trichoderma tmkA mutants were capable of colonizing the cucumber seedling roots to the same extent as the wild type. Trichoderma mycelium could be recovered on selective plates from cucumber roots after surface sterilization, and no significant difference could be recorded between the WT and different mutant lines in a colony dilution assay (Fig. 1A). Trichoderma-specific gene expression could be detected in planta in the roots 48 h after inoculation (Fig. 1B), thus reconfirming the presence of active mycelia in the plant roots.
FIG. 1.
Cucumber root colonization by T. virens wild type and tmkA mutant. (A) Roots were detached 48 h postinoculation and washed. After sterilization in 1% NaOCl for 2 min, the roots were homogenized in water and serial dilutions were plated on Trichoderma selective medium at 30°C. Bars: 1, WT; 2, ΔMAPK-72; 3, ΔMAPK-95; 4, ΔMAPK-7. (B) Expression of Trichoderma tmkA and the β-tubulin gene (βtub) was assessed 48 h after inoculation in cucumber roots by RT-PCR with specific primers. Actin of C. sativus was used as a reference for RNA equal loading. Noninoculated roots (R) were used as controls for primer specificity.
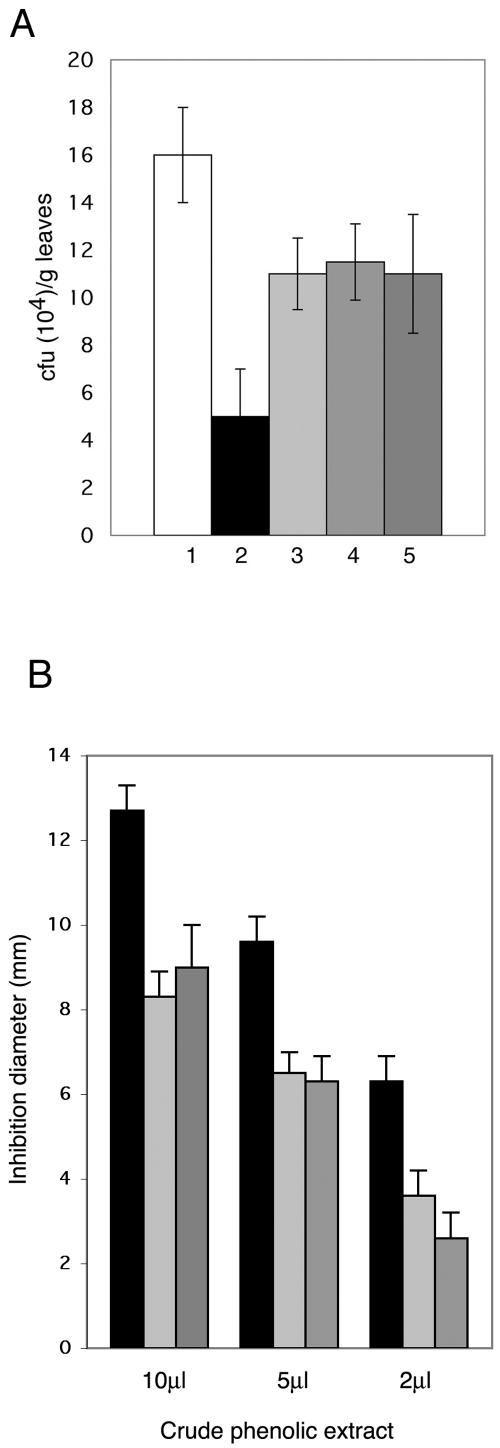
Multiplication of P. syringae pv. lachrymans in infected cucumber cotyledons (Fig. 2A) was 70% lower in the plants pretreated with the wild type than in untreated plants and only 31% lower in the plants pretreated with three tmkA mutants, indicating that inhibition of bacterial multiplication had not occurred to the same extent in this case and that the ability to afford systemic protection against bacterial leaf infection is severely reduced in the mutants with impaired MAPK signaling.
FIG. 2.
Effect of Trichoderma root inoculation on multiplication of P. syringae pv. lachrymans in challenged cotyledons, 96 h after infection. (A) Bars: 1, challenged plants, no Trichoderma inoculation; 2, inoculation with WT; 3, ΔMAPK-72; 4, ΔMAPK-95; 5, ΔMAPK-7. Columns represent means ± standard deviations of three independent experiments from which 10 cotyledons were randomly selected. (B) Bioassay comparing the antimicrobial activity of 2 to 10 μl of the aglycone fraction obtained by acid hydrolysis of crude phenolic extract of cucumber leaves 48 h postchallenge with P. syringae pv. lachrymans. Plants elicited with T. virens (black columns) or elicited with mutants ΔMAPK-95 (light gray columns) and ΔMAPK-7 (dark gray columns). The antimicrobial activity was assayed on P. syringae pv. lachrymans. Columns represent the mean inhibition diameters of three independent experiments ± standard deviations.
Previous work demonstrated that the bulk of the antimicrobial activity induced by T. asperellum (26) was found in the acid-hydrolyzed extract containing the phenolics in their aglycone form. Therefore, we compared the antimicrobial activities against P. syringae pv. lachrymans of the active conjugated fractions extracted from leaves of plants pretreated with the T. virens wild type and two different tmkA mutants. We could register a much stronger inhibitory activity in the fractions obtained from plants elicited by the T. virens wild type (Fig. 2B) than in those elicited by tmkA mutants. In accordance with previous results (26), extracts from plants challenged with P. syringae pv. lachrymans only were not active at these concentrations (data not shown).
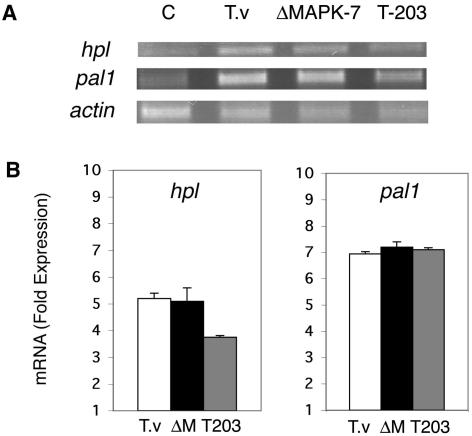
An activation in cucumber leaves of the defense-related genes pal1 and hpl upon root inoculation with T. asperellum has been shown (26). T. virens wild type is also able to systemically induce hpl gene expression fivefold and pal1 gene expression sevenfold. No significant difference in induction of the genes by a tmkA mutant could be detected (Fig. 3).
FIG. 3.
Systemic induction of pal1 and hpl by T. asperellum (T-203), T. virens (T.v) wild type, and a T. virens tmkA mutant (ΔM). (A) Expression of hpl and pal1 genes in cucumber leaves was assessed 48 h after Trichoderma inoculation of roots by RT-PCR with specific primers (24). Actin of C. sativus was used as a reference for RNA equal loading. (B) Quantification of mRNA fold induction. The values obtained for each band were normalized to actin. A value of 1 indicates no induction relative to basal expression in noninoculated plants. The results are averages of three independent experiments, and the error bars indicate standard deviations. Similar results were obtained with mutants ΔMAPK-72 and ΔMAPK-95.
Isolation of a tmkA homologue from T. asperellum.
We cloned the tmkA homologue task1 from T. asperellum and studied its expression along with tmkA to see if the same homologue from two different species responds to plant stimuli the same way. The tmkA homologue from T. asperellum was isolated using primers designed according to conserved sequences. Comparison of the deduced amino acid sequence (356 amino acids with a calculated molecular mass of 41.1 kDa) indicated a 96% identity with TmkA and 94% identity with Tvk1 from T. virens IMI 304061 and Gv-28-9, respectively. The coding sequence was interrupted by three introns, as reported for other MAPK encoding genes (17).
TmkA/Task1 transcripts and Thr-Tyr-phosphorylated protein levels increase during plant root colonization by Trichoderma.
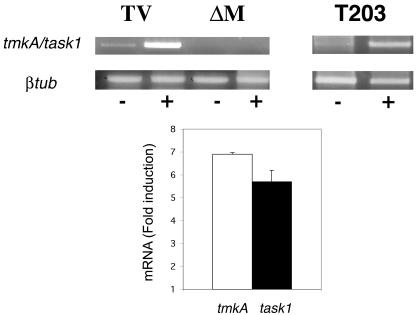
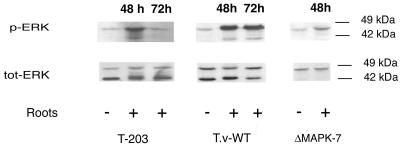
RT-PCR analysis shows that there is a six- to sevenfold induction of tmkA/task1 transcripts from the two Trichoderma species during plant root interaction (Fig. 4). When fungal protein extracts responding to selective anti-total and anti-phospho-p42/p44MAPK (Thr202/Tyr204) were analyzed, we observed a significant increase in the phosphorylation of two MAPK proteins at around 42 and 44 kDa during root interaction (Fig. 5) while we could not see a significant increase in the protein level. Analysis of the mutants at the protein level showed that the tmkA product corresponds to the 42-kDa MAPK and that the higher-molecular-mass form of MAPK is still activated upon plant root interaction.
FIG. 4.
tmkA/task1 transcipt induction by root contact. T. virens (TV, wild type; ΔM, mutants) and T. asperellum (T-203) were washed out from cucumber seedlings (+) 48 h postinoculation, and expression of tmkA/task1 was determined by RT-PCR with specific primers. RNA extracted from mycelia grown in SM with 0.05% glucose on nylon fibers was used as control (−). For the quantification of mRNA fold induction, the values obtained for each band were normalized to β-tubulin. A value of 1 indicates no induction relative to basal expression in the Trichoderma control. The results are the averages of three independent experiments.
FIG. 5.
Activation of Trichoderma MAPK by plant root interaction. Mycelia interacting with cucumber seedlings were harvested after 48 h (+). Mycelia grown in SM with 0.05% glucose were used as control (−). Cell lysates of T. asperellum (T-203), of the T. virens wild type (T.v-WT), and of the tmkA mutant (ΔMAPK-7) were analyzed by Western blotting using antibodies that specifically recognize either the dually phosphorylated forms of ERK1/2 or total ERK2 polypeptide.
Biocontrol activity against soilborne pathogens.
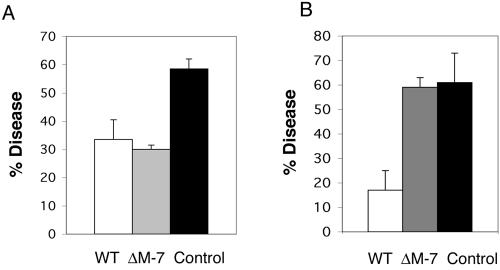
The biocontrol activity of the tmkA mutants was tested in greenhouse experiments against two root pathogens, R. solani and S. rolfsii, using bean plants as a model system. The MAPK mutants were as effective as the WT in biocontrol activity against the mycelial inoculum of R. solani (Fig. 6A). However, the tmkA mutant was unable to completely protect the plants against S. rolfsii infection (Fig. 6B). Nevertheless, the mutant did provide partial biocontrol. While large S. rolfsii lesions could be recorded in the untreated (no Trichoderma) plants (disease index 5 on a scale from 0 to 6), in the tmkA-mutant-treated plants, although infected at the same percentage (Fig. 6B), only small spots were visible (disease index 2), indicating a significant difference in the severity of the symptoms.
FIG. 6.
Biocontrol activity of T. virens and tmkA mutant against R. solani (A) and S. rolfsii (B). The disease percentage was recorded 10 days after seed planting and inoculation. The results are averages of three independent experiments with 60 plants in each treatment; bars indicate standard deviations between experiments.
DISCUSSION
Apart from direct mycoparasitism, induced systemic resistance plays a very important role in the suppression of plant pathogens by Trichoderma spp. (11). Understanding the molecular mechanisms underlying this phenomenon would help in genetically altering the pathways for improved induced resistance.
Our results indicate that, unlike in many plant-pathogen interactions (3, 6, 11, 14, 16, 27), the Trichoderma MAPK TmkA is not essential for hyphal growth on the root surface and penetration of the epidermal cells. T. virens tmkA mutants could penetrate the root epidermis and colonize the tissue as well as the wild type in both the plant systems used. Recently, it was shown (20) that the leaf pathogen Magnaporthe grisea can penetrate roots by means of simple hypopodia-like structures. Mutants impaired in the cyclic AMP signaling, unable to form functional appressoria and defective in the penetration of leaf surfaces, were not compromised in their ability to penetrate intact roots. This indicates that critical differences exist in the cAMP signal transduction pathway during infection of leaf and roots that might also apply to the MAPK signaling pathway in different fungi. The orthologue of tmkA from the soilborne fungus F. oxysporum, fmk1 was found to be indispensable for root colonization (6). This is also in contrast to our present findings, but many differences between the two soilborne fungi could account for this different behavior.
Despite colonizing the roots like the wild type, the tmkA mutants did not induce full systemic resistance. Loss of TmkA function and associated impairment of MAPK signaling is the simplest explanation for this phenotype, as well as the other developmental and biocontrol-related consequences of the mutations (18). Based on data from animal and yeast cells (1, 7), it is theoretically possible that a truncated, N-terminal MAPK protein produced in the mutant could interfere with signaling in the TmkA or other MAPK cascades. If this is true, the loss of systemic resistance might be the result of defects not only in TmkA but also in a combination of one or more of the Trichoderma MAPK pathways. Regardless of whether the similar phenotype of the three mutants arises from the specific loss of TmkA or from dominant-negative interference, our data show that Trichoderma MAPK signaling pathways are required for the induction of systemic resistance in the plant.
Yedidia et al. (26) demonstrated that the inhibition of Pseudomonas growth proliferation in cucumber leaves by T. asperellum correlates with the accumulation of phenolic compounds. Concomitantly, expression of pal1 and hpl, genes related to the phytoalexin synthesis pathway, was increased by Trichoderma root application and by pathogen challenge itself. However, the increase in P. syringae pv. lachrymans-induced pal1 expression and the related accumulation of phenolic compounds were not accompanied by an elevated level of antimicrobial capacity (26). Such an increase in antimicrobial potential was recorded only in the preelicited and challenged plants, suggesting the involvement of other biosynthetic pathways.
The capacity for affording systemic resistance is severely reduced in the mutants but not completely abolished (Fig. 2). These data suggest that some defense responses are still elicited. In the present work, we have found that pal1 and hpl genes were equally activated in plants pretreated with wild-type T. virens or in plants treated with the mutants, although a strong antimicrobial activity was recorded only in the T. virens wild-type-elicited plants. The elicitors involved in the process are still unknown, but it could be that elicitors of a different nature can act through different pathways. Our data indicate that tmkA is not required for the activation of these two genes in the plant, but its signaling is involved in the activation of other defense responses. Increased expression of pal1 and hp1 is clearly not the only factor responsible for the growth inhibition of the bacteria in the leaves, again suggesting the involvement of other enzymes that may be affected directly or indirectly by the fungal symbiont and its MAPK signaling pathway. In an attempt to isolate the active compound(s), we found that a highly active fraction can be concentrated after the addition of dichloromethane to the methanol extract of the aglycone fraction of crude phenolics and recovered by fractionation on an SI-1 silica column (Strata; Phenomenex, Torrance, Calif.) by 10% methanol. These properties indicate a highly polar nature of the active compounds. Furthermore, the absorption spectra were not typical for a phenolic compound (detailed chemical work in progress, unpublished data).
In the gene expression studies using RT-PCR, we could demonstrate that tmkA transcript levels increase during the Trichoderma-plant interaction. This increase is less evident in the corresponding translated protein, perhaps due to protein turnover aimed at a steady-state level of MAPK protein. On the other hand, there was a considerable increase in phosphorylation of the corresponding protein. We could also show that another MAPK of higher molecular mass (44 kDa) was activated during interaction with plant roots, activation that was detectable also in the mutants. This shows that the loss of TmkA does not affect the activation level of the 44-kDa species.
In the biocontrol assay, the mutant was as effective as the wild type in controlling infection due to R. solani but was less effective against S. rolfsii. Mukherjee et al. (18) predicted less biocontrol potential of tmkA mutants based on the fact that they were less active against the resting structures of the pathogens, i.e., the sclerotia. The fact that the mutants are as effective as the wild-type strain against R. solani can be explained by the fact that tmkA mutants were equally effective with respect to hyphal parasitism (18) and in the present experiment, like Mendoza-Mendoza et al. (17), we used hyphal inoculum for a biocontrol assay against R. solani.
The tmkA mutant was not able to reduce the disease incidence of S. rolfsii. In vitro experiments already proved the inability to parasitize sclerotia and also rapidly growing hyphae (18). Thus, a simple explanation for the finding shown in Fig. 6B could be that the parasitism of sclerotia would greatly reduce the effective inoculum, so the percentage of diseased plants is much lower in the wild type than in mutant-treated, pathogen-inoculated plants. However, infected plants grown in mutant-amended soil showed reduced lesion size compared to those of plants grown only with the pathogen, implying that the mutant has some, albeit reduced, biocontrol ability against S. rolfsii. The mutant can colonize the roots of cucumber seedlings (Fig. 1) and beans (not shown), and as was previously shown (23), intercellular colonization induces local mechanical defense responses that could reduce the symptoms caused by the soil pathogen.
In this study, we have shown for the first time that signal transduction by Trichoderma spp. is important for the induction of systemic induced resistance in the plant. The signals remain to be identified, but it seems that fungal MAPK pathways may transduce plant signals, telling the fungus to release factors (2, 10, 22) that are in turn detected by the plant. Plants recognize microorganisms by a variety of mechanisms, and the Trichoderma-plant interaction might be similar to plant-pathogen interactions that are governed by pathogen-derived avr genes and corresponding resistance genes in the plant host. A gene-for-gene model is certainly not the only possible one. Plant defense responses can be induced, for example, by oligosaccharide elicitors, both of plant origin (pectin and cellulose or hemicellulose oligomers) and of fungal origin (chitin or glucan oligomers). The signals of plant origin are produced by cell-wall-degrading hydrolases of fungi or bacteria, while those of fungal origin are produced by plant pathogenesis-related proteins (chitinases and glucanases) or fungal enzymes secreted during mycelial growth (9). Trichoderma species secrete a variety of hydrolytic enzymes, which could release oligosaccharide signals inciting plant defense.
Regardless of the mechanism responsible, we have shown here that a conserved fungal signal transduction pathway is involved in the three-way interaction between biocontrol fungus, plant pathogen, and plant. Furthermore, different signals control mycoparasitic activity and the ability to induce plant systemic resistance.
Acknowledgments
This work was supported by the Alexander and Myrna Strelinger Endowment Fund and a United States-Israel Binational Agricultural Research and Development Fund (US-3507-04).
REFERENCES
- 1.Bardwell, A. J., L. J. Flatauer, K. Matsukuma, J. Thorner, and L. Bardwell. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276:10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohnert, H. U., I. Fudal, W. Dioh, D. Tharreau, J. L. Notteghem, and M. H. Lebrun. 2004. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16:2499-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann, A., J. Schirawski, P. Muller, and R. Kahmann. 2003. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C., A. Harel, R. Gorovits, O. Yarden, and M. B. Dickman. 2004. MAPK regulation of sclerotial development in Sclerotinia sclerotiorum is linked with pH and cAMP sensing. Mol. Plant-Microbe Interact. 17:404-413. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, R. C., C. B. Nichols, G. M. Cox, J. R. Perfect, and J. Heitman. 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49:469-485. [DOI] [PubMed] [Google Scholar]
- 6.Di Pietro, A., F. I. Garcia-Maceira, E. Meglecz, and M. I. G. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140-1152. [PubMed] [Google Scholar]
- 7.Eblen, S. T., A. D. Catling, M. C. Assanah, and M. J. Weber. 2001. Biochemical and biological functions of the N-terminal, noncatalytic domain of extracellular signal-regulated kinase 2. Mol. Cell. Biol. 21:249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elad, Y., I. Chet, and Y. Henis. 1981. A selective medium for improving quantitative isolation of Trichoderma spp. from soil. Phytoparasitica 9:59-67. [Google Scholar]
- 9.Hahn, M. G. 1996. Microbial elicitors and their receptors in plants. Annu. Rev. Phytopathol. 34:387-412. [DOI] [PubMed] [Google Scholar]
- 10.Hanson, L. E., and C. R. Howell. 2004. Elicitors of plant defense responses from biocontrol strains of Trichoderma virens. Phytopathology 94:171-176. [DOI] [PubMed] [Google Scholar]
- 11.Harman, G. E., C. R. Howell, A. Viterbo, I. Chet, and M. Lorito. 2004. Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2:43-56. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, I., F. Namiki, and T. Tsuge. 2002. Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell 14:1869-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenczmionka, N. J., F. J. Maier, A. P. Losch, and W. Schafer. 2003. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 43:87-95. [DOI] [PubMed] [Google Scholar]
- 14.Kojima, K., T. Kikuchi, Y. Takano, E. Oshiro, and T. Okuno. 2002. The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 15:1268-1276. [DOI] [PubMed] [Google Scholar]
- 15.Lev, S., and B. A. Horwitz. 2003. A mitogen-activated protein kinase pathway modulates the expression of two cellulase genes in Cochliobolus heterostrophus during plant infection. Plant Cell 15:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayorga, M. E., and S. E. Gold. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485-497. [DOI] [PubMed] [Google Scholar]
- 17.Mendoza-Mendoza, A., M. J. Pozo, D. Grzegorski, P. Martinez, J. M. Garcia, V. Olmedo-Monfil, C. Cortes, C. Kenerley, and A. Herrera-Estrella. 2003. Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 100:15965-15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2003. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller, P., G. Weinzierl, A. Brachmann, M. Feldbrugge, and R. Kahmann. 2003. Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2:1187-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sesma, A., and A. E. Osbourn. 2004. The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 4311:582-586. [DOI] [PubMed] [Google Scholar]
- 21.Tucker, S. L., and N. J. Talbot. 2001. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39:385-417. [DOI] [PubMed] [Google Scholar]
- 22.Viterbo, A., M. Harel, and I. Chet. 2004. Isolation of two aspartyl proteases from Trichoderma asperellum expressed during colonization of cucumber roots. FEMS Microbiol. Lett. 238:151-158. [DOI] [PubMed] [Google Scholar]
- 23.Xu, J. R., and J. E. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10:2696-2706. [DOI] [PubMed] [Google Scholar]
- 24.Xu, J. R. 2000. Map kinases in fungal pathogens. Fungal Genet. Biol. 31:137-152. [DOI] [PubMed] [Google Scholar]
- 25.Yedidia, I., N. Benhamou, and I. Chet. 1999. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 65:1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yedidia, I., M. Shoresh, Z. Kerem, N. Benhamou, Y. Kapulnik, and I. Chet. 2003. Concomitant induction of systemic resistance to Pseudomonas syringae pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl. Environ. Microbiol. 69:7343-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng, L., M. Campbell, J. Murphy, S. Lam, and J. R. Xu. 2000. The bmp1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 13:724-732. [DOI] [PubMed] [Google Scholar]