Abstract
A light cycler-based real-time PCR (LC-PCR) assay that amplifies the F57 sequence of Mycobacterium avium subsp. paratuberculosis was developed. This assay also includes an internal amplification control template to monitor the amplification conditions in each reaction. The targeted F57 sequence element is unique for M.avium subsp. paratuberculosis and is not known to exist in any other bacterial species. The assay specificity was demonstrated by evaluation of 10 known M. avium subsp. paratuberculosis isolates and 33 other bacterial strains. The LC-PCR assay has a broad linear range (2 × 101 to 2 ×106 copies) for quantitative estimation of the number of M. avium subsp. paratuberculosis F57 target copies in positive samples. To maximize the assay's detection sensitivity, an efficient strategy for isolation of M. avium subsp. paratuberculosis DNA from spiked milk samples was also developed. The integrated procedure combining optimal M. avium subsp. paratuberculosis DNA isolation and real-time PCR detection had a reproducible detection limit of about 10 M. avium subsp. paratuberculosis cells per ml when a starting sample volume of 10 ml of M. avium subsp. paratuberculosis-spiked milk was analyzed. The entire process can be completed within a single working day and is suitable for routine monitoring of milk samples for M. avium subsp. paratuberculosis contamination. The applicability of this protocol for naturally contaminated milk was also demonstrated using milk samples from symptomatic M. avium subsp. paratuberculosis-infected cows, as well as pooled samples from a dairy herd with a confirmed history of paratuberculosis.
Mycobacterium avium subsp. paratuberculosis is the etiological agent of ruminant paratuberculosis (Johne's disease), as well as a specific pathogen in several other animal species (for reviews see references 4, 7, and 24). There have also been numerous reports suggesting that there is a potential association between M. avium subsp. paratuberculosis and human Crohn's disease (for reviews see references 4 and 25). However, due to the complex nature of human Crohn's disease, as well as conflicting experimental evidence, a definitive link between M. avium subsp. paratuberculosis and Crohn's disease can be neither confirmed nor discarded at present (1, 4, 23, 25). The possible M. avium subsp. paratuberculosis involvement in human disease has obviously raised significant public health concerns, and measures to minimize public exposure are encouraged. Indeed, there are several types of opportunities for human exposure to M. avium subsp. paratuberculosis, and the main focus has been on dairy products obtained from infected animals. M. avium subsp. paratuberculosis infections occur in the world's dairy herds. Infected cows shed M. avium subsp. paratuberculosis primarily in feces, as well as directly into milk. Milk may be contaminated directly within the udder or indirectly as a result of fecal contamination. The level of direct shedding of M. avium subsp. paratuberculosis into milk is less than the level of fecal shedding. Levels of less than 100 CFU per ml have been documented in symptomatic M. avium subsp. paratuberculosis-infected cows, and levels of 2 to 8 CFU per 50 ml have been documented in M. avium subsp. paratuberculosis-infected but asymptomatic cows (17, 39). The level of fecal shedding, on the other hand, can exceed 108 CFU per g, and thus fecal shedding may be a significant factor contributing to M. avium subsp. paratuberculosis contamination in raw milk (5, 29, 20). The fate of viable M. avium subsp. paratuberculosis found in raw milk is not fully understood yet. Several experimental studies have shown that viable M. avium subsp. paratuberculosis cells can survive the current standard pasteurization and cheese production processes when high numbers are present (10, 20, 21). Moreover, in some surveys it has been shown that viable M. avium subsp. paratuberculosis cells can occasionally be isolated from commercial pasteurized retail milk (2, 19; http://www.johnes.org/newsfiles/109216471862392.html). However, in other surveys the workers failed to isolate viable M. avium subsp. paratuberculosis cells in pasteurized milk samples despite the fact that the samples were positive for M. avium subsp. paratuberculosis DNA as determined by PCR, suggesting that the current pasteurization protocols may be effective in controlling M. avium subsp. paratuberculosis found in milk (16, 33). As long as the public impact of M. avium subsp. paratuberculosis remains uncertain, it is important that precautionary approaches concerning M. avium subsp. paratuberculosis in the human food chain be adopted. This means that all possible measures aimed at reducing or eradicating M. avium subsp. paratuberculosis in human food have to be encouraged. Routine screening of milk and other dairy products would be one way of monitoring for M. avium subsp. paratuberculosis contamination in the human food chain. At present, this approach is hampered primarily by difficulties in the detection of M. avium subsp. paratuberculosis when it is present in milk. The classical detection method is based on isolation of M. avium subsp. paratuberculosis from samples by culture techniques. However, this approach is labor-intensive and time-consuming, making it impractical for routine monitoring in milk given the current short shelf life of milk products. Moreover, this approach may underestimate the level of M. avium subsp. paratuberculosis or may fail to detect the organism as a result of the chemical decontamination step that is used to prevent culture overgrowth by competing microflora in milk. This step may also be deleterious to the M. avium subsp. paratuberculosis cells in the sample (11, 22). Immunology-based detection methods are faster than culture methods but are hampered by low sensitivity and cross-reactivity problems (30). PCRs provide a rapid alternative for qualitative and sensitive detection of M. avium subsp. paratuberculosis in milk and other clinical samples. Hence, a number of conventional PCR assays designed for M. avium subsp. paratuberculosis detection in milk have been described (8, 18, 34). Most of these PCR protocols target the IS900 insertion elements, which have generally been accepted as a standard marker for M. avium subsp. paratuberculosis (8, 18, 28, 34, 42). IS900 elements are multicopy insertion elements that are found at levels of 14 to 18 copies per M. avium subsp. paratuberculosis genome and therefore provide the added advantage of enhanced sensitivity for M. avium subsp. paratuberculosis detection (3). However, a drawback of using IS900 as a marker for M. avium subsp. paratuberculosis has emerged over the past few years. Sequences that are highly homologous to M.avium subsp. paratuberculosis IS900 are found in other environmental Mycobacterium species. So far, such genetic elements have been described for strains isolated from bovine feces which are positive with most of the current IS900 PCR systems used for standard M. avium subsp. paratuberculosis detection (9, 14). Therefore, other M. avium subsp. paratuberculosis-specific genetic elements have to be evaluated to improve the reliability of PCR detection of M. avium subsp. paratuberculosis. The alternative target elements in M. avium subsp. paratuberculosis include the F57, ISMav2, and Hsp X sequences (12, 35, 38). The F57 and Hsp X sequences occur as single copies, while at least three copies of ISMav2 are present in the M. avium subsp. paratuberculosis genome (12, 35, 38). Although these markers may not be as sensitive as the multicopy IS900 elements, they are highly specific for M. avium subsp. paratuberculosis and thus are less prone to false-positive results (6, 12, 13, 35, 37, 40, 41). The utility of such markers for M. avium subsp. paratuberculosis detection in milk is not known, and hence further evaluation of the markers is needed. Moreover, highly specific PCR detection systems mean that the benefits of rapid qualitative and quantitative detection offered by real-time PCR systems can be maximally exploited with slow-growing and difficult-to-culture organisms, such as M. avium subsp. paratuberculosis. A number of M. avium subsp. paratuberculosis real-time PCR detection assays are available, but most of them are IS900 based, making them prone to potential false-positive signals associated with some non- M. avium subsp. paratuberculosis Mycobacterium species (15, 26, 27, 31, 32). As an alternative, only one assay based on real-time nucleic acid sequence-based amplification detection of the dnaA gene has been described (36). We therefore sought to develop a real-time PCR assay for detection of M. avium subsp. paratuberculosis that targets a more specific genetic marker for this organism and is based on the light cycler system. We selected the M. avium subsp. paratuberculosis F57 sequence because it has been found only in M. avium subsp. paratuberculosis, which makes it a potentially highly specific marker for M. avium subsp. paratuberculosis (35). The function of the F57 sequence in M. avium subsp. paratuberculosis is not known yet, but the specificity of F57 for the organism has been supported by studies of other groups, as well as in our lab, with conventional PCR protocols (6, 14, 40, 41). Our objective was to develop a diagnostic PCR system for routine evaluation of milk samples. Therefore, taking into consideration the known potential PCR inhibition associated with milk samples, we incorporated an internal amplification control (IC) in the M. avium subsp. paratuberculosis real-time PCR assay which we developed to enable monitoring of the conditions of each reaction. Moreover, in order to maximize the sensitivity of detection of M. avium subsp. paratuberculosis with the new real-time M. avium subsp. paratuberculosis PCR assay, an efficient strategy for isolation of M. avium subsp. paratuberculosis DNA from milk was developed in parallel.
MATERIALS AND METHODS
Bacterial strains used.
To evaluate the specificity of the M. avium subsp. paratuberculosis light cycler-based real-time PCR (LC-PCR) assay which we developed, purified DNA templates from 10 M. avium subsp. paratuberculosis isolates, 21 non-M. avium subsp. paratuberculosis Mycobacterium spp., and nine non-Mycobacterium spp. were used (Table 1).
TABLE 1.
Evalution of LC-PCR specificity with M. avium subsp. paratuberculosis and non-M. avium subsp. paratuberculosis bacterial isolatesa
| Taxon | Source | LC-PCR result |
|---|---|---|
| M. avium subsp. paratuberculosis | ATCC 1698 | + |
| M. avium subsp. paratuberculosis | ATCC 43544 | + |
| M. avium subsp. paratuberculosis | ATCC 6783 | + |
| M. avium subsp. paratuberculosis 101/60/02 | Bovine (Switzerland) | + |
| M. avium subsp. paratuberculosis strain II | MAP vaccine strain | + |
| M. avium subsp. paratuberculosis K 43 | Bovine (United Kingdom) | + |
| M. avium subsp. paratuberculosis K 47 | Bovine (United Kingdom) | + |
| M. avium subsp. paratuberculosis K 57 | Bovine (United Kingdom) | + |
| M. avium subsp. paratuberculosis US7 | Bovine (United States) | + |
| M. avium subsp. paratuberculosis SN6 | Human (United States) | + |
| M. avium | ATCC 19421 | − |
| M. avium | DSM 44156 | − |
| M. intracellulareb | − | |
| M. intracellulare | DSM 43223 | − |
| M. bovisb | − | |
| M. smegmatis | DSM 43756 | − |
| M. gastri | DSM 43505 | − |
| M. fortuitum | DSM 43074 | − |
| M. kansasii | DSM 44162 | − |
| M. scrofulaceumb | − | |
| M. scrofulaceum | DSM 43992 | − |
| M. marinum | DSM 43518 | − |
| M. phlei | DSM 750 | − |
| Mycobacterium strain 2333 | Bovine (Sweden) | − |
| M. cookiib | − | |
| M. vaccaeb | − | |
| M. terraeb | − | |
| M. simiaeb | − | |
| M. gordonaeb | − | |
| M. xenopib | − | |
| M. chelonaeb | − | |
| M. szulgaib | − | |
| M. ulzeransb | − | |
| M. haemophilumb | − | |
| Lactobacillus acidophilus | ATCC 13651 | − |
| Salmonella enterica serovar Enteritidis | Wild strain | − |
| Listeria monocytogenes | Wild strain | − |
| Staphylococcus aureus | ATTC 25923 | − |
| Streptococcus uberis | Wild strain | − |
| Streptococcus agalactiae | ATCC 33019 | − |
| Bacillus cereus | ATCC 10876 | − |
| Enterobacter sakazkaii | Wild strain | − |
| E. coli | ATCC 25922 | − |
Duplicates of 1-ng genomic DNA templates purified from each bacterial isolate were amplified by the LC-PCR assay with the light cycler as outlined in Materials and Methods. The samples were monitored in channel 4 (640 nm) for the IC template and in channel 6 (705 nm) for M. avium subsp. paratuberculosis F57 sequence amplification. A positive result indicates detection of M. avium subsp. paratuberculosis F57 amplification, and a negative result indicates that no F57 amplification was observed. Appropriate amplification conditions were confirmed for all the reactions by monitoring IC template amplification in the LC Red-640 channel.
Clinical isolates obtained from the Swiss National Centre for Mycobacteria collection.
Purification of DNA templates from bacterial cultures.
A total genomic DNA template was purified directly from each bacterium by starting with either a colony or a cell suspension of the isolate that had been grown under appropriate culture and medium conditions. The DNA was purified using a High Pure PCR template preparation kit used in accordance with the manufacturer's guidelines, as outlined in the kit protocol (Roche Molecular Diagnostics, Penzberg, Germany). The concentration of each purified DNA template was determined by spectrophotometry.
Generation of the quantification standards and internal control template.
To generate M. avium subsp. paratuberculosis genome quantification standards, the 254-bp target region in the M. avium subsp. paratuberculosis F57 sequence was amplified with primers MAPf57p1 and MAPf57p2 (Table 2), using a conventional PCR (3 min at 95°C, followed by 30 cycles of 15 s at 95°C, 15 s at 56°C, and 30 s at 72°C and then 5 min at 72°C). The amplicon was excised from the agarose gel, purified with spin columns (QIAGEN), and quantified. The purified amplicon was then used to prepare the standard dilutions used throughout this study. For storage, dilutions of the standards were frozen in aliquots, and when needed, the aliquots were thawed before use and then stored at 4°C during use. An IC template was developed for monitoring PCR amplification conditions and detecting PCR inhibition in each reaction. The IC template system was based on a 400-bp fragment of plasmid pUC19 DNA. This fragment was generated by PCR amplification of the target region using the PuC19fw-PuC19rv primer pair (Table 2) under the following conditions: 3 min at 95°C, followed by 30 cycles of 15 s at 95°C, 15 s at 56°C, 30 s at 72°C and then 5 min at 72°C. The resulting PCR amplicon was purified on spin columns and quantified. Appropriate dilutions of the IC template were also prepared and frozen in aliquots for storage. Aliquots were thawed and added to the LC-PCR assays.
TABLE 2.
Oligonucleotide primers and LC probes used in this studya
| Target | Oligonucleotide | Sequence (5′-3′) | Location | Product size (bp) |
|---|---|---|---|---|
| M. avium subsp. paratuberculosis | MAPf57p1 | TTG GAC GAT CCG AAT ATG T | 126-144 | 254 |
| F57 sequence (accession no. | MAPf57p2 | AGT GGG AGG CGT ACC A | 365-380 | |
| X70277) | MAPf57-3iFluo | CAC GCA GGC ATT CCA AGT | 250-267 | |
| MAPf57-5iRed705 | TGA CCA CCC TTC CCG TCG | 270-287 | ||
| pUC19 plasmid DNA (accession | PuC19fw | CGG AGA CGG TCA CAG CT | 49-65 | 400 |
| no. L09137) | PuC19rv | TTG CAT GCC TGC AGG T | 433-448 | |
| PuC19-3iFluo | GCA AGG CGA TTA AGT TGG GTA AC | 330-350 | ||
| PuC19-5iRed640 | CAG GGT TTT CCC AGT CAC GAC | 355-375 |
All oligonucleotides were synthesized by Microsynth (Balgach, Switzerland), and all probes were selected using the LC Probe Design software and were synthesized by TIB-Molbiol (Berlin, Germany).
LC-PCR conditions.
Real-time PCRs were performed with a Light Cycler 2.0 instrument (Roche Molecular Diagnostics) by using a total reaction volume of 20 μl in glass capillary tubes. Optimization reactions were first performed until the best primer, probe concentrations, and cycling conditions that allowed coamplification of the M. avium subsp. paratuberculosis F57 sequence and IC template in one reaction were established. The optimized reaction mixture contained 1× LightCycler-Faststart DNA master mixture plus hybridization probe mixture (Roche Molecular Diagnostics), each primer (MAPf57p1, MAPf57p2, PuC19fw, and PuC19rv) at a concentration of 800 nM, each LC probe (Table 2) at a concentration of 200 nM, and 2,000 copies of PCR-generated IC template. The amplification started with an initial preincubation step at 95°C for 10 min to activate the DNA polymerase, and this was followed by 45 cycles of 95°C for 10 s, 56°C for 20 s, and 72°C for 18 s. Fluorescence signal generation corresponding to each amplicon was monitored during the 56°C annealing step in channel 4 (640 nm) for the IC and in channel 6 (705 nm) for M. avium subsp. paratuberculosis F57 sequence amplification.
Evaluation of LC-PCR assay analytical detection limit and specificity.
To determine the lower limits of detection of the M. avium subsp. paratuberculosis F57 sequence target, several replicates of purified M. avium subsp. paratuberculosis genomic DNA dilutions were analyzed by the LC-PCR assay. For inclusivity studies, the M. avium subsp. paratuberculosis LC-PCR assay was performed with 1-ng portions of total genomic DNA templates extracted from different M. avium subsp. paratuberculosis isolates. For the exclusivity studies, 1-ng total genomic DNA templates extracted from 21 non-M. avium subsp. paratuberculosis Mycobacterium spp. and nine non-Mycobacterium spp. were analyzed in duplicate in the LC-PCR assay. The influence of a nonspecific DNA background on the performance of the LC-PCR assay was investigated by monitoring amplification of different amounts of the M. avium subsp. paratuberculosis F57 target in the presence of 200 ng nonspecific background nucleic acids. These background nucleic acids were based on mixtures of total genomic DNA from the different non-M. avium subsp. paratuberculosis Mycobacterium spp. or non-Mycobacterium spp. listed in Table 1.
Estimation of M. avium subsp. paratuberculosis counts.
Counting of mycobacteria in the M. avium subsp. paratuberculosis stock culture (strain ATCC 19698) was performed by light microscopy as described by Wittenbrink et al. (44). Briefly, suspensions of mycobacterium cultures were serially diluted in phosphate-buffered saline (PBS) in 10-fold steps. A 1-μl drop of each dilution was placed on a glass slide in a 1-cm2 area and stained by the Ziehl-Neelsen method. Mycobacterium particles were counted by light microscopy using an ocular standard screen delimiting a 0.01-mm2 quadratic microscopic field at a magnification of ×1,000. Using this magnification, the ratio of one field of the screen to the whole area of the smear was 1:10,000. Counting of mycobacteria was done in triplicate; in each trial mycobacteria were counted in 25 randomly selected microscopic fields, and the number of mycobacteria per ml was determined by using the following equation: total counts/ml = mean no. of mycobacteria in 25 fields × 10,000 × 1/dilution × 1,000.
The accuracy of the M. avium subsp. paratuberculosis counts obtained by this approach was confirmed by culturing the M. avium subsp. paratuberculosis stocks on solid media on HEYM slants. After this, the M. avium subsp. paratuberculosis stocks were used to prepare M. avium subsp. paratuberculosis dilutions to spike raw milk samples as described below.
Preparation of M. avium subsp. paratuberculosis-spiked raw milk samples.
Bulk tank raw milk samples were collected from dairy herds with no known history of paratuberculosis. The M. avium subsp. paratuberculosis-negative status of each milk sample was confirmed by PCR analysis before the sample was used in these experiments. Tenfold serial dilutions of viable M. avium subsp. paratuberculosis cells were prepared from a stock suspension containing 108 cells per ml (see above for the method used to verify M. avium subsp. paratuberculosis stock concentrations). Dilutions containing from 107 to 101 M. avium subsp. paratuberculosis cells per ml were prepared in PBS. An aliquot from each dilution step was diluted 10-fold in raw milk to obtain spiked samples containing from 106 to 100 M. avium subsp. paratuberculosis cells per ml of raw milk. As a negative control, an unspiked milk aliquot from the same batch was included in each M. avium subsp. paratuberculosis detection run.
Evaluation of M. avium subsp. paratuberculosis DNA isolation methods for M. avium subsp. paratuberculosis-spiked raw milk samples.
Several commercial DNA isolation protocols for isolation of M. avium subsp. paratuberculosis DNA from 10-ml raw milk samples artificially contaminated with M. avium subsp. paratuberculosis ATCC 19698 were evaluated. Ten-milliliter raw milk samples spiked with 106, 102, and no M. avium subsp. paratuberculosis cells per ml were prepared from the same batch of M. avium subsp. paratuberculosis-free raw milk. One hundred microliters of Triton X-100 (Calbiochem, Germany) was added to each mixture, and the mixtures were centrifuged for 30 min at 4,500 rpm to obtain pellets. Most of each supernatant was discarded, leaving about 0.5 ml. The pellets were resuspended in the remaining supernatant and transferred to Eppendorf tubes. A second centrifugation step was performed (10 min at 14,000 rpm), the supernatants were discarded, and the pellets were stored at −20°C until they were used for isolation of M. avium subsp. paratuberculosis DNA. This was the starting point for evaluating most of the different DNA isolation strategies. The only exceptions were the samples processed using the bead-based DNA isolation protocols with the Bugs'n Beads and ChlamCAP kits (see below). In all cases for each M. avium subsp. paratuberculosis concentration analyzed, triplicate samples were processed.
(i) DNA precipitation I (First-DNA kit).
The procedure used for DNA precipitation I (First-DNA kit; GEN-IAL Ltd., Troisdorf, Germany) included minor modifications of the kit provider's protocol, which was designed for isolation of DNA from food samples. Briefly, the pellets obtained from centrifugation of 10-ml milk samples were suspended in 500 μl Lyse 1 buffer and 50 μl Lyse 2 buffer. Ten microliters of proteinase K provided in the kit was added to each preparation, and this was followed by incubation at 65°C for about 30 min or more with regular mixing until all of the pellet was dissolved. A protein precipitation step was performed by adding 375 μl Lyse 3 buffer, mixing the preparation, incubating it at −20°C for 5 min, and centrifuging it (13,000 rpm, 10 min). Ethanol precipitation of DNA from the supernatant was performed as outlined in the kit protocol, and the DNA template was redissolved in 100 μl of water. The eluted DNA samples were heated for 10 min at 65°C, the DNA yield was determined, and then 5-μl aliquots of the template were used in reactions for LC-PCR analysis.
(ii) Genomic DNA purification kit.
The second DNA precipitation method that was evaluated was the Puregene DNA purification system (Gentra Systems, Minneapolis, Minn.) procedure for DNA isolation from body fluids. The milk pellets were resuspended in 1 ml of the supernatant after the first centrifugation step. The rest of the protocol (i.e., protein and DNA precipitation steps) was performed as recommended by the kit supplier. In the final step the DNA pellet was redissolved in 100 μl of the DNA hydration solution provided in the kit and heated for 10 min at 65°C. The nucleic acid concentrations of the samples were determined, and 5-μl aliquots of the template were used in reactions for the LC-PCR assays.
(iii) High Pure template preparation kit.
As a modification, a mechanical lysis step was included in the original High Pure template preparation kit (Roche Diagnostics GmbH, Penzberg, Germany) protocol. Briefly, the milk pellets were resuspended in 240 μl of an M. avium subsp. paratuberculosis lysis buffer (20 mM Tris-HCl [pH 8.0], 400 mM NaCl, 0.6% sodium dodecyl sulfate, 2 mM EDTA). A proteinase K lysis step was performed as outlined in the kit protocol by adding 60 μl of the proteinase K solution supplied in the kit, followed by incubation at 65°C until the milk pellets were dissolved. Then 300 μl of the kit binding buffer was added. The mixtures were transferred onto the ribolysing matrix in Ribolyser tubes, and a mechanical lysis step (6.5 ms−1 for 45 s) was performed using a Ribolyser (Hybaid, Ashford, United Kingdom). The samples were immediately incubated at 70°C for 10 min. Then the mixtures were briefly centrifuged, added to the DNA binding columns, and processed as described in the kit protocol. Finally, the DNA templates were eluted in 100 μl of the elution buffer supplied in the kit. The nucleic acid concentrations of the samples were determined, and 5-μl aliquots were used as templates in the LC-PCR assays.
(iv) Generation capture column kit.
The procedure outlined in the Generation capture column kit (Gentra Systems, Minneapolis, Minn.) protocol was followed, with minor modifications. The milk pellets were resuspended in 200 μl of PBS, and each well-mixed sample was applied to the DNA binding column provided with the kit. The DNA purification steps were performed as outlined in the kit protocol, and finally the purified DNA was eluted in 200 μl of the DNA elution solution (solution 2) provided in the kit. The nucleic acid concentration in the eluant was determined, and 5-μl aliquots of the template were used in the LC-PCR assays.
(v) Bacteria and DNA-binding magnetic beads.
In principle, the bacterial and DNA-binding magnetic bead protocols are based on bacterial immobilization on beads. The bead-bound bacteria are lysed to release nucleic acids, which are also bound by the same bead matrix. After washing, the captured DNA templates are released from the beads by heating, and aliquots of the template are directly used in PCR assays. A Bugs'n Beads kit and a ChlamCAP kit (Genpoint AS, Oslo, Norway) were both used to isolate M. avium subsp. paratuberculosis DNA from the spiked milk samples. The milk pellets obtained after the first centrifugation step were resuspended in 300 μl (Bugs'n Beads kit) and 700 μl (ChlamCAP kit) of the supernatant. Each preparation was then mixed with the appropriate bead suspension, and DNA isolation was performed as outlined in the protocol for each kit. In the final step the DNA was eluted from the beads by resuspending the bead-DNA complex in 50 μl water and heating the mixture at 85°C for 10 min. After this, 5-μl aliquots of the released nucleic acids were used as templates in the LC-PCR assays.
Detection limit and reproducibility of LC-PCR assay with M. avium subsp. paratuberculosis-spiked raw milk.
The detection limits of the integrated optimal template preparation and LC-PCR analysis protocols for direct M. avium subsp. paratuberculosis detection in artificially contaminated raw milk samples were evaluated. Ten replicates of raw milk spiked with 1,000, 100, 10, and 1 M. avium subsp. paratuberculosis cells per ml were prepared as outlined above. DNA templates were prepared from these samples using the High Pure template preparation kit protocol, including the mechanical lysis step outlined above. The numbers of positive and negative results for the 10 replicates of each group of M. avium subsp. paratuberculosis-spiked samples were recorded, and the percentages of positive results were determined.
Negative controls.
Several steps were used during the analysis in order to avoid potential sample cross contamination and false-positive results. DNA extraction, PCR mixture preparation, and post-PCR analysis were carried out in separate rooms. Filter-protected pipette tips were used in all experiments. DNA extraction process-negative controls containing buffer or water were included in each DNA extraction run and analyzed by PCR for cross contamination. In the M. avium subsp. paratuberculosis spiking experiments a negative control consisting of unspiked milk from the same batch was included during each template purification run and subsequently analyzed in the LC-PCR run. Finally in each LC-PCR run, a negative control consisting of water instead of DNA template was also included.
RESULTS
Optimizing the assay conditions and evaluation of specificity.
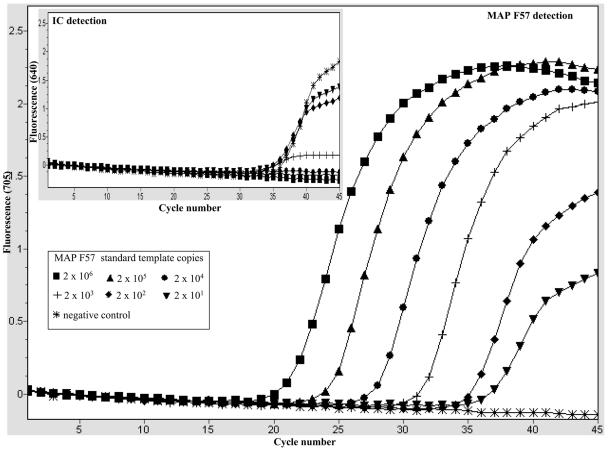
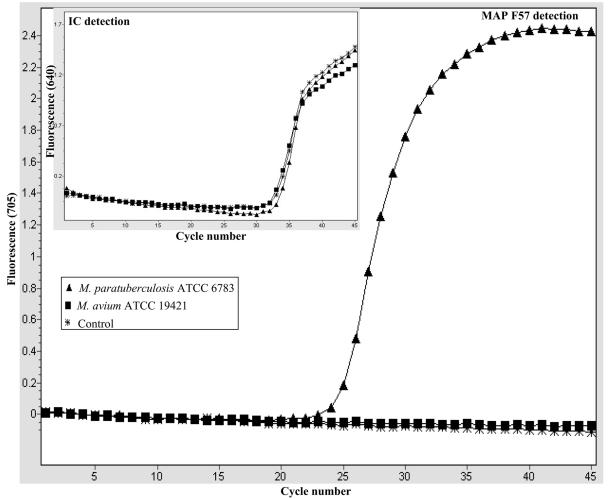
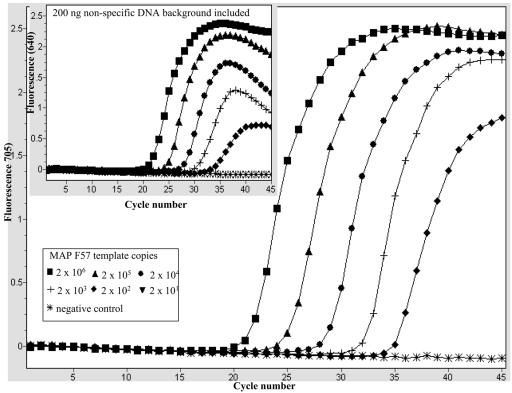
Optimal PCR assay conditions that allowed efficient coamplification of the F57 target sequence and the IC template in the same reaction were established. For the IC template, we used a purified PCR fragment generated by amplification of a 400-bp region of the pUC19 plasmid, as described in Materials and Methods. We began by determining the optimal amount of IC template needed in each reaction, and we found by titration that 2,000 copies of the IC template was sufficient to give a clear amplification signal, without severely compromising the detection of small amounts of the M. avium subsp. paratuberculosis target in the assay (data not shown). As an example, Fig. 1 shows the amplicon detection dynamics during coamplification of M. avium subsp. paratuberculosis F57-based standard dilutions (2 × 101 to 2 × 106 copies) and a fixed amount of the IC (2,000 copies) per reaction mixture. The reactions were monitored for amplification of the IC template (LC Red-640) and M.avium subsp. paratuberculosis F57 target (LC Red-705) in their light cycler detection channels (Fig. 1). As shown by the light cycler graphic plot in the inset in Fig. 1, under these conditions clear IC template amplification signals were observed in the presence of 0 to 2×103 copies of the M. avium subsp. paratuberculosis F57 target. IC template amplification did not occur when more than 2×103 copies of the M. avium subsp. paratuberculosis F57 target were coamplified in the same assay, and it was assumed that there was indirect competition with the F57 target for the reaction reagents under these conditions (Fig. 1, inset). These conditions allowed effective monitoring of PCR conditions at low M. avium subsp. paratuberculosis target concentrations. At the same time, low levels of PCR inhibition that might otherwise have been missed with high levels of the IC template could also be detected. This optimal amount of IC was used for all the LC-PCR analyses throughout this study. Next, the analytical specificity of the LC-PCR assay for M. avium subsp. paratuberculosis was confirmed. This was done by testing purified DNA templates from M. avium subsp. paratuberculosis reference strains and M. avium subsp. paratuberculosis isolates of different origins (Table 1). The assay's specificity was further demonstrated by its ability to exclude all non-M. avium subsp. paratuberculosis bacterial species listed in Table 1. For all the reactions appropriate amplification conditions were confirmed by monitoring IC amplification. A typical example is shown in Fig. 2, which shows amplification of DNA templates from M. avium subsp. paratuberculosis ATCC 6783 and M. avium ATCC 19421. Amplification of the M. avium subsp. paratuberculosis F57 sequence (LC-Red 705) was detected only with the M. avium subsp. paratuberculosis DNA template (Fig. 2). Meanwhile, monitoring the same reactions for IC amplification (LC-Red 640) confirmed that there was amplification in all the reactions (Fig. 2, inset), which showed that appropriate PCR amplification conditions were present for all the samples, and the negative results were due to a lack of the M. avium subsp. paratuberculosis F57 sequence. Thus, using such selectivity studies, we demonstrated that the LC-PCR assay which we developed is highly specific for M. avium subsp. paratuberculosis, and no cross-reactivity was observed with the bacterial species listed in Table 1. The complex nature of milk meant that derived PCR samples typically had low levels of M. avium subsp. paratuberculosis target molecules and high background levels of nonspecific DNA contributed by the other milk components. Therefore, as a next step, the LC-PCR assay's detection and amplification performance was evaluated in the presence of defined nonspecific DNA backgrounds. The nonspecific DNA backgrounds tested consisted of mixtures of genomic DNA templates derived from the non-M. avium subsp. paratuberculosis Mycobacterium spp. or non-Mycobacterium spp. shown in Table 1. This was done by amplifying 10-fold serial dilutions (2 × 102 to 2 × 106 copies) of an M. avium subsp. paratuberculosis F57 standard alone or in the presence of 200 ng of nonspecific DNA as a background in the assay. An example is shown in Fig. 3, in which the amplification profiles of the M. avium subsp. paratuberculosis F57 standards alone or in the presence of a 200-ng DNA background (Fig. 3, inset) are shown. The amplification profiles of M. avium subsp. paratuberculosis F57 target titrations were not significantly altered by inclusion of 200 ng of background DNA made up of a mixture of DNA templates from the non-M. avium subsp. paratuberculosis Mycobacterium spp. This was confirmed when the LC-PCR assay's amplification efficiencies for these experiments were compared, and the efficiencies did not differ significantly under the two experimental conditions (data not shown). Similar results were obtained when a non-Mycobacterium spp. DNA template mixture was evaluated as a background (data not shown). Therefore, these experiments indicated that the LC-PCR assay's performance was not significantly altered by the high-level nonspecific DNA backgrounds.
FIG. 1.
Dynamics of F57 sequence and IC template coamplification under optimized LC-PCR assay conditions: graphic plot from the light cycler. The data show the results for coamplification of the F57 sequence standard dilutions and a constant amount of the IC template (2,000 copies) in the LC-PCR assay. The large graph shows M. avium subsp. paratuberculosis F57 amplification as monitored in the LC-Red 705 channel, and the inset shows IC template amplification as monitored in the LC-Red 640 channel. The inset shows that the amount of IC template used is large enough to effectively monitor amplification in the presence of small amounts of the M. avium subsp. paratuberculosis F57 target (0 to 2×103 copies). No IC amplification was detected in the presence of large numbers of M. avium subsp. paratuberculosis F57 molecules (2 × 104 to 2 × 106 copies) due to competition. MAP, M. avium subsp. paratuberculosis.
FIG. 2.
Evaluation of the specificity of the M. avium subsp. paratuberculosis LC-PCR assay. Genomic DNA templates (1 ng) isolated from M. avium subsp. paratuberculosis ATCC 6783 and M. avium ATCC 19421 were amplified, and the amplicons were detected as outlined in Materials and Methods. The large graph shows F57 sequence amplification monitored by LC-Red 705 detection, and the inset shows IC amplification monitored by LC-Red 640 detection. In each reaction, the IC control template was included, and its amplification was confirmed (inset). MAP, M. avium subsp. paratuberculosis.
FIG. 3.
Influence of nonspecific DNA background on amplification of the F57 sequence by the LC-PCR assay. Tenfold serial dilutions of F57 sequence standard dilutions (2 × 102 to 2 × 106) and the IC template (2,000 copies) were amplified alone (large graph) or after they were spiked into reaction mixtures containing 200 ng of nonspecific DNA (inset). The inset shows the influence of a DNA background mixture consisting of DNA templates from the following non-M. avium subsp. paratuberculosis mycobacteria: M. avium, M. bovis, M. intracellulare, M. phlei, M. scrofulaceum, M. terrae, and Mycobacterium strain 2333. MAP, M. avium subsp. paratuberculosis.
Analytical sensitivity and quantitative detection.
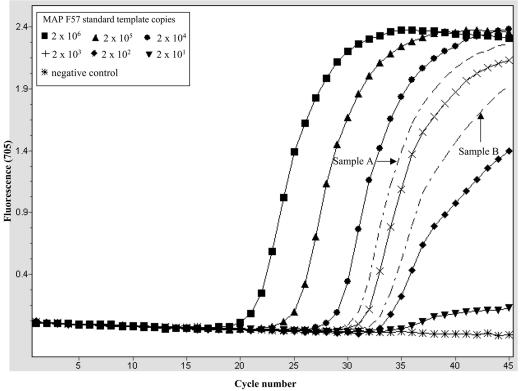
The smallest amount of M. avium subsp. paratuberculosis genomic DNA that could be detected by the LC-PCR assay system was determined next. This was done using purified genomic DNA isolated from a pure culture of an M. avium subsp. paratuberculosis reference strain (ATCC 6783). The LC-PCR assay was applied to decreasing amounts of the M. avium subsp. paratuberculosis DNA template (140 fg to 10 fg). A minimum detection limit of 10 fg M. avium subsp. paratuberculosis DNA (equivalent to about two to three genome copies) per reaction was determined. To determine the reproducibility of the assay's performance, several replicates containing the various amounts of M. avium subsp. paratuberculosis DNA were tested. The results of these tests are summarized in Table 3. M. avium subsp. paratuberculosis was detected with high reproducibility (100%) in the presence 35 fg or more of the M. avium subsp. paratuberculosis DNA template. The reproducibility of M. avium subsp. paratuberculosis detection was poor (38%) at lower M. avium subsp. paratuberculosis DNA concentrations (10 fg), which was expected due to random target particle distribution at low concentrations. The assay's ability to estimate the number of M. avium subsp. paratuberculosis genome copies in samples was tested next. Quantification standards based on the M. avium subsp. paratuberculosis F57 sequence and quasi-unknown samples consisting of spectrophotometer-quantified M. avium subsp. paratuberculosis genomic DNA were amplified (see Materials and Methods). This was followed by quantitative estimation of the original target copy numbers in the unknown samples using the F57 sequence-based standard curves generated by the light cycler software program. An example of a quantitative detection run is shown in Fig. 4, in which target copy numbers in 15 pg and 1.5 pg of M. avium subsp. paratuberculosis DNA were determined. These quantitative runs were performed on four separate occasions, and each sample was analyzed in triplicate. A summary of the results of these experiments is shown in Table 4. A comparison of the calculated numbers of M. avium subsp. paratuberculosis genome copies in the samples tested and the actual LC-PCR estimates is shown in Table 4. The means and standard deviations of the quantitative estimates and sample crossing points obtained in the LC-PCR assay indicate that there was low experiment-to-experiment variation (Table 4). Based on these results, this approach is more accurate for quantitatively estimating high M. avium subsp. paratuberculosis target copy numbers (i.e., the numbers in 15 pg of M. avium subsp. paratuberculosis DNA) than for quantitatively estimating low target copy numbers. In the latter case the assay seemed to overestimate the number of target copy numbers in 1.5 pg of M. avium subsp. paratuberculosis DNA compared to the number theoretically expected. This may have been a result of variation in the overall PCR system performance at low and high target molecule concentrations or the result of the influence of other components present in the sample. For example, the M. avium subsp. paratuberculosis genomic DNA templates contained the F57 target region in addition to the rest of the M. avium subsp. paratuberculosis genome DNA and other copurifying bacterial components. Our assumption is that the crowding effects of other sample components in the reaction mixture may have an overall stimulatory effect on the LC-PCR assay. This could lead to earlier crossing points in the samples compared to the crossing points for similar amounts of the PCR-generated quantification standard used and hence overestimation of the actual copy number. Similarly, the opposite effect would be expected in the presence of an inhibitor. Such effects may be more pronounced at low copy numbers because the target is a limiting factor in the reaction under these conditions compared to when large amounts of target are present in the reaction mixture.
TABLE 3.
Evaluation of analytical sensitivity and reproducibility using purified M. avium subsp. paratuberculosis genomic DNA templatesa
| Amt of DNA (fg) | No. of replicates | No. positive | No. negative | % Positive |
|---|---|---|---|---|
| 140 | 10 | 10 | 0 | 100 |
| 70 | 10 | 10 | 0 | 100 |
| 35 | 10 | 10 | 0 | 100 |
| 10 | 13 | 5 | 8 | 38 |
Replicates containing different amounts (140 fg to 10 fg) of M. avium subsp. paratuberculosis genomic DNA isolated from a pure culture of the M. avium subsp. paratuberculosis reference strain (ATCC 6783) were subjected to LC-PCR amplification as outlined in Materials and Methods. The overall positive and negative results were recorded, and the number of positive hits was expressed as a percentage.
FIG. 4.
Quantitative estimation of M. avium subsp. paratuberculosis target copy numbers in defined amounts of purified M. avium subsp. paratuberculosis genomic DNA using the LC-PCR assay. F57 sequence-based quantification standards (2 × 101 to 2 × 106 copies) and quasi-unknown samples A and B containing 15 pg and 1.5 pg of M. avium subsp. paratuberculosis DNA, respectively, were amplified. Based on theoretical calculations, samples A and B contained about 3,000 and 300 M. avium subsp. paratuberculosis genome copies, respectively. The target copy numbers in these samples were estimated from the standard curve generated from amplification of the F57 standards. The results obtained from these quantitative assays are summarized in Table 4. MAP, M. avium subsp. paratuberculosis.
TABLE 4.
Quantitative estimation of target copy numbers in defined amounts of M. avium subsp. paratuberculosis genomic DNA samples using the LC-PCR assaya
| Run | Calculated target copy no. expected in 15 pg of genomic DNA template | LC-PCR quantitative estimate of target copy no. in 15 pg of genomic DNA template | Crossing point | Calculated target copy no. expected in 1.5 pg of genomic DNA template | LC-PCR quantitative estimate of target copy no. in 1.5 pg of genomic DNA template | Crossing point |
|---|---|---|---|---|---|---|
| 1 | 3,000 | 4,000 | 29.19 | 300 | 792 | 31.37 |
| 2 | 3,000 | 4,330 | 29.08 | 300 | 669 | 31.60 |
| 3 | 3,000 | 2,900 | 29.34 | 300 | 552 | 31.55 |
| 4 | 3,000 | 3,210 | 29.18 | 300 | 550 | 31.56 |
| Mean | 3,610 | 29.12 | 648 | 31.52 | ||
| SD | 666 | 0.11 | 108 | 0.10 |
Triplicate samples containing 15 pg and 1.5 pg purified M. avium subsp. paratuberculosis genomic DNA template and F57 standard dilutions were amplified by the LC-PCR assay, followed by quantitative determination of target copy numbers as shown in Fig. 4. The quantitative analysis was repeated on four separate occasions, and each time the means of target copy number estimates and sample crossing points were recorded. Theoretical estimates of the target copy numbers expected in the samples analyzed are also shown. To show the variation in the assay's performance and estimates on different occassions, overall means and standard deviations for the repeated assays are also shown.
Optimization of extraction of M. avium subsp. paratuberculosis DNA from milk samples.
To maximize the sensitivity of detection of M. avium subsp. paratuberculosis by LC-PCR, we tested different commercial DNA template preparation protocols. Total genomic DNA templates were prepared from triplicate samples of milk that had been spiked with 106 and 102 M. avium subsp. paratuberculosis cells per ml (see Materials and Methods). To begin with, 10-ml samples of M. avium subsp. paratuberculosis-spiked raw milk were similarly processed by centrifugation until the pellet stage. After this, genomic DNA templates were prepared from the milk pellets using the different template isolation protocols for the different kits. The DNA yield and purity (based on the ratio of optical density at 260 nm to optical density at 280 nm) were determined for each template preparation protocol. This was followed by analysis of the templates for the presence of the M. avium subsp. paratuberculosis F57 sequence by the LC-PCR assay. A summary of this comparative evaluation of template preparation protocols is shown in Table 5. The most efficient DNA isolation protocols were determined based on DNA template quality and detection sensitivity based on sample crossing point means from an LC-PCR analysis of samples spiked with the same M. avium subsp. paratuberculosis concentrations. The best performances were obtained with the DNA precipitation protocols (First-tissue DNA kit and Genomic DNA purification kit) and a combination of extensive lysis treatment (ribolysis and proteinase K treatment) with nucleic acid binding column purification of the template using the High Pure PCR template preparation kit (Roche Molecular Diagnostics). The latter procedure was selected for use in combination with the LC-PCR assay for direct detection of M. avium subsp. paratuberculosis in milk samples.
TABLE 5.
Comparative evaluation of different template purification kits for performance in isolation of M. avium subsp. paratuberculosis DNA template from M. avium subsp. paratuberculosis-spiked milk samples<-kn;-0.5q>a
| Template preparation protocol | M. avium subsp. paratuberculosis concn (cells/ml) | Crossing point (mean) |
|---|---|---|
| DNA precipitation I | 106 | 17.40 |
| (First-DNA kit) | 102 | 27.35 |
| DNA precipitation II | 106 | 18.84 |
| (Genomic DNA purification kit) | 102 | NTb |
| DNA binding column I | 106 | 16.50 |
| (High Pure template preparation kit) | 102 | 27.47 |
| DNA binding column II | 106 | 24.77 |
| (Gentra column) | 102 | NT |
| DNA binding beads I | 106 | 21.60 |
| (Bugs’n Beads kit) | 102 | Negc |
| DNA binding beads II | 106 | 19.35 |
| (ChlamCAP kit) | 102 | Neg |
Total genomic DNA templates were prepared by starting with 10-ml raw milk samples spiked with 106, 102, and 0 M. avium subsp. paratuberculosis cells per ml using different template preparation kits. This was followed by LC-PCR analysis of triplicate samples of 5-μl aliquots of the template isolated using each kit. The mean sample crossing points of the two M. avium subsp. paratuberculosis concentrations tested for each DNA isolation procedure are shown.
NT, not tested.
Neg, negative LC-PCR result.
Evaluation of the sensitivity of the LC-PCR assay with raw milk.
Next, we tested the detection limit of an optimized DNA isolation regimen followed by LC-PCR assay detection using M. avium subsp. paratuberculosis-spiked raw milk samples. Table 6 shows the results of an analysis of 10 replicates each of 10-ml raw milk samples spiked with approximately 104, 103, 102, 101, and 100 M. avium subsp. paratuberculosis cells per ml. The overall numbers of positive and negative results from this evaluation were recorded, and the percentage of positive results was determined. The percentage of positive results for samples spiked with 102 M. avium subsp. paratuberculosis cells or more was 100%. For samples containing about 10 M. avium subsp. paratuberculosis cells per ml, 7 of 10 samples (70%) were positive, and no M. avium subsp. paratuberculosis was detected in the samples spiked with less than 10 cells per ml. Therefore, the detection routine which we developed has a detection limit in the range from 10 to 100 M. avium subsp. paratuberculosis cells per ml of raw milk, if a 10-ml starting sample is processed.
TABLE 6.
Evaluation of LC-PCR sensitivity with M. avium subsp. paratuberculosis-spiked raw milk samplesa
| M. avium subsp. paratuberculosis concn (cells/ml) | No. of replicates | No. positive | No. negative | % Positive |
|---|---|---|---|---|
| 1,000 | 10 | 10 | 0 | 100 |
| 100 | 10 | 10 | 0 | 100 |
| 10 | 10 | 7 | 3 | 70 |
| 1 | 10 | 0 | 10 | 0 |
Genomic DNA templates were prepared by starting with 10-ml raw milk samples spiked with 1,000, 100, 10, 1, and 0 M. avium subsp. paratuberculosis cells per ml, using the optimized M. avium subsp. paratuberculosis DNA template isolation procedure. This was followed by analysis of 5-μl template aliquots in 20-μl LC-PCR assays as described in Materials and Methods. Template isolation and LC-PCR analysis were repeated on 10 seperate occasions, and the M. avium subsp. paratuberculosis detection results were recorded. The overall total number of positive hits from these experiments was expressed as a percentage.
Detection of M. avium subsp. paratuberculosis in naturally contaminated milk samples by LC-PCR assay.
Finally, the optimized M. avium subsp. paratuberculosis detection regimen was evaluated to determine its ability to detect M. avium subsp. paratuberculosis in naturally contaminated milk. First, the method was applied to raw milk samples collected from two clinical cases involving symptomatic M. avium subsp. paratuberculosis-infected cows. These cows were confirmed by fecal culture and microscopy to be infected with M. avium subsp. paratuberculosis (data not shown). In both cases, the raw milk samples were found to be M. avium subsp. paratuberculosis positive by LC-PCR. Next, 80 milk samples were collected from a 100-cow dairy herd with a confirmed history of paratuberculosis. In this herd there had also recently been two confirmed cases of symptomatic M. avium subsp. paratuberculosis infection, and some of the cows included in the sample were paratuberculosis seropositive based on a commercial enzyme-linked immunosorbent assay kit test with serum samples. Sixteen pooled samples were prepared from the 80 raw milk samples; each 10-ml sample was made up of 2-ml samples from five different cows. These samples were subjected to the M. avium subsp. paratuberculosis detection procedure described above, and 2 of the 16 pooled samples were found to be positive for M. avium subsp. paratuberculosis (data not shown). One of the PCR-positive pooled samples also contained milk from cows confirmed to be serologically paratuberculosis positive by the enzyme-linked immunosorbent assay with serum samples. Thus, with this analysis we confirmed that the M. avium subsp. paratuberculosis detection protocol can successfully detect M. avium subsp. paratuberculosis in naturally contaminated milk.
DISCUSSION
M. avium subsp. paratuberculosis remains a significant problem for the dairy industry around the world. Infection with M. avium subsp. paratuberculosis leads to chronic disease in the affected cattle, as well as significant losses in productivity. The affected animals shed M. avium subsp. paratuberculosis primarily in their feces, as well as directly in milk. The suggestion that M. avium subsp. paratuberculosis may be linked to human Crohn's disease has focused a spotlight on milk products as potential vehicles for transmission of this animal pathogen to humans. Sensitive and specific PCR assays for detection of M. avium subsp. paratuberculosis could contribute immensely to research efforts aimed at understanding the pathogenicity of this organism in ruminant paratuberculosis, its potential role in human Crohn's disease, and its transmission via foods of animal origin. Several M. avium subsp. paratuberculosis detection PCR systems are now widely available; however, several practical limitations remain. In the current study, we sought to address current detection specificity as well as sensitivity problems associated with PCR detection of M. avium subsp. paratuberculosis pertaining to milk. In our lab, as well as in other labs, confirmation of the presence of M. avium subsp. paratuberculosis isolates or surveillance work with milk has been based mainly on IS900 PCR systems (8, 19, 28, 34). However, evidence showing that IS900-positive genetic elements that are highly homologous to M. avium subsp. paratuberculosis IS900 exist in other non-M. avium subsp. paratuberculosis environmental Mycobacterium species has resulted in doubts concerning the current M. avium subsp. paratuberculosis detection systems (9, 14). We therefore sought to develop a new light cycler-based real-time PCR assay for detecting M. avium subsp. paratuberculosis. The assay developed targets the F57 sequence of M. avium subsp. paratuberculosis, a genetic element that is currently known to exist only in M. avium subsp. paratuberculosis and that so far has been found to be highly specific for this organism (14, 6, 35, 40, 41). In order to use this M. avium subsp. paratuberculosis PCR detection system as a routine diagnostic tool for milk, an IC template system was incorporated into the assay. This enables amplification conditions and PCR inhibition to be monitored in each sample tested. Our evaluation of this assay confirmed that F57 targeting is highly specific for M. avium subsp. paratuberculosis. No false-positive signals were observed with several non-M. avium subsp. paratuberculosis bacterial species tested. DNA templates derived from milk or other food matrices usually contain very low levels of the contaminating pathogen DNA along with high background levels of nonspecific DNA. Therefore, the influence of the sample matrix on the performance of the new LC-PCR system was evaluated, and the assay's performance was not significantly influenced by the presence of large amounts of nonspecific background DNA in the reaction mixture.
To benefit from the rapid specific detection of M. avium subsp. paratuberculosis by the LC-PCR assay, it was essential that the assay's detection sensitivity be maximized through optimal preparation of M. avium subsp. paratuberculosis DNA templates from milk samples. Generally, milk is a difficult matrix from which to isolate M. avium subsp. paratuberculosis DNA, due to its complex matrix that contains a lot of other substances that may inhibit subsequent PCR detection (43). These components need to be efficiently removed during the template isolation step to minimize their influence on the LC-PCR assay. Moreover, the M. avium subsp. paratuberculosis cells themselves are also structurally complex and must be efficiently lysed to release as much of the target DNA molecules as possible. We determined the most efficient regimen for isolation of the M. avium subsp. paratuberculosis DNA template from milk samples. To begin with, we found that in templates prepared by traditional procedures (i.e., mechanical lysis followed by phenol-chloroform extraction, and ethanol precipitation) there were high levels of residual PCR inhibition (data not shown). As a result of a comparative evaluation of several commercial DNA isolation techniques with M. avium subsp. paratuberculosis-spiked raw milk samples, we chose a procedure that was rapid, yielded high-quality M. avium subsp. paratuberculosis DNA templates, and was less prone to cross contamination. This protocol was a modified protocol that combined mechanical sample lysis in a Ribolyser and subsequent template purification with nucleic acid binding spin columns using the High Pure PCR template preparation kit protocol of Roche Diagnostics.
By integrating the optimized M. avium subsp. paratuberculosis template preparation procedure and the LC-PCR detection system, we evaluated the applicability of this approach to actual milk samples. We applied these protocols to 10-ml raw milk samples that had been spiked with defined concentrations of M. avium subsp. paratuberculosis. The highly reproducible M. avium subsp. paratuberculosis detection limits achieved with these studies were between 10 and 100 M. avium subsp. paratuberculosis cells per ml of milk. The analytical sensitivity obtained here also falls within the range that was reported in studies in which M. avium subsp. paratuberculosis IS900 elements were targeted (18, 26, 32, 34). In addition, the targeting of ISMav2 as an alternative marker for M. avium subsp. paratuberculosis in one study also gave a detection limit of 10 cells per ml of milk (37). We found that targeting of the F57 sequence, a single-copy element, can also enable highly sensitive detection of M. avium subsp. paratuberculosis in milk. However, it is important to emphasize that the M. avium subsp. paratuberculosis concentrations reported to date are only approximations. It is not possible with currently used techniques to determine definite concentrations of M. avium subsp. paratuberculosis in milk samples. The detection limits obtained in this study should be sufficient for detection of M. avium subsp. paratuberculosis in heavily to mildly contaminated milk samples. Indeed, this was demonstrated here using naturally contaminated raw milk samples. First, milk samples were obtained from two clinical cases involving M. avium subsp. paratuberculosis-infected symptomatic cows, and these samples were found to be positive for M. avium subsp. paratuberculosis by LC-PCR analysis. In pooled raw milk samples from a herd with a confirmed history of paratuberculosis, we found that 2 of 16 samples were positive for M. avium subsp. paratuberculosis as determined by the M. avium subsp. paratuberculosis detection procedure developed.
In summary, a new real-time PCR assay for detection of M. avium subsp. paratuberculosis that targets the F57 sequence and includes an IC template was developed and evaluated. The new assay is an ideal diagnostic PCR tool for routine large-scale analysis of milk samples. We hope that the procedures which we developed will contribute to the routine detection of M. avium subsp. paratuberculosis-contaminated milk products as one way of minimizing public exposure while the debate on the public health significance of this organism is being resolved.
Acknowledgments
We thank S. Englund (Department of Bacteriology, National Veterinary Institute, Sweden) for providing Mycobacterium strain 2333 DNA, as well as F. Baggi (Swiss National Center for Mycobacteria, Department of Medical Microbiology, University of Zurich) and J. Hermon-Taylor and K. Sidi-Boumedine (Department of Surgery, St. George's Medical School, United Kingdom) for providing various mycobacterial strains or genomic DNA templates used in this work.
REFERENCES
- 1.Anonymous. 2000. Possible links between Crohn's disease and paratuberculosis. European Commission draft report SANCO/B3/R16/2000.1-76. Directorate, General Health and Consumer protection, Brussels, Belgium. [Online.] http://archive.food.gov.uk/pdf_files/papers/acm493.pdf.
- 2.Ayele, W. Y., P. Svastova, P. Roubal, M. Bartos, and I. Pavlik. 2005. Mycobacterium avium subsp. paratuberculosis cultured from locally and commercially pasteurized cow's milk in the Czech Republic. Appl. Environ. Microbiol. 71:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull. T. J., J. Hermon-Taylor, I. Pavlik, F. El-Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146:3285. [DOI] [PubMed] [Google Scholar]
- 4.Chacon, O., L. E. Bermudez, and R. G. Barletta. 2004. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu. Rev. Microbiol. 58:329-363. [DOI] [PubMed] [Google Scholar]
- 5.Cocito, C., P. Gilot, M. Coene, M. De Kesel, P. Poupart, and P. Vannuffel. 1994. Paratuberculosis. Clin. Microbiol. Rev. 7:328-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coetsier, C., P. Vannuffel, N. Blondeel, J. F. Denef, C. Cocito, and J. L. Gala. 2000. Duplex PCR for differential identification of Mycobacterium bovis, M. avium, and M. avium subsp. paratuberculosis in formalin-fixed paraffin-embedded tissues from cattle. J. Clin. Microbiol. 38:3048-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, M. T. 2003. Paratuberculosis: review of present knowledge. Acta Vet. Scand. 44:217-221. [PubMed] [Google Scholar]
- 8.Corti, S., and R. Stephan. 2002. Detection of Mycobacterium avium subspecies paratuberculosis specific IS900 insertion sequences in bulk-tank milk samples obtained from different regions throughout Switzerland. BMC Microbiol. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 10.Donaghy, J. A., N. L Totton, and M. T. Rowe. 2004. Persistence of Mycobacterium paratuberculosis during manufacture and ripening of cheddar cheese. Appl. Environ. Microbiol. 70:4899-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dundee, L., I. R. Grant, H. J. Ball, and M. T. Rowe. 2001. Comparative evaluation of four decontamination protocols for the isolation of Mycobacterium avium subsp. paratuberculosis from milk. Lett. Appl. Microbiol. 33:173-177. [DOI] [PubMed] [Google Scholar]
- 12.Ellingson, J. L., C. A. Bolin, and J. R. Stabel. 1998. Identification of a gene unique to Mycobacterium avium subspecies paratuberculosis and application to diagnosis of paratuberculosis. Mol. Cell. Probes 12:133-142. [DOI] [PubMed] [Google Scholar]
- 13.Ellingson, J. L., J. R. Stabel, W. R. Bishai, R. Frothingham, and J. M. Miller. 2000. Evaluation of the accuracy and reproducibility of a practical PCR panel assay for rapid detection and differentiation of Mycobacterium avium subspecies. Mol. Cell. Probes 14:153-161. [DOI] [PubMed] [Google Scholar]
- 14.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 15.Fang, Y., W. H. Wu, J. L. Pepper, J. L. Larsen, S. A. Marras, E. A. Nelson, W. B. Epperson, and J. Christopher-Hennings. 2002. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine fecal samples. J. Clin. Microbiol. 40:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, A., L. Mutharia, S. Chen, K. Rahn, and J. Odumeru. 2002. Effect of pasteurization on survival of Mycobacterium paratuberculosis in milk. J. Dairy Sci. 85:3198-3205. [DOI] [PubMed] [Google Scholar]
- 17.Giese, S. B., and P. Ahrens. 2000. Detection of Mycobacterium avium subsp. paratuberculosis in milk from clinically affected cows by PCR and culture. Vet. Microbiol. 77:291-297. [DOI] [PubMed] [Google Scholar]
- 18.Grant, I. R., C. M. Pope, L. M. O'Riordan, H. J. Ball, and M. T. Rowe. 2000. Improved detection of Mycobacterium avium subsp. paratuberculosis in milk by immunomagnetic PCR. Vet. Microbiol. 77:369-378. [DOI] [PubMed] [Google Scholar]
- 19.Grant, I. R., H. J. Ball, and M. T. Rowe. 2002. Incidence of Mycobacterium paratuberculosis in bulk raw and commercially pasteurized cows' milk from approved dairy processing establishments in the United Kingdom. Appl. Environ. Microbiol. 68:2428-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant, I. R., E. I. Hitchings, A. McCartney, F. Ferguson, and M. T. Rowe. 2002. Effect of commercial-scale high-temperature, short-time pasteurization on the viability of Mycobacterium paratuberculosis in naturally infected cows' milk. Appl. Environ. Microbiol. 68:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant, I. R. 2003. Mycobacterium paratuberculosis and milk. Acta Vet. Scand. 44:261-266. [PubMed] [Google Scholar]
- 22.Grant, I. R., and M. T. Rowe. 2004. Effect of chemical decontamination and refrigerated storage on the isolation of Mycobacterium avium subsp. paratuberculosis from heat-treated milk. Lett. Appl. Microbiol. 38:283-288. [DOI] [PubMed] [Google Scholar]
- 23.Greenstein, R. J., and M. T. Collins. 2004. Emerging pathogens: is Mycobacterium avium subspecies paratuberculosis zoonotic? Lancet 364:396-397. [DOI] [PubMed] [Google Scholar]
- 24.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermon-Taylor, J., T. J. Bull, J. M. Sheridan, J. Cheng, M. L. Stellakis, and N. Sumar. 2000. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can. J. Gastroenterol. 14:521-539. [DOI] [PubMed] [Google Scholar]
- 26.Khare, S., T. A. Ficht, R. L. Santos, J. Romano, A. R. Ficht, S. Zhang, I. R. Grant, M. Libal, D. Hunter, and L. G. Adams. 2004. Rapid and sensitive detection of Mycobacterium avium subsp. paratuberculosis in bovine milk and feces by a combination of immunomagnetic bead separation-conventional PCR and real-time PCR. J. Clin. Microbiol. 42:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, S. G., S. J. Shin, R. H. Jacobson, L. J. Miller, P. R. Harpending, S. M. Stehman, C. A. Rossiter, and D. A. Lein. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Investig. 14:126-131. [DOI] [PubMed] [Google Scholar]
- 28.Millar, D., J. Ford, J. Sanderson, S. Withey, M. Tizard, T. Doran, and J. Hermon-Taylor. 1996. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows' milk in England and Wales. Appl. Environ. Microbiol. 62:3446-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nauta, M. J., and J. W. van der Giessen. 1998. Human exposure to Mycobacterium paratuberculosis via pasteurised milk: a modelling approach. Vet. Rec. 143:293-296. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen, S. S., S. M. Thamsborg, H. Houe, and V. Bitsch. 2000. Bulk-tank milk ELISA antibodies for estimating the prevalence of paratuberculosis in Danish dairy herds. Prev. Vet. Med. 44:1-7. [DOI] [PubMed] [Google Scholar]
- 31.O'Mahony, J., and C. Hill. 2002. A real time PCR assay for the detection and quantitation of Mycobacterium avium subsp. paratuberculosis using SYBR Green and the Light Cycler. J. Microbiol. Methods 51:283-293. [DOI] [PubMed] [Google Scholar]
- 32.O'Mahony, J., and C. Hill. 2004. Rapid real-time PCR assay for detection and quantitation of Mycobacterium avium subsp. paratuberculosis DNA in artificially contaminated milk. Appl. Environ. Microbiol. 70:4561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Reilly, C. E., L. O'Connor, W. Anderson, P. Harvey, I. R. Grant, J. Donaghy, M. Rowe, and P. O'Mahony. 2004. Surveillance of bulk raw and commercially pasteurized cows' milk from approved Irish liquid-milk pasteurization plants to determine the incidence of Mycobacterium paratuberculosis. Appl. Environ. Microbiol. 70:5138-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pillai, S. R., and B. M. Jayarao. 2002. Application of IS900 PCR for detection of Mycobacterium avium subsp. paratuberculosis directly from raw milk. J. Dairy Sci. 85:1052-1057. [DOI] [PubMed] [Google Scholar]
- 35.Poupart, P., M. Coene, H. Van Heuverswyn, and C. Cocito. 1993. Preparation of a specific RNA probe for detection of Mycobacterium paratuberculosis and diagnosis of Johne's disease. J. Clin. Microbiol. 31:1601-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Lazaro, D., J. Lloyd, A. Herrewegh, J. Ikonomopoulos, M. D'Agostino, M. Pla, and N. Cook. 2004. A molecular beacon-based real-time NASBA assay for detection of Mycobacterium avium subsp. paratuberculosis in water and milk. FEMS Microbiol. Lett. 237:119-126. [DOI] [PubMed] [Google Scholar]
- 37.Stratmann, J., B. Strommenger, K. Stevenson, and G. F. Gerlach. 2002. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J. Clin. Microbiol. 40:4244-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strommenger, B., K. Stevenson, and G. F. Gerlach. 2001. Isolation and diagnostic potential of ISMav2, a novel insertion sequence-like element from Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 196:31-37. [DOI] [PubMed] [Google Scholar]
- 39.Sweeney R. W., R. H. Whitlock, and A. E. Rosenberger. 1992. Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J. Clin. Microbiol. 30:166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tasara, T., L. E. Hoelzle, and R. Stephan. 24June2005. Development and evaluation of a Mycobacterium avium subspecies paratuberculosis (MAP) specific multiplex PCR assay. Int. J. Food Microbiol. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15982769&query_hl=1. [DOI] [PubMed]
- 41.Vansnick, E., P. De Rijk, F. Vercammen, D. Geysen, L. Rigouts, and F. Portaels. 2004. Newly developed primers for the detection of Mycobacterium avium subspecies paratuberculosis. Vet. Microbiol. 100:197-204. [DOI] [PubMed] [Google Scholar]
- 42.Vary, P. H., P. R. Anderson, E. Green, and J. Hermon-Taylor. 1990. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J. Clin. Microbiol. 28:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wittenbrink, M. M., D. Thiele, and H. Krauss. 1994. Comparison of dark-field microscopy, culture, and polymerase chain reaction (PCR) for detection of Borrelia burgdorferi in field-collected Ixodes ricinus ticks. Zentrbl. Bakteriol. 281:183-191. [DOI] [PubMed] [Google Scholar]