Abstract
Herpes simplex virus (HSV) persists in its human host and evades the immune response by undergoing a latent infection in sensory neurons, from which it can reactivate periodically. HSV expresses >80 gene products during productive (“lytic”) infection, but only the latency-associated transcript (LAT) gene is expressed at abundant levels during latent infection. The LAT gene has been shown to repress lytic-gene expression in sensory neurons. In this study, we use chromatin immunoprecipitation to show that HSV lytic-gene promoters become complexed with modified histones associated with heterochromatin during the course of establishment of latent infection. Experiments comparing LAT-negative and LAT-positive viruses show that a function encoded by the LAT gene increases the amount of dimethyl lysine 9 form of histone H3 or heterochromatin and reduces the amount of dimethyl lysine 4 form of histone H3, a part of active chromatin, on viral lytic-gene promoters. Thus, HSV, and in particular the HSV LAT gene, may manipulate the cellular histone modification machinery to repress its lytic-gene expression and contribute to the persistence of its genome in a quiescent form in sensory neurons.
Keywords: chromatin modification, gene regulation, herpes simplex virus
Herpes simplex virus (HSV) productively infects the mucosal epithelium and spreads within the mucosal site. Thereafter, HSV infects sensory neurons and establishes a latent infection, from which the virus can periodically reactivate and cause recurrent infections. During productive (“lytic”) infection, HSV expresses >80 viral gene products whereas, during latent infection, the only viral gene products expressed at abundant levels are the latency-associated transcripts or LATs (1). The molecular mechanisms allowing abundant expression of HSV gene products in epithelial and other nonneuronal cells versus the limited gene expression in sensory neurons are not well understood. Due to the significant disease burden caused by HSV in encephalitis, keratitis, neonatal infection, and disseminated infection in immunocompromised individuals (2), and promotion of HIV transmission (3), an understanding of the mechanisms of latent infection and derivation of means to block or interrupt it could help relieve this medical burden.
One viral gene postulated to affect viral infection of neurons encodes the LATs (4). LATs are expressed from a neuron-specific promoter/enhancer (5), and various studies have reported that the LATs promote viral reactivation from latent infection (6), promote viral establishment of latent infection (7), cause a repression of productive viral gene expression in sensory neurons during acute infection (8) and during latent infection (9), and protect infected neurons from death by apoptotic (10) or other mechanisms (11). During productive infection, viral DNA is relatively free of nucleosomes (12), and only relatively low levels of viral DNA are associated with histone H3 (13). In contrast, during latent infection, viral DNA is assembled into nucleosomal chromatin (14). Histone modifications, particularly methylation and acetylation, play an important role in defining the type of regulation imposed on eukaryotic genes by nucleosomal chromatin, with particular modifications resulting in chromatin-mediated gene activation (active or euchromatin) or repression (inactive or heterochromatin) (15). Little is known about the form of chromatin on the viral chromosome except that the LAT promoter and 5′ exon are found in active chromatin but lytic-gene promoters are not associated with active chromatin (16). Based on this background, we hypothesized that heterochromatin is assembled onto viral lytic-gene promoters during latent infection and that the LAT gene functions to promote this process.
Materials and Methods
Viruses and Infection Procedures. HSV-1 WT KOS strain virus, the KdlLAT LAT gene deletion mutant virus, the FSLAT LAT+ repaired virus strain, and their propagation and titration have been described (8). For animal infections, 7-week-old male CD-1 mice (Charles River Laboratories) were anesthetized and infected with 2 × 106 plaque-forming units (pfu) of the indicated strain of HSV-1 after corneal scarification (17). Mice were housed in accordance with institutional and National Institutes of Health (NIH) guidelines on the care and use of animals in research, and all procedures were approved by the Institutional Animal Use Committee of Harvard Medical School.
Real-Time PCR Analysis. Real-time PCR was performed by using SYBR Green PCR Master Mix and an ABI Prism 7700 sequence detection system (Applied Biosystems). PCRs were amplified for 40 cycles (30 s at 95°C, then 60 s at 60°C). The specificity of each reaction was verified by electrophoretic separation of exponential-phase PCR products on an agarose gel. Each PCR was run in duplicate. Primers were designed according to the manufacturer's guidelines. Their sequences are as follows: infected cell protein (ICP) 4 (nucleotides 146,500–146,557; -304 to -247), (5′-TAGCATGCGGAACGGAAGC-3′ and 5′-CGCATGGCATCTCATTACCG-3′); ICP8 (nucleotides 622,90–62,418; -100 to +28), (5′-CCACGCCCACCGGCTGATGAC-3′ and 5′-TGCTTACGGTCAGGTGCTCCG-3′); ICP27 (nucleotides 113,377–113,471; -2119 to -125), (5′-ACCCAGCCAGCGTATCCACC-3′ and 5′-ACACCATAAGTACGTGGCATGT-3′); thymidine kinase (TK) (nucleotides 47,855–48,111; -200 to +56), (5′-CAGCTGCTTCATCCCCGTGG-3′ and 5′-AGATCTGCGGCACGCTGTTG-3′); and GAPDH (GenBank accession no. NM_008084, nucleotides 781–900), (5′-CAATGTGTCCGTCGTGGATCT-3′ and 5′-TTGAAGTCGCAGGAGACAACC-3′).
Chromatin Immunoprecipitation (ChIP) Analysis. Trigeminal ganglia (TG) were dissected from mice at different time intervals postinfection and dissociated at 37°C in DMEM containing 9 mM NaHCO3, 20 mM Hepes, 5% 10× trypsin, and 0.02% collagenase type 1A for 3 h. Formaldehyde [final concentration 1% (vol/vol)] was added to the cell suspension to cross-link the chromatin at room temperature with shaking for 10 min. The reaction was stopped by addition of glycine to a final concentration of 125 mM, and the suspension was incubated for an additional 5 min at room temperature with shaking. The cells were collected by centrifugation, and the pellet was washed three times with ice-cold PBS with protease inhibitor (Roche Diagnostics), resuspended in SDS lysis buffer (1% SDS/10 mM EDTA/50 mM Tris, pH 8.1), and incubated on ice for 10 min.
The cell lysates were sonicated for six 10-s pulses on setting 2.5 with a Heat Systems XL Sonicator to shear the chromatin into lengths of ≈500 bp. The sheared chromatin was diluted by 10-fold in radioimmunoprecipitation assay (RIPA) lysis buffer (0.1% SDS/1% sodium deoxycholate/150 mM NaCl/10 mM Na2PO4/2 mM EDTA/0.2 mM NaVO3/1% Nonidet P-40) with protease inhibitor (Roche), and then was precleared by incubation with salmon sperm DNA/protein A agarose slurry (Upstate Biotechnology, Lake Placid, NY) with rotation at 4°C for 2 h and centrifugation. The chromatin supernatant was then incubated with the appropriate antibody overnight at 4°C with rotation. Anti-histone H3 antibody (Abcam, Inc., Cambridge, MA) and anti-dimethyl Lys-4 H3 antibody (Upstate Biotechnology) were used at a 1:200 dilution whereas the anti-dimethyl Lys-9 H3 antibody (Upstate Biotechnology) was used at 1:100 dilution.
Immunocomplexes were collected by incubation with salmon sperm DNA/protein A agarose slurry for 2 h at 4°C with rotation. Beads were washed three times on a rotating platform with 1 ml of low-salt wash buffer (150 mM NaCl/20 mM Tris·Hcl, pH 8.1/2 mM EDTA/1% Triton X-100/0.1% SDS), followed by one wash with 1 ml of high-salt wash buffer (500 mM NaCl/20 mM Tris·HCl, pH 8.1/2 mM EDTA/1% Triton X-100/0.1% SDS). Immunocomplexes were eluted by incubation at 65°C with freshly prepared elution buffer (0.1% SDS/0.1 M NaHCO3) for 30 min, followed by rotating for 15 min at room temperature. NaCl was added to a final concentration of 0.2 M, and the eluates were incubated at 65°C overnight to reverse the cross-linking. The eluates were then digested with proteinase K, and the DNA was purified by phenol:chloroform extraction and ethanol precipitation.
Quantitative Analysis. Threshold cycle number, Ct, was measured as the PCR cycle at which the amount of amplified target reaches the threshold value. Quantification was determined by the 2-ΔΔCt method as described in Applied Biosystems User Bulletin No. 2 (http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf). The amount of viral DNA relative to that at day 1 postinfection (Fig. 1A) was calculated as 2-(ΔΔCt) where ΔΔCt = ΔCt (viral) - ΔCt (GAPDH) and ΔCt (viral) = Ct (time point) - Ct (day 1) and ΔCt (GAPDH) = Ct (time point) - Ct (day 1). The relative amounts (fold-enrichment) of viral DNA sequences associated with the indicated histones as presented in the other figures were calculated as 2-(ΔΔCt) where ΔΔCt = ΔCt (viral) - ΔCt (GAPDH), ΔCt (viral) = Ct (IP) - Ct (input), and ΔCt (GAPDH) = Ct (IP) - Ct (input). Statistical comparisons of the significance of the differences between LAT+ and LAT- samples were conducted by using the Friedman two-way analysis of rank.
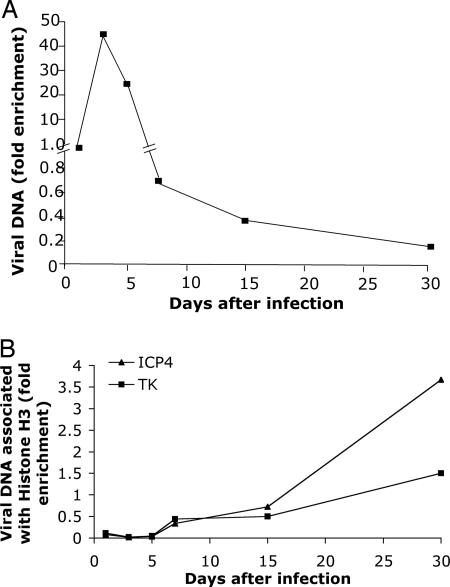
Fig. 1.
Time course of histone H3 assembly on HSV lytic-gene promoters during infection of murine trigeminal ganglion neurons. (A) Total HSV DNA levels in ganglia. At the times indicated, mice were euthanized, trigeminal ganglia were removed, lysates were prepared, and real-time PCR was performed. The values for HSV ICP4 gene sequences shown were determined for each time point and normalized to the amount in the ganglia at day 1 and then normalized to the levels of the cellular GAPDH gene, which was quantified in a separate PCR. (B) ChIP of viral DNA associated with histone H3. ChIP was conducted on the lysates in A by using anti-histone H3 antibody as the primary antibody. Viral DNA sequences were quantified by using primers for the ICP4 or TK gene promoters for real-time PCR. The fraction of the viral DNAs immunoprecipitated was normalized to the fraction of GAPDH DNA immunoprecipitated in the same immunoprecipitate to give the fold-enrichment values.
Results
To examine the assembly of chromatin on viral lytic-gene promoters during infection of murine trigeminal ganglia after corneal infection (17), we infected mice by introduction of HSV-1 WT virus onto the cornea and euthanized the mice at various times postinfection, removed the trigeminal ganglia, and prepared lysates for ChIP and DNA quantification by real-time PCR by using primers for viral promoters. As observed previously (18), viral thymidine kinase (TK) gene DNA content in the ganglia relative to a host gene (GAPDH) peaked at ≈3 days postinfection (dpi) and then declined by 10- to 100-fold to a plateau level by 15–30 dpi (Fig. 1 A). We measured the amount of HSV DNA assembled into chromatin by ChIP using an anti-histone H3 antibody. Viral and GAPDH DNAs in each immunoprecipitate and lysate were quantified by real-time PCR for the ICP4 and TK gene and GAPDH promoter sequences, respectively. To quantify the relative amounts of the different viral promoters associated with the different histones, we calculated the values as the fraction of the viral promoter immunoprecipitated with a specific antibody divided by the fraction of GAPDH DNA immunoprecipitated with that antibody in the same reaction. The viral DNA was normalized to GAPDH to control for variability in efficiency of immunoprecipitation. The fraction of GAPDH immunoprecipitated with each of the antibodies was equivalent for LAT+ versus LAT- virus-infected ganglia (results not shown); therefore, this was a valid normalization. Relative to host GAPDH, little viral DNA was associated with histone H3 before 5 dpi, but this association increased from 5 days through 30 dpi for both the ICP4 and TK gene promoters (Fig. 1B).
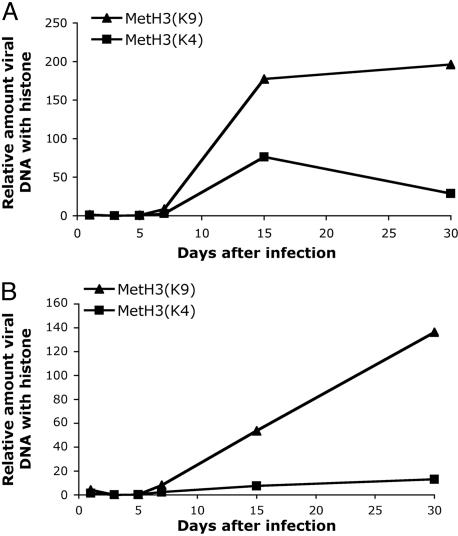
To examine the form of chromatin associated with the viral promoters, we performed ChIP by using antibodies specific for histone H3 methylated at the lysine 4 (K4) or lysine 9 (K9) residues. Histone H3 methylated at the K4 position, Met-H3(K4), is associated with active or euchromatin (19, 20) whereas histone H3 methylated at the K9 position, Met-H3(K9), is associated with inactive or heterochromatin (19–21). As with total H3, relatively low levels of methylated histone H3 were associated with either the ICP4 or the TK gene promoters through 5 dpi. K9 and K4 methylation of H3 on the ICP4 gene promoter increased by 15 days and was maintained through 30 days (Fig. 2A). K9 and K4 methylation of H3 on the TK gene promoter increased through 30 dpi (Fig. 2B). Higher relative levels of K9 methylation than K4 methylation were observed consistently (Figs. 2 and 5 and results not shown). Therefore, we concluded that histone modification and assembly of heterochromatin were likely to play a role in repression of viral lytic gene expression during latent infection of murine trigeminal ganglia.
Fig. 2.
Kinetics of assembly of methylated histone H3 forms on HSV lyticgene promoters. ChIP analyses as described in Fig. 1b were conducted with either anti-dimethyl-histone H3 (Lys-9) or anti-dimethyl-histone H3 (Lys-4) antibody. Quantification of DNA was performed by using real-time PCR. (A) ICP4 promoter. (B) TK promoter.
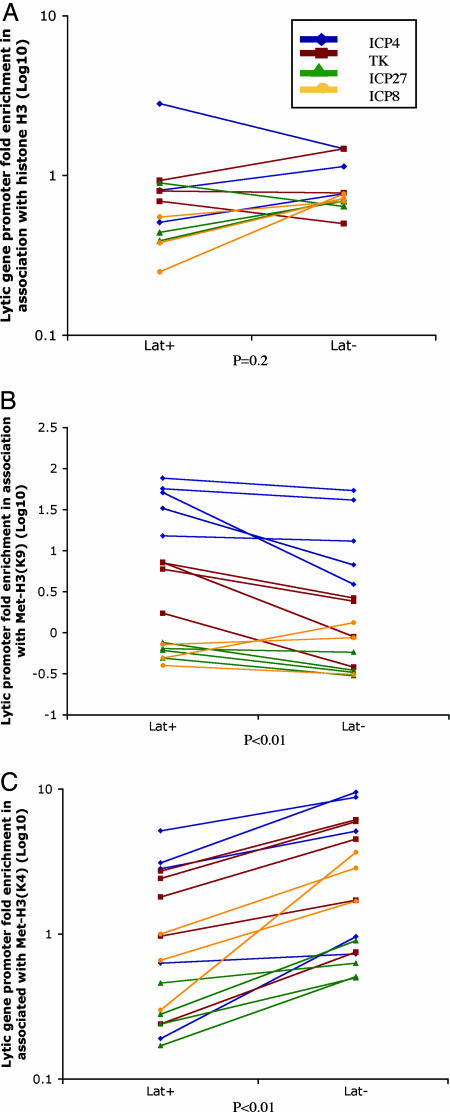
Fig. 5.
Comparison of immunoprecipitated viral DNA sequences from LAT+ versus LAT- virus-infected ganglia samples using ChIP assays for different forms of H3 histone. After establishment of latent infection of mice with LAT+ or LAT- virus, ganglia were harvested, and ChIP was conducted with antibodies specific for histone H3 (A), dimethyl lysine 9 form of histone H3 (B), or the dimethyl lysine 4 form of histone H3 (C). Real-time PCR analysis was performed in duplicate on the immunoprecipitates, and the fold-enrichment of viral promoter sequences was calculated as described in Materials and Methods. Statistical analysis was done by using the Friedman two-way analysis of rank.
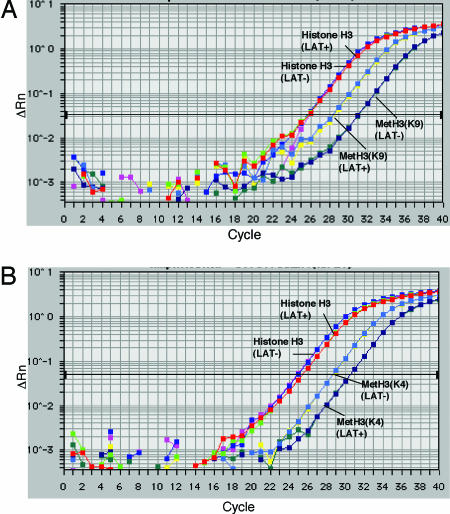
To examine the role of the LAT gene in the assembly and modification of chromatin on viral lytic gene promoters, we infected mice by the corneal route with the KdlLAT LAT- virus, in which sequences -792 to +1015 of the LAT gene transcriptional unit have been deleted (Fig. 3), or the KFSLAT LAT+ virus, in which the KdlLAT deletion has been repaired (8). Control experiments showed similar levels of histone H3 forms on KOS WT virus and KFSLAT virus lytic-gene promoters (results not shown) so we compared the LAT+ and LAT- viruses in these studies. Previous studies had shown that this LAT gene deletion results in higher levels of acute (8) and latent viral gene expression in ganglia (8, 18). At 30 dpi, the mice were euthanized, trigeminal ganglia were recovered, and ChIP was performed to determine the total amount of ICP27 gene promoter DNA associated with various forms of histone H3 (Fig. 4). In this experiment, the amounts of ICP27 gene promoter sequences associated with histone H3 were equivalent for the LAT+ versus LAT- infected ganglia (Fig. 4). Immunoprecipitation with an antibody specific for Met-H3 (K9) yielded ICP27 promoter DNA with a Ct value that was decreased by more than two cycles in the LAT+ sample relative to the LAT- virus sample (Fig. 4A), showing that >4-fold increased amounts of this promoter were associated with this form of H3 in LAT+ virus-infected ganglia. In contrast, immunoprecipitation with antibody specific for Met-H3(K4) showed DNA with a Ct value increased by two cycles in the LAT+ virus-infecting ganglia relative to the LAT- virus-infected sample (Fig. 4B), showing 4-fold decreased amounts of the Met-H3(K4) on this viral promoter in LAT+ virus-infected ganglia.
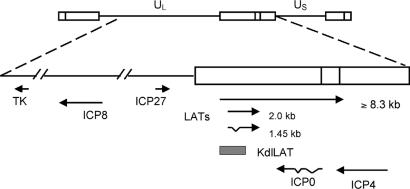
Fig. 3.
Map of the HSV-1 genome and the HSV genes involved in this study. At the top of the figure, the HSV-1 genome is shown in the prototype orientation. UL, unique long (L) region; US, unique short (S) region; open boxes, repeat regions. At the bottom are shown the locations and orientations of the TK, ICP8, ICP27, LAT, ICP0, and ICP4 genes. The hatched bar represents the region deleted in the KdlLAT viral genome.
Fig. 4.
Real-time PCR detection of HSV ICP27 gene promoter sequences associated with histone H3, dimethyl K4 histone H3, and dimethyl K9 histone H3. Ganglia from mice at 30 dpi (latent infection) were harvested, and ChIP was conducted with antibodies specific for histone H3 or the dimethyl lysine 4 or dimethyl lysine 9 forms of histone H3. Real-time PCR analysis was performed on the immunoprecipitates. ΔRn, fluorescence difference from background determined by using sequence detection system software (Applied Biosystems).
These results suggested that LAT increased the amount of heterochromatin and decreased the amount of active chromatin on viral lytic-gene promoters. We therefore conducted multiple, independent ChIP experiments to look at a group of representative immediate early and early viral gene promoters, the ICP4, ICP8, ICP27, and TK gene promoters. Comparisons of amounts of viral DNAs immunoprecipitated from LAT+ versus LAT- ganglia samples in assays for different histones and different promoters are shown in Fig. 5. For total histone 3, there was no significant difference in association with the lytic-gene promoters as a group between LAT+ and LAT- ganglia (P = 0.248). However, for Met-H3(K9), there was increased association lytic-gene promoters as a group in the LAT+ ganglia as compared with the LAT- ganglia (14/16 comparisons; P = 0.003). Equally importantly, for Met-H3(K4), there was decreased association of the lytic-gene promoters in the LAT+ ganglia as compared with the LAT- ganglia (17/17 comparisons; P < 0.0001). Thus, when LAT was expressed, lytic-gene promoters as a group showed increased amounts of associated Met-H3(K9) or heterochromatin and decreased amounts of Me-H3(K4) or active chromatin, consistent with the proposed role of LAT in repressing viral lytic-gene expression (8, 9). Nevertheless, there is likely to be gene-specific association with different forms of histones. The ICP8 gene promoter may be unique in being associated with higher amounts of H3 histone in the LAT- virus-infected ganglia (Fig. 5A). In addition, the ICP27 gene promoter may be associated with limited amounts of both K4 and K9 dimethylated forms of histone H3 (Fig. 5 B and C). Further studies are needed to better document the gene-specific differences in histone association.
Discussion
This study shows that HSV lytic-gene promoters become progressively associated with histones as infection of trigeminal ganglion neurons progresses. Low levels of histones were associated with viral DNA during the acute viral infection period from 1–5 dpi, but viral DNA became progressively associated with histone H3 through 15–30 dpi as latent infection is established. The amount of Met-H3(K9), a component of heterochromatin or inactive chromatin, also accumulated on viral lytic gene promoters after 5 dpi. These results are consistent with chromatin assembly on viral DNA and/or histone modification playing a role in down-regulation of HSV gene infection during viral latent infection of sensory neurons.
We observed that expression of LATs correlated with a 2- to 4-fold increased association of Met-H3(K9) with lytic-gene promoters and a decreased association of Met-H3(K4) with viral promoters of a similar magnitude. In three experiments, we saw a 4- to 8-fold shift to inactive chromatin when the LAT gene was expressed. Considering the lytic-gene promoters as a group, the increased association of Met-H3(K9) and decreased association of Met-H3(K9) in the LAT+ ganglia as compared with the LAT- ganglia were statistically significant. The magnitude of the effects may be underestimated because, in these studies, we observed increased lethality with the LAT- virus-infected animals as compared with the LAT+ virus-infected animals (29% versus 18%; results not shown). The increased lethality is consistent with our hypothesis that LAT serves to control lytic-gene expression in neurons. The effect of LAT may also be under-estimated due to death of neurons in the LAT- virus-infected, surviving animals, as documented by Thompson and Sawtell (11). The magnitude of the effects that we observed is consistent with previously observed effects of LAT: 3-fold higher numbers of latently infected cells (7), 3-fold higher levels of latent viral genomes (11), 4-fold lower levels of neurons expressing ICP4 at 3 dpi (8), 5-fold lower levels of ICP4 gene transcripts, and 10-fold lower levels of TK gene transcripts at 30 dpi (9).
Although the process of heterochromatin assembly on the HSV genome could, in principle, be a passive process that occurs as a result of the relative inactivity of the viral lytic genes in sensory neurons, our results demonstrate that the HSV LAT transcriptional unit or a gene product from this region plays an active role in this process. It is formally possible that LAT-mediated repression occurs by mechanisms unrelated to chromatin, and that the methylation of histone H3 K9 and the demethylation of H3 K4 are indirect consequences of this repression. Nevertheless, an attractive hypothesis is that LAT acts directly on chromatin structure, which leads to a repression of lytic-gene expression. Because (i) the LATs are not generally thought to encode any protein products and (ii) the histone modification that we have shown to be promoted by LAT, methylation of histone H3 at the K9 residue, is stimulated by RNA-silencing mechanisms (22), it is tempting to speculate that the LAT RNA, or some portion of it, induces a small interfering RNA (siRNA)-like effect that causes heterochromatin formation on viral lytic-gene promoters. Because several HSV genes are affected by LAT expression, this could be a mechanism different from the gene-specific silencing mechanisms that have been documented. Alternatively, the major (2-kb) LAT, which overlaps ICP0 (Fig. 3) and minor species (>8.3 kb) from the transcription unit that may overlap the ICP4 gene (8), could silence these two genes needed for activation of expression of the other viral genes. There is no evidence that LAT affects the levels of expression of ICP0 gene transcripts in ganglia (23), but ICP0 has been shown to associate with a complex that inhibits formation of active chromatin (24). Thus, ICP0 may promote formation of active chromatin on viral promoters whereas LAT promotes formation of heterochromatin by down-regulating expression of ICP0, at the transcriptional and/or translational level. As an alternative or additional possibility, a LAT deletion mutant virus does show reduced expression of a transcript complementary to ICP4 called Anti-4 (23), which might arise from the LAT promoter. The Anti-4 transcript could play a role in down-regulating ICP4, which in turn could up-regulate other viral genes in neurons, perhaps by initiating formation of dsRNA and thus siRNA that could participate in heterochromatin formation (22). Finally, it is conceivable that the LAT gene promoter could exert cis-acting effects on lytic-gene promoters in the viral genome.
HSV LAT has been reported to exert several effects on viral latent infection, including promotion of reactivation, establishment of latent infection (6), repression of lytic-gene expression during acute (8, 9) and latent infection (9), and prevention of neuronal cell death by apoptotic (10) or other mechanisms (11). Bloom and coworkers (16) have shown that the LAT promoter and coding sequences have histone H3 acetylated at lysines 9 and 14 assembled onto them, consistent with the active nature of the LAT transcriptional unit. There may be barriers that keep the LAT transcriptional unit assembled into euchromatin whereas the rest of the genome-containing lytic-gene promoters are assembled into heterochromatin. The limited viral gene expression could prevent induction of apoptosis in the neurons and/or prevent recognition of infected cells and induction of apoptosis by cytotoxic T lymphocytes. Thus, the numerous effects ascribed to LAT could be reconciled if LAT promotes deposition of inactive chromatin on lytic-gene promoters, which leads to a repression of lytic-gene expression.
HSV establishes a latent infection in sensory neurons to persist in humans and has evolved mechanisms that allow it to maintain a quiescent form that is not detected by our immune system. Little is known about the mechanisms by which viral gene expression is down-regulated in neurons. The results in this study point to histone modifications as a critical mechanism in regulation of HSV gene expression during latent infection in sensory neurons. Viruses often target important cellular regulatory mechanisms to promote their own replication, spread, and survival. We hypothesize that HSV uses the LAT gene or one of its products to manipulate the cellular histone modification machinery to assemble heterochromatin in place of euchromatin on viral lytic-gene promoters and shut down the expression of these viral genes and allow the virus to persist as a latent infection in a quiescent form in its human host.
Acknowledgments
We thank Steve Buratowski, Elliott Kieff, and members of the D.M.K. laboratory for their helpful discussions on this work and manuscript and Lisa Holik for assistance in preparation of the manuscript. This research was supported by National Institutes of Health Grant P01 NS35138.
Author contributions: Q.-Y.W., D.M.C., and D.M.K. designed research; Q.-Y.W. and C.Z. performed research; Q.-Y.W., C.Z., K.E.J., R.C.C., D.M.C., and D.M.K. analyzed data; and Q.-Y.W., C.Z., K.E.J., R.C.C., D.M.C., and D.M.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSV, herpes simplex virus; LAT, latency-associated transcript; ChIP, chromatin immunoprecipitation; TK, thymidine kinase; dpi, days postinfection; Ct, threshold cycle number; ICP, infected cell protein.
References
- 1.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Williams & Wilkins, Philadelphia), 4th Ed., pp. 2399-2460.
- 2.Whitley, R. J. & Roizman, B. (2001) Lancet 357, 1513-1518. [DOI] [PubMed] [Google Scholar]
- 3.Wald, A. & Link, K. (2002) J. Infect. Dis. 185, 45-52. [DOI] [PubMed] [Google Scholar]
- 4.Stevens, J. G., Wagner, E. K., Devi-Rao, G. B., Cook, M. L. & Feldman, L. T. (1987) Science 235, 1056-1059. [DOI] [PubMed] [Google Scholar]
- 5.Zwaagstra, J. C., Ghiasi, H., Slanina, S. M., Nesburn, A. B., Wheatley, S. C., Lillycrop, K., Wood, J., Latchman, D. S., Patel, K. & Wechsler, S. L. (1990) J. Virol. 64, 5019-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leib, D. A., Bogard, C. L., Kosz-Vnenchak, M., Hicks, K. A., Coen, D. M., Knipe, D. M. & Schaffer, P. A. (1989) J. Virol. 63, 2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson, R. L. & Sawtell, N. M. (1997) J. Virol. 71, 5432-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garber, D. A., Schaffer, P. A. & Knipe, D. M. (1997) J. Virol. 71, 5885-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. H., Kramer, M. F., Schaffer, P. A. & Coen, D. M. (1997) J. Virol. 71, 5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perng, G.-C., Jones, C., Ciacci-Zanella, J., Stone, M., Henderson, G., Yukht, A., Slanina, S. M., Hofman, F. M., Ghiasi, H., Nesburn, A. B. & Wechsler, S. L. (2000) Science 287, 1500-1503. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, R. L. & Sawtell, N. M. (2001) J. Virol. 75, 6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leinbach, S. S. & Summers, W. C. (1980) J. Gen. Virol. 51, 45-59. [DOI] [PubMed] [Google Scholar]
- 13.Herrera, F. J. & Triezenberg, S. J. (2004) J. Virol. 78, 9689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshmane, S. L. & Fraser, N. W. (1989) J. Virol. 63, 943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- 16.Kubat, N. J., Amelio, A. L., Giordani, N. V. & Bloom, D. C. (2004) J. Virol. 78, 12508-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leib, D. A., Coen, D. M., Bogard, C. L., Hicks, K. A., Yager, D. R., Knipe, D. M., Tyler, K. L. & Schaffer, P. A. (1989) J. Virol. 63, 759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer, M. F., Chen, S. H., Knipe, D. M. & Coen, D. M. (1998) J. Virol. 72, 1177-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noma, K., Allis, C. D. & Grewal, S. I. (2001) Science 293, 1150-1155. [DOI] [PubMed] [Google Scholar]
- 20.Litt, M. D., Simpson, M., Gaszner, M., Allis, C. D. & Felsenfeld, G. (2001) Science 293, 2453-2455. [DOI] [PubMed] [Google Scholar]
- 21.Boggs, B. A., Cheung, P., Heard, E., Spector, D. L., Chinault, A. C. & Allis, C. D. (2002) Nat. Genet. 30, 73-76. [DOI] [PubMed] [Google Scholar]
- 22.Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I. & Martienssen, R. A. (2002) Science 297, 1833-1837. [DOI] [PubMed] [Google Scholar]
- 23.Chen, S.-H., Lee, L. Y., Garber, D. A., Schaffer, P. A., Knipe, D. M. & Coen, D. M. (2002) J. Virol. 76, 4764-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu, H., Liang, Y., Mandel, G. & Roizman, B. (2005) Proc. Natl. Acad. Sci. USA 102, 7571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]