Abstract
Our understanding of pathways leading to antitumor immunity may depend on an undistorted knowledge of the primary antigenic targets of patients' autologous T cell responses. In the melanoma model derived from patient DT, we applied cryopreserved short-term autologous mixed lymphocyte–tumor cell cultures (MLTCs) in combination with an IFN-γ enzyme-linked immunospot (ELISPOT) assay to cDNA expression screening. We identified three previously unknown peptides processed from melanosomal proteins tyrosinase (presented by HLA-A*2601 and -B*3801) and gp100 (presented by HLA-B*07021) and five neoantigens generated by somatic point mutations in the patient's melanoma. The mutations were found in the genes SIRT2, GPNMB, SNRP116, SNRPD1, and RBAF600. Peptides containing the mutated residues were presented by HLA-A*03011, -B*07021, and -B*3801. Mutation-induced functional impairment was so far demonstrated for SIRT2. Within MLTC responder populations that were independently expanded from the patient's peripheral blood lymphocytes of different years, T cells against mutated epitopes clearly predominated. These results document a high degree of individuality for the cellular antitumor response and support the need for individualizing the monitoring and therapeutic approaches to the primary targets of the autologous T cell response, which may finally lead to a more effective cancer immunotherapy.
Keywords: expression cloning, peptide, tumor antigen, mixed lymphocyte–tumor cell culture, cytotoxic T lymphocytes
During the last decade, human tumor antigens that were recognized by autologous T lymphocytes were identified and assigned to distinct categories according to their expression profile and to the presence or absence of structural alterations (1, 2). The knowledge of these antigens renewed the enthusiasm to develop strategies for specific immunotherapy. Structurally unaltered, common antigens are favored targets for immune intervention, because they are not restricted to individual patients and are expressed on different tumor types. However, depending on their tissue distribution, common antigens might be subjected to complex tolerance mechanisms efficiently counteracting autoaggression at the expense of antitumor responses. In systematic animal studies, rejection responses and protective immunity were primarily induced by unique tumor antigens (3), at least some of which were generated by somatic mutations in normal gene products during the oncogenic process (4). Analogous, individually distinct mutated antigens were until now identified only in few patients, perhaps because of methodological bias (5). As a consequence, efforts to use and understand antitumor T cell reactivity were so far largely limited to the interaction with common tumor antigens and mostly ignored the individuality of tumor–host interaction.
Current T cell-based techniques for the identification of new antigens rely on the availability of clonal T cells. However, even T cell clones with the same specificity differ considerably in their expansion potential (6, 7), which is possibly due to their state of maturation. Therefore, it is likely that the spectrum of tumor antigens detected with highly selected clonal T cells does not adequately reflect the situation in vivo.
We modified the cDNA expression cloning of tumor antigens by applying cryopreserved, multispecific mixed lymphocyte–tumor cell cultures (MLTCs) in addition to clonal T cells. In the example of a long-term surviving melanoma patient with metastatic disease, we identified eight antigens, three of which were previously unknown peptide antigens processed from structurally unaltered melanosomal proteins, and five of which were neoantigens generated by somatic mutations in tumor cells. T cells reactive with mutated peptides clearly predominated in independent MLTCs.
Materials and Methods
Supporting Information. For further details, see Figs. 7–10 and Tables 2–5, which are published as supporting information on the PNAS web site.
Patient and Cell Lines. In 1985, 23-year-old patient DT was found to have a malignant melanoma. From 1987 on, skin, lymphatic, and visceral spread including ovarian, peritoneal, and spleen metastases became apparent, and she finally died from progressive disease in 1992 (Table 2, which is published as supporting information on the PNAS web site). Peripheral blood mononuclear cells (PBMCs) were collected in April 1988, March 1989, April 1989, September 1989, March 1990, and January 1992 and cryopreserved in liquid nitrogen. The autologous melanoma line MZ7-MEL was established from a splenic metastasis in January 1988. Melanoma cells, autologous Epstein–Barr virus-transformed B cells (MZ7-EBV-B), and COS-7 cells were maintained in RPMI medium 1640 supplemented with 10% FCS, 1% l-glutamine and 100 units/ml penicillin-streptomycin (GIBCO/BRL, Karlsruhe, Germany).
MLTC and T Cell Clones. Autologous MLTC were generated by coculturing on 24-well plates 1.5 × 106 PBMC and 105 irradiated (100 Gy) MZ7-MEL cells per well in 2 ml of AIM-V (GIBCO/BRL) supplemented with 10% human serum (AIM-V/HS) and recombinant human IL-2 (from day 3 on, 250 units/ml, Chiron-Behring, Marburg, Germany). When indicated, tumor cells were pretreated with IFN-γ (100 units/ml; Boehringer-Mannheim) for 3 days (Fig. 1). MLTC were stimulated once per week until day 21 with irradiated melanoma cells and IL-2 before CD8+ T lymphocytes were isolated by using the CD8+ T cell Isolation kit (Miltenyi Biotech). CD8+ MLTC were continued by weekly stimulating 106 T cells per well with 105 irradiated melanoma cells and 2 × 105 irradiated CD8- autologous PBMC. CD8+ MLTC responders were regularly frozen in aliquots at different time points 3–4 days after a final stimulation with irradiated tumor cells. Some MLTCs were cloned by limiting dilution, and T cell clones were propagated as described (8). Notably, all functional assays in this study were carried out with CD8+ MLTC or CD8+ T cell clones (cytotoxic T lymphocytes, CTLs) stimulated with autologous tumor cells.
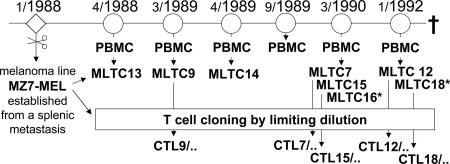
Fig. 1.
Origin of MLTC and CTL clones. MLTC were performed by stimulating PBMC taken from patient DT at indicated time points (month/year) with autologous tumor cells (MZ7-MEL) as detailed in Materials and Methods. CTL clones were derived by limiting dilution from some of the MLTC (designation x/y for CTL clones: x was the serial number of the MLTC, y was the serial number of the T cell clone). *, Stimulator cells were pretreated with IFN-γ.
T Cell Receptor (TCR) Vβ Chain Analysis. The TCR Vβ usage of CTL clones was analyzed by using the IOTest Beta Mark TCR Vβ Repertoire kit (Beckman Coulter) or by RT-PCR with TCR Vβ chain-specific primers (9).
Plasmids. cDNAs encoding HLA-A*03011, HLA-A*2601, HLA-B*07021, HLA-B*3801, and HLA-Cw*1203 were cloned by RT-PCR from melanoma cells into pcDNAI/Amp or pcDNA3 (Invitrogen) as described (10). Plasmids MAGE-A3/pcDSRα and gp100/pB7-Neo were kindly provided by P. van der Bruggen (Ludwig Institute for Cancer Research, Brussels, Belgium). Plasmids 123.B2 and Aa1.2 encoded full-length tyrosinase and Melan-A/MART-1, respectively (ref. 11 and T.W., unpublished data).
Construction and Screening of a cDNA Library. A cDNA library was constructed from MZ7-MEL cells in pcDNAI/Amp (Invitrogen) and divided into pools of ≈100 cDNAs (11). For the identification of DT melanoma-specific antigens, cDNA library screening was performed with cyropreserved MLTC or clonal T cells in IFN-γ ELISPOT assays. COS-7 cells (20,000 per well) were cotransfected on MultiScreen ELISPOT HP plates (Millipore, Eschborn, Germany) with pooled cDNAs (≈100–200 ng per well) and appropriate HLA class I cDNAs (100 ng per well) using SuperFect or PolyFect transfection reagents (Qiagen, Hilden, Germany). After 20–24 h, COS-7 transfectants were assayed for recognition by T cells with IFN-γ ELISPOT assays.
IFN-γ ELISPOT Assays. IFN-γ ELISPOT assays were performed and evaluated as described (12). Target cells for T cell recognition were melanoma cells (30,000 per well), COS-7 transfectants (20,000 per well), or peptide-loaded EBV-B cells (75,000 per well) in 50 μl of AIM-V/HS. CD8+ T cells (MLTC responders at 5,000–10,000 per well, CTL clones at 1,000–5,000 per well) were added in 50 μl of AIM-V/HS containing 500 units/ml IL-2. T cells from continuous cultures were tested 4–5 days after stimulation with tumor cells. Frozen MLTC or T cell clones were thawed and kept for 2 days in AIM-V/HS containing IL-2 (250 units/ml) before testing. Assays were incubated for 16–24 h at 37°C in 5% CO2 and were then developed.
For frequency analyses on ex vivo lymphocytes, mature dendritic cells (mDCs) were generated from PBMC of patient DT as described (13). mDCs were loaded with synthetic peptides and tested in 48-h IFN-γ ELISPOT assays for recognition by CD8+ T cells isolated from the patient's cryopreserved PBMC.
Recognition of cDNA Fragments and Synthetic Peptides by T Cells. Fragments of antigen-coding cDNAs were amplified by PCR and cloned into pcDNA3.1/V5-His TOPO (Invitrogen). COS-7 cells were transiently cotransfected with cDNA fragments and HLA cDNAs and tested for recognition by T cells. Peptides were predicted from positive fragments with the help of public databases (www.uni-tuebingen.de/uni/kxi and http://bimas.dcrt.nih.gov/molbio/hla_bind). Peptides (synthesized by J. W. Drijfhout, University of Leiden, Leiden, The Netherlands) were solubilized in PBS/5% DMSO and stored at -20°C; they were tested for recognition by T cells with IFN-γ ELISPOT assays (see above) or with 51Cr release assays performed as described (12).
Peptide/HLA Tetramer Staining. Ex vivo PBMC were in parallel analyzed in IFN-γ ELISPOT assays and stained with peptide/MHC tetramers. Tetramers were purchased from Proimmune Limited (Oxford) and applied according to the manufacturer's recommendations. PBMCs (4 × 106) were incubated with PE-labeled tetramers for 45 min on ice in 100 μl of buffer (PBS/1% BSA, pH 7.4), then anti-CD8-FITC (Caltag) was added. After 15 min, the cells were washed three times and resuspended in buffer with propidium iodide (PI) (2 μg/ml). PI-negative cells were gated, and 500,000 events were counted by using a Coulter Epics XL (Beckman Coulter).
Gene Abbreviations and EMBL/GenBank Accession Numbers. Gene abbreviations of reference sequences and mutated cDNA sequences identified in this study, and their EMBL/GenBank accession numbers are given in Tables 1 and 4.
Table 1. Identification of antigens recognized by T cells reactive with DT melanoma cells.
| T cells*
|
Restricting HLA allele
|
Detection frequency‡
|
Nucleic acid exchange
|
Amino acid exchange
|
Peptides§ (amino acid residues)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Antigen | MLTC | CTL clones | Method† | Gene | |||||
| Tyr/A26 | MLTC 18 | III | TYR¶ | HLA-A*2601 | – | – | – | QCSGNFMGF (90–98) | |
| Tyr/B38 | MLTC 16 | III | TYR | HLA-B*3801 | – | – | – | LHHAFVDSIF (388–397) | |
| gp100/B7 | (MLTC 12) | CTL12/45 | II | SILV∥ | HLA-B*07021 | – | – | – | SSPGCQPPA (529–537)** |
| SIRT2mut/A3 | (MLTC 9) | CTL9/89 | I | SIRT2_v3mut‡‡ | HLA-A*03011 | 1/700 | C → T | P199L | KIFSEVTLK (192–200) |
| GPNMBmut/A3 | MLTC 18 | IV | GPNMB_v2mut‡‡ | HLA-A*03011 | 11/1.920 | G → A | G181D | TLDWLLQTPK (179–188) | |
| SNRP116mut/A3 | MLTC 18 | IV | SNRP116mut‡‡ | HLA-A*03011 | 2/1.920 | G → A | E677K | KILDAVVAQK (668–677) | |
| RBAF600mut/B7 | MLTC 16 | IV | RBAF600mut‡‡ | HLA-B*07021 | 7/2.300 | G → A | G329R | RPHVPESAF (329–337) | |
| SNRPD1mut/B38 | (MLTC 15) | CTL15/165 | V | SNRPD1mut‡‡ | HLA-B*3801 | 38/1.920 | C → T | T16I | SHETVIIEL (11–19) |
CD8+ MLTC responder cell populations and CTL clones used for antigen identification (see also Fig. 1)
cDNAs encoding DT melanoma antigens and peptide coding regions were identified in different ways. I, the MZ7-MEL cDNA library was screened with CTL9/89 as published (11); II, MLTC12 and MLTC16 responders were found to contain T cells reactive with COS-7 cells cotransfected with SILV (gp 100) cDNA and HLA-B*07021 cDNA (Figs. 2 and 4), peptide gp 100/B7 was identified with CTL12/45; III, MLTC16 and MLTC18 were found to contain T cells reactive with COS-7 cells cotransfected with TYR (tyrosinase) cDNA and HLA-A*2601 or HLA-B*3801 cDNA (Figs. 2 and 4); the respective peptides were identified with non-clonal MLTC responders (MLTC18 responders frozen between days 46 and 64 for Tyr/A26 and MLTC16 responders frozen on d28 for Tyr/B38); IV, cryopreserved MLTC and the IFN-γ ELISPOT assay as readout were applied for screening of the cDNA library prepared from MZ7-MEL cells, cloning of antigen-coding cDNA, and identification of peptides (MLTC18 responders frozen on d32 for antigens GPNMBmut/A3 and SNRP116mut/A3, MLTC16 responders frozen on d32 for RBAF600mut/B7; example given in Fig. 3); V, the MZ7-MEL cDNA library was screened with the same procedure as in IV, except that effectors were cryopreserved MLTC15-derived clonal T cells (CTL 15/165)
Frequencies (x/y) indicate the number of positive pools (x) per total number of pools screened (y). Each pool contained ≈ 100 cDNA clones of the MZ7-MEL cDNA library and was transfected into COS-7 cells together with the respective HLA I allele
The sequences of peptides that were recognized by T cells at lowest concentrations are indicated (see also Table 3). Mutated residues are printed in bold letters and are underlined
Encodes human tyrosinase (GenBank accession no. NM_000372)
Encodes human silver homolog (mouse) gp 100 (GenBank accession no. NM_006928)
In titration assays, the gp100/B7 peptide induced half-maximal lysis by independently generated CTL clones only at concentrations of 0.1–1.0 μM. Such high peptide requirement together with the need for an intact N-terminal region (see Fig. 8) suggest that peptide 529–537 might be posttranslationally modified
Results
Derivation of DT Melanoma-Reactive T Cells. PBMCs collected from patient DT between 1988 and 1992 were stimulated in autologous MLTC. Measurable lymphocyte expansion regularly started after two to three stimulations (Fig. 7). Expansion depended on the presence of autologous tumor cells, suggesting that it was antigen-driven (not shown). Responder lymphocytes were tested by IFN-γ ELISPOT assays from day 25 on. All MLTC responder cell populations used herein recognized autologous melanoma cells, but not autologous EBV-B cells. Reactivity to tumor cells was confined to CD8+ T cells in all MLTC tested (data not shown). CD8+ T cells were purified from MLTC responders after two to four stimulations, further expanded, and cryopreserved at different time points. Aliquots were subsequently thawed for testing and screening procedures. Clonal tumor-reactive CTL were derived by limiting dilution from some MLTC (Fig. 1).
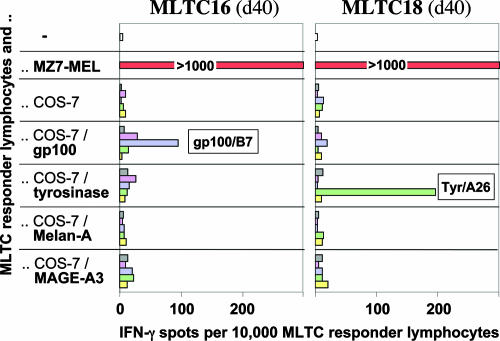
DT Melanoma-Reactive T Cells Recognized Previously Unknown Peptides from Common Nonmutated Melanoma Antigens. According to RT-PCR analyses, the melanoma line MZ7-MEL expressed MAGE-A3, tyrosinase, Melan-A/MART-1, and gp100 (data not shown). COS-7 cells were cotransfected with plasmid pairs encoding one of these four antigens and one of the five HLA class I alleles cloned from MZ7-MEL cells. Transfectants were tested for their ability to induce IFN-γ spot production in independently generated MLTC responder populations. No response was seen toward Melan-A/MART-1 and MAGE-A3 with any of the HLA I alleles. However, some MLTCs contained lymphocytes recognizing tyrosinase in association with HLA-A*2601 (Tyr/A26) or HLA-B*3801 cDNA (Tyr/B38), and gp100 in association with HLA-B*07021 (gp100/B7). As representative examples, data obtained with MLTC16 (d40) and MLTC18 (d40) are shown in Fig. 2. Approximately 1% of the MLTC16 (d40) responders recognized gp100/B7, and 2% of MLTC18 (d40) responders reacted with Tyr/A26. Notably, the reactivity to melanoma cells by far exceeded the reactivity to known common melanoma antigens. The appearance of T cell responses to Tyr/A26, Tyr/B38, and gp100/B7 in independent MLTC is summarized in Fig. 4.
Fig. 2.
Reactivity of MLTC responders to known common antigens. Responder lymphocytes of MLTC16(d40) and MLTC18(d40) (10,000 lymphocytes per well) were tested in IFN-γ ELISPOT assays for recognition of antigens MAGE-A3, tyrosinase, gp100, and Melan-A/MART-1. Data are means of duplicates. Stimulators were melanoma cells (MZ7-MEL, 30,000 per well) and COS-7 cells (20,000 per well) cotransfected with antigen-coding cDNA (indicated) and HLA class I cDNA cloned by RT-PCR from MZ7-MEL cells: yellow, HLA-A*03011; green, HLA-A*2601; purple, HLA-B*07021; pink, HLA-B*3801; gray, HLA-Cw*1203.
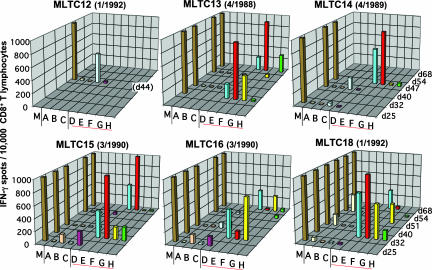
Fig. 4.
Specificity of tumor-reactive T cells enriched in DT MLTC. Independent autologous MLTC were performed with PBMC collected from patient DT during a 4-year period (Fig. 1). CD8+ MLTC responders were isolated and cryopreserved at various time points and later tested in 20-h IFN-γ ELISPOT assays for recognition of autologous melanoma cells (30,000 per well) (M) and COS-7 cells (20,000 per well) transiently transfected with expression plasmids encoding the antigen/HLA combinations tyrosinase/HLA-A*2601 (A), tyrosinase/HLA-B*3801 (B), gp100/HLA-B*07021 (C), SIRT2_v3mut/HLA-A*03011 (D), GPNMB_v2mut/HLA-A*03011 (E), SNRP116mut/HLA-A*03011 (F), RBAF600mut/HLA-B*07021 (G), SNRPD1mut/HLA-B*3801 (H). Data are means of duplicates and represent the number of spot forming cells per 15,000 (day 25) or 10,000 (later than day 25) CD8+ MLTC responders.
The peptide-coding regions for Tyr/A26, Tyr/B38, and gp100/B7 were identified by cDNA fragmentation (data not shown). Synthetic peptides encoded by these fragments and causing T cell activation at lowest concentrations are listed in Table 1 (Table 3 lists all of the synthetic peptides tested herein). Special requirements for presentation and recognition of the gp100/B7 peptide are communicated in Fig. 8.
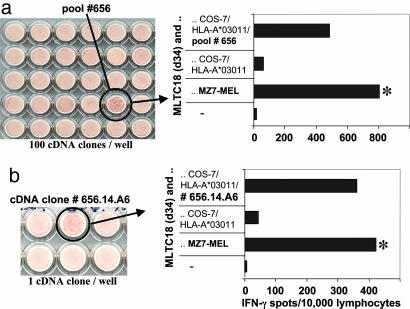
DT Melanoma-Reactive T Cells Recognized at Least Five Mutated Neoantigens. Screening of a cDNA library from MZ7-MEL cells with autologous tumor-reactive T cells resulted in the identification of five mutated antigens (summarized in Table 1). Whereas antigen SIRT2mut/A3 was identified with clonal T cells as described before (11), antigens GPNMBmut/A3, SNRP116mut/A3 and RBAF600mut/B7 were identified with cryopreserved MLTC16 and MLTC18 responder populations by IFN-γ ELISPOT assays. As a representative example, the identification of antigens GPNMBmut/A3 and SNRP116mut/A3 with MLTC18 is detailed in Fig. 3 and its legend. This procedure led also to the identification of antigen SNRPD1mut/B38, except that clonal T cells, CTL15/165, were applied. CTL15/165 was derived by limiting dilution from MLTC15 and was found to recognize an hitherto unknown antigen (see below).
Fig. 3.
Cloning of antigen-coding cDNA with cryopreserved MLTC18 responders. MLTC18(d25) contained T cells against Tyr/A26 at low frequencies, but was strongly reactive with autologous melanoma cells (Fig. 2). Its antitumor reactivity was partially inhibited by an anti-HLA-A3 monoclonal antibody (data not shown). MLTC18 was further expanded. On day 32, 2.78 × 108 CD8+ T cells were frozen in aliquots 4 days after the last stimulation with autologous tumor cells. For cDNA library screening, aliquots were thawed and T cells were kept for 2 days in medium with IL-2. In parallel, COS-7 cells were cotransfected directly in MultiScreen ELISPOT HP plates with pools of ≈100 cDNA clones and with HLA-A*03011 cDNA. After 24 h, MLTC responders were added at 10,000 lymphocytes per well. Each cDNA pool was then tested for its ability to induce IFN-γ spot formation. Thirteen of 1,920 cDNA pools were positive. A filter section with positive pool 656 is shown next to the spot evaluation data (a). From pool 656, we identified cDNA clone 656.14.A6 as inducer of spot formation after cotransfection with HLA-A*03011 (b). This cDNA clone encoded GPNMB_v2mut (Table 1). Only 11 of 13 positive “pools of 100” identified in this screening experiment contained GPNMB_v2mut cDNA as verified by PCR. From the two remaining pools, we isolated a cDNA clone encoding SNRP116mut (Table 1). *, Reactivity to melanoma cells was tested with the same MLTC population in parallel (30,000 MZ7-MEL cells per well; a, 1,000 lymphocytes per well; b, 500 lymphocytes per well).
At the genomic DNA level MZ7-MEL cells were heterozygous for all five mutations, whereas autologous MZ7-EBV-B cells carried only wild-type alleles (data not shown). This finding indicated that the mutations were of somatic origin and had occurred in the DT melanoma or its precursor lesion.
Homologous wild-type cDNAs were cloned by RT-PCR from MZ7-EBV-B cells. Even in the high-efficiency COS expression system, none of the wild-type cDNAs induced T cell recognition except for nonmutated SIRT2. Wild-type SIRT2 was recognized after transfection into COS-7 cells, but recognition was clearly inferior to its mutated counterpart (data not shown). Moreover, CTL clones against SIRT2mut/A3 did not recognize autologous EBV-B cells in a 4-h 51Cr release assay at effector-to-target ratios up to 40:1, which indicated that the affinity of these T cells toward the homologous wild-type peptide was too low to mediate lysis of naturally processing and presenting cells expressing wild-type SIRT2 only (data not shown). These observations suggested that the mutations generated immunogenic peptides. In all cases, T cell-recognized peptides contained the mutated residues (Table 1; Table 3 lists synthetic peptides tested herein).
Functional analyses were performed only for mutated SIRT2. Introduction of the P219L mutation into SIRT2_v1 protein reduced its enzymatic activity by 80–90% compared to the wild-type protein (Fig. 9).
T Cells Against Mutated Antigens Prevailed in Independently Generated MLTC. MLTC responder populations generated from PBMCs of different years were tested at various time points for reactivity against the DT melanoma antigens reported herein (Table 1). Targets were COS-7 cells transiently transfected with the antigens and their presenting HLA alleles. Enrichment of T cells specific for single antigens to at least 1% of the total CD8+ count was found at least once in one of six MLTCs for Tyr/A26, one of six MLTCs for Tyr/B38, two of six MLTCs for gp100/B7, two of six MLTCs for SIRT2mut/A3, five of five MLTCs for GPNMBmut/A3, five of five MLTCs for SNRP116mut/A3, five of five MLTCs for RBAF600mut/B7, and three of five MLTC for SNRPD1mut/B38. These data documented that, among lymphocytes stimulated with autologous tumor cells, especially those against mutated antigens, GPNMBmut/A3, SNRP116mut/A3, and RBAF600mut/B7 regularly prevailed over T cells against common antigens (Fig. 4).
Within continuously propagated MLTCs, the relative contribution of distinct T cell specificities to overall tumor cell recognition was not always stable or increasing over time, but rather vanished in some cultures (Fig. 4). This finding might be explained by different expansion capacities of individual clones.
Analyses on MLTC-Derived Clonal T Cells. MLTC7, 9, 12, 15, and 18 were cloned by limiting dilution. Stable T cell clones were obtained against gp100/B7 and all five mutated antigens. For example, from MLTC 15, cloned on day 35, we derived 125 T cell clones, not all of which were further expandable after initial testing on COS transfectants. Thirty-five clones were directed against GPNMBmut/A3, 34 clones were directed against SNRP116mut/A3, 18 clones were directed against either gp100/B7, RBAF600mut/A3, SIRT2mut/A3, or Tyr/A26, and 38 clones recognized none of the DT antigens known by then (data not shown). With one of the latter clones (CTL15/165), antigen SNRPD1mut/B38 was identified (Table 1). Taken together, ≈70–80% of melanoma-reactive T cell clones established by limiting dilution from MLTC recognized one of the antigens listed in Table 1 (data not shown).
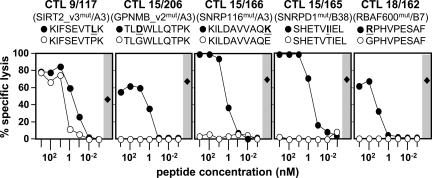
Clonal T cells against mutated DT melanoma antigens were tested for their functional avidity. Target cells (MZ7-EBV-B) were pulsed with graded doses of homologous mutated and wild-type peptides (Table 1) and tested for recognition in a 4-h 51Cr release assay. Lysis induced by the mutated peptides was half-maximal at concentrations ranging from 0.11 to 11 nM (Fig. 5). Among wild-type peptides, only the SIRT2 192–200 peptide induced lysis at 10-fold higher concentrations compared with the corresponding mutated peptide, which is in line with the results seen with COS transfectants (see above).
Fig. 5.
Titration of peptides. Data of a 4-h 51Cr release assay are shown. Peptides (sequences and concentrations as indicated, mutated residues bold and underlined, see Table 1) were directly added to 51Cr-labeled MZ7-EBV-B cells (2,000 per well). CTL clones (specificity indicated in parentheses) were added at an effector-to-target (E/T) cell ratio of 20:1. Filled circles, mutated peptides; open circles, homologous wild-type peptides; filled diamonds, lysis of MZ7-MEL cells (E/T = 20:1, 2,000 melanoma cells per well).
TCR Vβ analyses were performed to study the clonality of T cell responses to epitopes gp100/B7, SIRT2mut/A3, GPNMBmut/A3, and SNRP116mut/A3. T cells against gp100/B7, derived from three independent MLTC, carried at least three distinct TCR Vβ chains. All T cell clones against SIRT2mut/A3, derived from two independent MLTC, expressed Vβ1. At least two distinct T cell receptors were involved in the recognition of GPNMBmut/A3 and at least four in the recognition of SNRP116mut/A3 (Table 5).
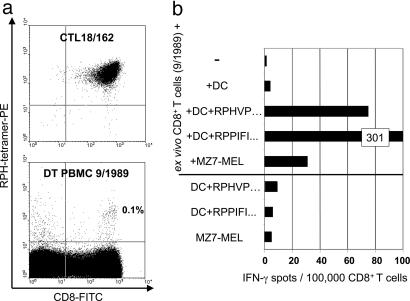
Analyses on ex Vivo PBMC. CD8+ T cells were isolated from freshly thawed PBMC taken in 1989 and 1992 and were directly, without prior stimulation, tested in 48-h IFN-γ ELISPOT assays for recognition of autologous melanoma cells and autologous blood-derived mDC loaded with peptides. T cells recognizing HLA-B7-restricted EBV-B peptide EBNA-3A 379–387 (14) were detectable at a frequency of ≈300 per 105 CD8+ cells in all samples tested. However, in PBMC from 1992, T cells against melanoma cells or melanoma peptide-loaded mDC were not found at a frequency above 20 per 105 CD8+ cells, which we regard as a detection threshold for this type of assay (data not shown). Only in PBMC from September 1989 did we observe anti-RBAF600mut/B7 T cells at a frequency of ≈70 per 105 of CD8+ T cells. In accordance, ≈0.1% of CD8+ T cells in the same PBMC sample were positively stained with RBAF600mut/B7 tetramers (Fig. 6). This finding demonstrated in principle the expansion of functional antimelanoma T cells in the patient's peripheral blood. The overall low frequency of tumor-reactive T cells was in accordance with the delayed expansion of MLTC responders in the course of continuous stimulation with autologous tumor cells (Fig. 7).
Fig. 6.
Detection of RBAF600mut/B7-reactive T cells in ex vivo PBMC. (a) PBMCs taken from patient DT in September 1989 and, as a control, the RBAF600mut/B7-specific T cell clone CTL18/162, were stained with phycoerythrin (PE)-conjugated tetrameric complex B7/RPHVPESAF containing the mutant RBAF600 peptide and HLA-B7 and with an antibody to CD8 conjugated to FITC. (b) In parallel, CD8+ T cells were positively selected from the same PBMC and were analyzed by a 48-h IFN-γ ELISPOT assay using peptide-pulsed autologous DCs (20,000 per well) and melanoma cells (30,000 per well) as antigen-presenting cells. Peptides were the RBAF600mut/B7 peptide RPH-VPESAF (Table 1) and, as a positive control, the HLA-B7-restricted EBNA 3A 379-387 peptide RPPIFIRRL (14).
Discussion
Although several lines of evidence point to the clinical relevance of host antitumor T cell responses, pathways leading to antitumor immunity or causing the immune system's failure to eliminate transformed cells are by far not understood (15); this may explain why therapeutic vaccination studies performed during the last decade led only to a limited and unsatisfying success rate. Regressions of metastases after therapeutic vaccination were observed only in a minority of patients (16), and there was only weak evidence that in vivo expansion of T cells directed against vaccine antigens correlated with tumor regression (17, 18). Clinical studies focused on the induction and priming of T cell responses and to the overcoming of immunological tolerance, whereas other important aspects could not be addressed in these studies (19). One of these aspects is that the patients were vaccinated regardless of their immune repertoire's ability to mount responses to the vaccine antigens. Studies attempting to dissect overall antitumor T cell responses demonstrated that only a rather small portion is covered by reactivity toward known common antigens (20, 21). In contrast to immune responses in transmittable and population-threatening infectious diseases, an evolutionary process favoring predominant responses against certain tumor-associated antigens cannot be assumed in cancer. Therefore, it appears reasonable to anticipate a high and, at present, unpredictable degree of individuality in tumor–host interactions. According to this, we consider the identification of the preferential targets of the individual antitumor repertoire an integral requirement for further development of therapeutic vaccination in cancer.
Cryopreserved DT MLTC responder populations were applied to cDNA expression screening in combination with a lymphokine spot readout assay, whose sensitivity is known to be superior to the measurement of cytokine concentrations in culture supernatants (22). Altogether, we identified three previously unknown tyrosinase and gp100 peptide epitopes and five neoantigens generated by somatic point mutations. Five of the eight DT melanoma peptide antigens were identified with nonclonal MLTC responders, three were found with the same MLTC, and two neoantigens were found in a single screening experiment (Table 1 and Fig. 2). In our experience, by using independently generated cryopreserved MLTC in combination with a sensitive readout for first-line screening experiments, antigen discovery was accelerated, became more consistent, and allowed to detect a wider and perhaps more representative spectrum of T cell target antigens.
We conclude from our observations that anti-DT melanoma T cells were induced by direct tumor–host interactions. T cells against mutated antigens dominated autologous MLTC responses in PBMC taken over years (Fig. 4). By determining the peptide requirements for induction of lysis, high-avidity patterns were verified (Fig. 5). The avidity of CTL against mutated DT melanoma antigens reached the strength of antiviral T cells indicating functional maturation in vivo (23, 24). This finding strongly suggested induction of T cell responses by the patient's tumor tissue, because these antigens arose de novo by somatic mutations and were therefore restricted to her tumor cells. Immune responses against de novo antigens arising in tumors are supposedly not subjected to central and extratumoral peripheral tolerance mechanisms (25), which may have added to the preponderance of T cells against at least some mutated DT antigens in independent MLTC.
Genetically altered oncoproteins are of particular interest in cancer immunotherapy because their expression is truly tumor-specific and supposedly stable throughout disease progression. In the melanoma model derived from patient SK29(AV), we had previously identified a neoantigen generated by mutant cyclin-dependent kinase allele CDK4-R24C (26), later shown to act as a dominant oncogene in vivo (27, 28). As seen in an SK29 MLTC, generated and analyzed in the same way as DT MLTC, reactivity toward CDK4mut/A2 clearly dominated the T cell response against autologous melanoma cells (Fig. 10). So far, formal evidence for a functional impact of a mutation found in the DT melanoma has been obtained for the SIRT2 mutation in enzymatic assays in vitro (Fig. 9) and by introducing it into the homologous yeast protein ySir2 (29). Also, the other proteins found to be mutated in DT melanoma cells have been implicated in vital pathways of eukaryotic cells (Tables 1 and 4), and the mutant alleles might therefore have contributed to the malignant cell phenotype.
The specificity analysis of T cell clones derived from various DT MLTC demonstrated that we had found most, although not all, of the antigens targeted by the patient's antitumor T cells. The spectrum of these antigens revealed a high degree of individuality, which is only in part imposed by the patient's HLA class I phenotype. DT melanoma cells strongly expressed melanosomal proteins for which no peptides were known to be presented by one of the patient's HLA class I alleles, with the exception of gp100 peptides presented by HLA-A3 (30–32). However, none of the DT MLTCs contained detectable reactivity toward gp100/A3, although HLA-A3 served as a restricting molecule for three distinct mutated epitopes and we observed T cell responses against gp100 in most MLTC.
In few human tumor models, multiple antigens recognized by autologous T cell responses were stepwise identified over time (26, 33–35). In each case, a rather individual set of both shared and mutated antigens was targeted by T cells, which is in line with the findings reported herein. Mutated antigens appeared to be among the preferred targets (33–36). Although their identification is currently not feasible in a large number of patients, the vaccination of patients with autologous tumor material such as whole tumor cells (37), heat shock proteins (38), and amplified RNA (39) is based on their existence.
The individuality of tumor–T cell interaction is determined by the antigenic tumor phenotype resulting from genetic and epigenetic alterations, but also by the natural HLA polymorphism and by the individual T cell repertoire. Technical progress, such as the procedure reported herein as well as advances in genome-wide screening technologies (40, 41), should help in the future to identify more rapidly and more efficiently the preferential target molecules of individual antitumor responses. Their knowledge will allow to comprehensively study tumor–T cell interactions and to improve their therapeutic potential.
Supplementary Material
Acknowledgments
We thank T. Kreer and T. Gareis for technical assistance and gratefully acknowledge helpful comments from H. Schreiber and T. Boon. This research was supported by Deutsche Forschungsgemeinschaft Grant SFB432/A1 (to T.W.). C.G. and C.W. were supported by a grant from the Deutsche Krebshilfe (Project 70-2428).
Author contributions: V.L., C.H., and T.W. designed research; T.W., V.L., M.F., C.G., R.A.F., A.L., D.F., and C.W. performed research; R.A.F. and C.W. contributed new reagents/analytic tools; V.L., M.F., C.G., R.A.F., A.L., D.F., C.H., and T.W. analyzed data; and V.L. and T.W. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MLTC, mixed lymphocyte–tumor cell culture; PBMC, peripheral blood mononuclear cell; CTL, cytotoxic T lymphocyte; TCR, T cell receptor; mDC, mature dendritic cell.
Data deposition: The sequences described in this paper have been deposited in the GenBank database (accession nos. AJ577268 and AJ505014–AJ505017).
References
- 1.Van den Eynde, B. J. & van der Bruggen, P. (1997) Curr. Opin. Immunol. 9, 684-693. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg, S. A. (1999) Immunity 10, 281-287. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava, P. J. & Old, L. J. (1988) Immunol. Today 9, 78-83. [DOI] [PubMed] [Google Scholar]
- 4.Beck-Engeser, G. B., Monach, P. A., Mumberg, D., Yang, F., Wanderling, S., Schreiber, K., Espinosa, R., III, Le Beau, M. M., Meredith, S. C. & Schreiber, H. (2001) J. Exp. Med. 194, 285-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilboa, E. (1999) Immunity 11, 263-270. [DOI] [PubMed] [Google Scholar]
- 6.Lim, D. G., Bieganowska Bourcier, K., Freeman, G. J. & Hafler, D. A. (2000) J. Immunol. 165, 6214-6220. [DOI] [PubMed] [Google Scholar]
- 7.Coulie, P. G., Karanikas, V., Colau, D., Lurquin, C., Landry, C., Marchand, M., Dorval, T., Brichard, V. & Boon, T. (2001) Proc. Natl. Acad. Sci. USA 98, 10290-10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wölfel, T., Hauer, M., Klehmann, E., Brichard, V., Ackermann, B., Knuth, A., Boon, T. & Meyer zum Büschenfelde, K.-H. (1993) Int. J. Cancer 55, 237-244. [DOI] [PubMed] [Google Scholar]
- 9.Peggs, K., Verfuerth, S., Pizzey, A., Ainsworth, J., Moss, P. & Mackinnon, S. (2002) Blood 99, 213-223. [DOI] [PubMed] [Google Scholar]
- 10.Ennis, P. D., Zemmour, J., Salter, R. D. & Parham, P. (1990) Proc. Natl. Acad. Sci. USA 87, 2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brichard, V., Van Pel, A., Wölfel, T., Wölfel, C., De Plaen, E., Lethé, B., Coulie, P. & Boon, T. (1993) J. Exp. Med. 178, 489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britten, C. M., Meyer, R. G., Kreer, T., Drexler, I., Wolfel, T. & Herr, W. (2002) J. Immunol. Methods 259, 95-110. [DOI] [PubMed] [Google Scholar]
- 13.Jonuleit, H., Kuhn, U., Müller, G., Steinbrink, K., Paragnik, L., Schmitt, E., Knop, J. & Enk, A. H. (1997) Eur. J. Immunol. 27, 3135-3142. [DOI] [PubMed] [Google Scholar]
- 14.Hill, A., Worth, A., Elliott, T., Rowland-Jones, S., Brooks, J., Rickinson, A. & McMichael, A. (1995) Eur. J. Immunol. 25, 18-24. [DOI] [PubMed] [Google Scholar]
- 15.Houghton, A. N., Gold, J. S. & Blachere, N. E. (2001) Curr. Opin. Immunol. 13, 134-140. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg, S. A., Yang, J. C. & Restifo, N. P. (2004) Nat. Med. 10, 909-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banchereau, J., Palucka, A. K., Dhodapkar, M., Burkeholder, S., Taquet, N., Rolland, A., Taquet, S., Coquery, S., Wittkowski, K. M., Bhardwaj, N., et al. (2001) Cancer Res. 61, 6451-6458. [PubMed] [Google Scholar]
- 18.Lonchay, C., van der Bruggen, P., Connerotte, T., Hanagiri, T., Coulie, P., Colau, D., Lucas, S., Van Pel, A., Thielemans, K., van Baren, N. & Boon, T. (2004) Proc. Natl. Acad. Sci. USA 101, 14631-14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinman, R. M. & Mellman, I. (2004) Science 305, 197-200. [DOI] [PubMed] [Google Scholar]
- 20.Anichini, A., Mortarini, R., Maccalli, C., Squarcina, P., Fleischhauer, K., Mascheroni, L. & Parmiani, G. (1996) J. Immunol. 156, 208-217. [PubMed] [Google Scholar]
- 21.van Elsas, A., Hurwitz, A. A. & Allison, J. P. (1999) J. Exp. Med. 190, 355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabilan, L., Andersson, G., Lolli, F., Ekre, H. P., Olsson, T. & Troye-Blomberg, M. (1990) Eur. J. Immunol. 20, 1085-1089. [DOI] [PubMed] [Google Scholar]
- 23.Derby, M., Alexander-Miller, M., Tse, R. & Berzofsky, J. (2001) J. Immunol. 166, 1690-1697. [DOI] [PubMed] [Google Scholar]
- 24.Slifka, M. K. & Whitton, J. L. (2001) Nat. Immunol. 2, 711-717. [DOI] [PubMed] [Google Scholar]
- 25.Gotter, J. & Kyewski, B. (2004) Curr. Opin. Immunol. 16, 741-745. [DOI] [PubMed] [Google Scholar]
- 26.Wölfel, T., Hauer, M., Schneider, J., Serrano, M., Wölfel, C., Klehmann-Hieb, E., De Plaen, E., Hankeln, T., Meyer zum Büschenfelde, K.-H. & Beach, D. (1995) Science 269, 1281-1284. [DOI] [PubMed] [Google Scholar]
- 27.Zuo, L., Weger, J., Yang, Q., Goldstein, A. M., Tucker, M. A., Walker, G. J., Hayward, N. & Dracopoli, N. C. (1996) Nat. Genet. 12, 97-99. [DOI] [PubMed] [Google Scholar]
- 28.Sotillo, R., Dubus, P., Martin, J., de la Cueva, E., Ortega, S., Malumbres, M. & Barbacid, M. (2001) EMBO J. 20, 6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrod, S., Cockell, M. M., Laroche, T., Renauld, H., Ducrest, A. L., Bonnard, C. & Gasser, S. M. (2001) EMBO J. 20, 197-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawakami, Y., Robbins, P. F., Wang, X., Tupesis, J. P., Parkhurst, M. R., Kang, X., K., S., Appella, E. & Rosenberg, S. A. (1998) J. Immunol. 161, 6985-6992. [PubMed] [Google Scholar]
- 31.Kawashima, I., Tsai, V., Southwood, S., Takesako, K., Celis, E. & Sette, A. (1998) Int. J. Cancer 78, 518-524. [DOI] [PubMed] [Google Scholar]
- 32.Yamshchikov, G., Thompson, L., Ross, W. G., Galavotti, H., Aquila, W., Deacon, D., Caldwell, J., Patterson, J. W., Hunt, D. F. & Slingluff, C. L., Jr. (2001) Clin. Cancer Res. 7, 909S-916S. [PubMed] [Google Scholar]
- 33.Coulie, P. G., Ikeda, H., Baurain, J. F. & Chiari, R. (1999) Adv. Cancer Res. 76, 213-242. [DOI] [PubMed] [Google Scholar]
- 34.Robbins, P. F., El-Gamil, M., Li, Y. F., Zeng, G., Dudley, M. & Rosenberg, S. A. (2002) J. Immunol. 169, 6036-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zorn, E. & Hercend, T. (1999) Eur. J. Immunol. 29, 592-601. [DOI] [PubMed] [Google Scholar]
- 36.Karanikas, V., Colau, D., Baurain, J. F., Chiari, R., Thonnard, J., Gutierrez-Roelens, I., Goffinet, C., Van Schaftingen, E. V., Weynants, P., Boon, T. & Coulie, P. G. (2001) Cancer Res. 61, 3718-3724. [PubMed] [Google Scholar]
- 37.O'Rourke, M. G., Johnson, M., Lanagan, C., See, J., Yang, J., Bell, J. R., Slater, G. J., Kerr, B. M., Crowe, B., Purdie, D. M., et al. (2003) Cancer Immunol. Immunother. 52, 387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belli, F., Testori, A., Rivoltini, L., Maio, M., Andreola, G., Sertoli, M. R., Gallino, G., Piris, A., Cattelan, A., Lazzari, I., et al. (2002) J. Clin. Oncol. 20, 4169-4180. [DOI] [PubMed] [Google Scholar]
- 39.Su, Z., Dannull, J., Heiser, A., Yancey, D., Pruitt, S., Madden, J., Coleman, D., Niedzwiecki, D., Gilboa, E. & Vieweg, J. (2003) Cancer Res. 63, 2127-2133. [PubMed] [Google Scholar]
- 40.Maecker, B., von Bergwelt-Baildon, M., Anderson, K. S., Vonderheide, R. H. & Schultze, J. L. (2001) Curr. Mol. Med. 1, 609-619. [DOI] [PubMed] [Google Scholar]
- 41.Qiu, P., Shandilya, H., D'Alessio, J. M., O'Connor, K., Durocher, J. & Gerard, G. F. (2004) BioTechniques 36, 702-707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.