Abstract
To test the hypothesis that inaccurate DNA synthesis by mammalian DNA polymerase η (pol η) contributes to somatic hypermutation (SHM) of Ig genes, we measured the error specificity of mouse pol η during synthesis of each strand of a mouse Ig κ light chain transgene. We then compared the results to the base substitution specificity of SHM of this same gene in the mouse. The in vitro and in vivo base substitution spectra shared a number of common features. A highly significant correlation was observed for overall substitutions at A-T pairs but not for substitutions at G-C pairs. Sixteen mutational hotspots at A-T pairs observed in vivo were also found in spectra generated by mouse pol η in vitro. The correlation was strongest for errors made by pol η during synthesis of the non-transcribed strand, but it was also observed for synthesis of the transcribed strand. These facts, and the distribution of substitutions generated in vivo, support the hypothesis that pol η contributes to SHM of Ig genes at A-T pairs via short patches of low fidelity DNA synthesis of both strands, but with a preference for the non-transcribed strand.
High affinity antibodies result from somatic hypermutation (SHM) of Ig genes followed by selection. The SHM process introduces base substitutions at a very high rate into DNA encoding the variable regions of immunoglobulins (1–4). Although the mechanism for introducing these sequence changes is currently unknown, several features of SHM specificity offer clues to the DNA transactions that might be involved. For example, SHM primarily occurs in two highly mutable DNA sequence motifs (5–9). One is the RGYW sequence (the underlined G is mutated, R = A or G, Y = T or C, and W = A or T), which is found in SHM substitution spectra in equal proportions in both DNA strands. The other is the WA motif, where substitutions are more likely in one strand than the other (5, 7–10). A clue to the origins of the substitutions in the WA motif is the observation that their type and location correlates with the base substitution error specificity of human DNA polymerase η when copying a bacterial gene sequence in a model system in vitro (8). Based on that correlation and additional considerations of the two mutable motifs (11), we suggested that errors at A-T base pairs by pol η may contribute to as much as one third of somatic mutations in Ig genes, preferentially during synthesis of the non-transcribed strand. This hypothesis is supported by the observation that XP-V patients lacking active polymerase η have a lower proportion of somatic substitutions at A-T base pairs in Ig genes (12). Because pol η error specificity does not correlate with substitutions in the RGYW sequence motif, and because those substitutions are distributed equally on both strands, we further suggested that SHM may involve more than one DNA transaction and more than one DNA polymerase (8). This finding is consistent with the two-phase model of SHM proposed earlier (9, 13). Other DNA polymerases suggested to participate in SHM include pol ι (14–16), pol ζ (17, 18) and pol μ (19).
The present study tests the hypothesis that pol η is involved by taking advantage of two previous accomplishments. One is the description of 916 base substitutions arising during SHM of the mouse VκOx1 transgene (7). As one of the largest published collections of somatic mutations in an Ig gene, this spectrum most likely represents the intrinsic basis of hypermutation. The other is the expression and purification of recombinant mouse pol η (20), which has very low base substitution fidelity in a model fidelity assay system in vitro (21). In the present study, we modify the DNA template used for that in vitro fidelity assay, to monitor the base substitution error specificity of mouse pol η when synthesizing either the transcribed strand or the non-transcribed strand of the mouse VκOx1 gene sequence. We then compare pol η error specificity to the specificity of unselected substitutions generated during SHM of this same sequence in the mouse.
Materials and Methods
Materials.
All phage and bacterial strains and other materials used for the fidelity assay were from previously described sources (22). Recombinant mouse DNA polymerase η was expressed and purified as described (20).
Construction of New M13mp2 Derivatives.
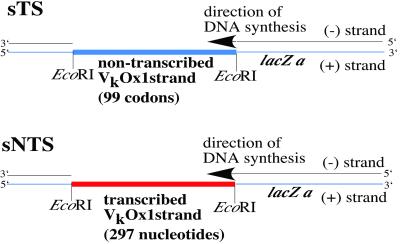
We constructed two new DNA substrates for fidelity assays by using the Ig κ light chain transgene VκOx1 in plasmid Lk-pSV2neo (MJS22Not) (23). The Ig gene (IG) was amplified by PCR by using the following primers with built-in EcoRI restriction sites: IG left 5′ GAT GAA TTC ACA AAT TGT TCT CAC CCA GT and IG right 5′ GAT GAA TTC AGT GGG TTA CTA CTC CAC T. The amplified product was cloned into the EcoRI site of the bacteriophage M13mp2 lacZ gene DNA, in both orientations (Fig. 1). The non-transcribed strand was inserted into the phage (+) strand DNA in frame with the lacZ gene. The resulting plaques (mp2-IG-sTS) are blue on 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) plates, although the intensity of blue color is less than for wild-type M13mp2 plaques because of the extra amino acids present at the N terminus of the β-galactosidase gene. In frame insertion permits confirmation of inaccurate synthesis by pol η by scoring plaque colors because of mutations in the transgene. Insertion of the transcribed strand into the phage (+) strand (construct mp2-IG-sNTS) produces six in-frame nonsense codons, resulting in colorless plaques. Gapped DNA substrates were prepared (22) by annealing single stranded phage DNA ((+) strand) with denatured, double stranded M13mp2 DNA that has been cut with EcoRI.
Figure 1.
Gapped DNA substrates used in this study. (Upper) sTS – the non-transcribed strand of VκOx1 is inserted in frame (therefore blue color) into the EcoRI site in the N terminus of the β-galactosidase α-complementation gene in M13mp2. In this 297-nt gapped substrate, a DNA polymerase synthesizes the 282 nucleotides of the transcribed strand of the Ig gene. (Lower) sNTS – the transcribed strand of the VκOx1 gene (depicted in red, colorless phenotype because of in-frame nonsense codons, see Methods) is inserted into the EcoRI site. In this 297-nt gapped substrate, a DNA polymerase synthesizes the 282 nucleotides of the non-transcribed strand of the Ig gene. For convenience, only part of the 7.2-kB M13 mp2 circular duplex is shown in both substrates.
Assay for Base Substitution Errors Generated During DNA Synthesis.
DNA synthesis reactions by pol η were performed as described (21, 22). Both gapped substrates were filled to apparent completion (data not shown, but see figure 1 in ref. 24 for a typical analysis). DNA products were introduced into Escherichia coli to obtain independent plaques derived from individual molecules of copied DNA. Phage DNA samples were prepared from independent isolates chosen without color selection and sequenced. Previous studies show that, when copied DNA molecules are introduced into E. coli to score errors by plaque color, the newly synthesized strand is expressed with 40 to 60% efficiency (21, 22). Consistent with this observation, about 50% the mp2-IG-sNTS and mp2-IG-sTS isolates sequenced here contained one or more sequence changes resulting from errors by mouse pol η (Table 1).
Table 1.
DNA sequence changes found in M13-IG plaques derived from products of gap-filling synthesis
| Substrate | Total sequences with mutations | Observed sequence change
|
||
|---|---|---|---|---|
| Single base substitution | Tandem double substitution | Single base frameshift | ||
| mp2-IG-sNTS | 128 | 704 | 36 | 94 |
| mp2-IG-sTS | 134 | 582 | 25 | 113 |
No sequence changes were present in DNA from plaques derived from uncopied mp2-IG-sNTS and mp2-IG-sTS DNAs (nine of each).
In Vivo SHM Spectrum.
The collection of in vivo mutations in VkOx transgene has been described before (6, 7). We consider that this large dataset reflects intrinsic bias in somatic hypermutation process. The compilation includes data derived from transgenic light chains with multiple copies of the transgene and from cells selected in gut Peyer's Patches (PP). The multiple copies are targeted in the same cell even when the light chain they encode is not part of the antigen binding antibody molecule. This result implies that the majority of the mutations accumulated are unselected. In the case of PP-derived cells, the selective pressure is multiple; therefore again, the common denominator of the biases observed would reflect the intrinsic biases (6, 25).
Statistical Analysis.
Monte Carlo modification of the Pearson χ2 test of spectra homogeneity (26) and the Kendall's tau correlation coefficient (27, 28) were used to compare spectra. Calculations were done by using the programs corr12 (27, 28) and hg-publ (29). To simultaneously examine the correlation between three spectra by multiple regression analysis, the “linear regression module” of the program statistica was used. Correlation between a distribution of mutable motifs and a distribution of mutations along a target sequence was measured by using a Monte Carlo procedure (5). Small values (< 0.05) of the probability P(W ≤ Wrandom) indicate a significant correlation between a mutable motif and mutations (5, 8). Hotspots were predicted as described (28).
Results
System to Monitor Error Specificity During Synthesis of Each DNA Strand of IG Gene.
We previously developed an assay to measure the fidelity with which DNA polymerases copy the E. coli lacZ α-complementation gene sequence that codes for β-galactosidase (22). The 407-nt, single-stranded template is present in a gap in otherwise double-stranded M13mp2 DNA. Correct synthesis to fill the gap generates DNA products that yield blue M13 plaques when introduced into E. coli cells that are then plated on indicator plates. Errors made during gap filling are scored as lighter blue or colorless plaques, and the nature of the polymerase error is determined by DNA sequence analysis of these mutant M13mp2 plaques. Previous studies (24) using this system showed that pol η has very low fidelity. Average base substitution error rates for the human and mouse enzymes were 3.2 × 10−2 and 2.2 × 10−2, respectively (21). Because of this, the vast majority of DNA products of gap filling contain multiple base substitution errors. Under this extraordinary circumstance, pol η error specificity can be obtained by simply sequencing M13mp2 plaques derived from the products of gap filling, without any phenotypic scoring. This approach provides a unique opportunity to investigate the mutagenic potential of pol η during copying of either strand of an actual IG gene sequence. To do this, we inserted the Ig κ light chain transgene VκOx1 into the unique EcoRI site in the LacZ sequence in two different orientations and prepared two gapped substrates (Fig. 1). In one instance (mp2-IG-sTS), pol η synthesizes 282 nt of the transcribed strand of the VκOx1 gene (plus a few nucleotides of the EcoRI linker), and, in the other case (mp2-IG-sNTS), it synthesizes the non-transcribed strand.
Pol η Error Specificity During Synthesis of the Transcribed and Non-Transcribed Strands of IG Gene.
Mouse pol η was used to synthesize the transcribed or the non-transcribed strand, and DNA products were introduced into E. coli and plated to recover M13 plaques from which DNA samples of unselected independent plaques were sequenced. As expected based on earlier studies (21, 24), about 50% of phage obtained after transfection by the products of inaccurate pol η gap filling contained a large number and a variety of sequence changes (Table 1). In both spectra, the predominant changes were single base substitutions, indicative of all twelve possible base-base mismatches, in a variety of sequence contexts throughout the 282 nt of VκOx1 gene (Table 2; Fig. 2). Several features of pol η error specificity are particularly relevant to SHM. Both here (Fig. 2, Tables 1 and 2) and in our earlier study (21), mouse pol η generated base pair substitutions much more frequently than frameshifts. Among base substitutions, mutations at A-T base pairs were produced more frequently than at G-C pairs (ref. 21, Fig. 2, Table 2). Pol η generated transversions at A-T pairs at lower rates than transitions; transversions comprised 40% of the sTS spectrum and 34% of the sNTS spectrum. The transitions mostly resulted from a higher error rate for misincorporation of dGMP opposite template T than for misincorporation of dCMP opposite template A (ref. 21, Fig. 2, Table 2). Misincorporation of dGMP opposite T preferentially occurred when template T was preceded by an A-T or T-A base pair (ref. 21, and see below), a sequence context that conforms to the WA substitution hotspot motif seen in SHM. The proportion of potential mutable sites is similar for two strands: the NTS has 24 WA sites whereas the TS has 19 WA sites. For the comparisons made below, note that, when displayed by using the sequence of the non-transcribed strand, T-dGMP errors are seen as A to G transitions for synthesis of the non-transcribed strand (Fig. 2, filled green circles, and Table 2, mp2-IG-sNTS). They are depicted as T to C transitions (Fig. 2, open blue circles and Table 2, mp2-IG-sTS) for synthesis of the transcribed strand.
Table 2.
No. of substitutions in mouse spectra in vivo and in vitro
| Spectrum | Total base substitutions | Types of base substitutions (percent from the total)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutations at A-T base pairs
|
Mutations at G-C base pairs
|
||||||||||||||
| Transitions
|
Transversions
|
Total at AT | Transitions
|
Transversions
|
Total at GC | ||||||||||
| A→G | T→C | A→T | A→C | T→A | T→G | G→A | C→T | G→T | G→C | C→G | C→A | ||||
| VκOx1 in vivo | 916 | 147 (16) | 74 (8) | 95 (10) | 56 (6) | 30 (3) | 18 (2) | 420 (46) | 205 (22) | 175 (19) | 25 (3) | 32 (4) | 32 (3) | 27 (3) | 496 (54) |
| mp2-IG-sTS | 632 | 15 (2) | 262 (41) | 80 (13) | 46 (7) | 39 (6) | 16 (3) | 458 (72) | 45 (7) | 77 (12) | 5 (1) | 18 (3) | 3 (<1) | 26 (4) | 174 (28) |
| mp2-IG sNTS | 776 | 364 (47) | 30 (4) | 37 (5) | 20 (3) | 90 (11) | 49 (6) | 590 (76) | 48 (6) | 75 (10) | 15 (2) | 1 (<1) | 29 (4) | 18 (2) | 186 (24) |
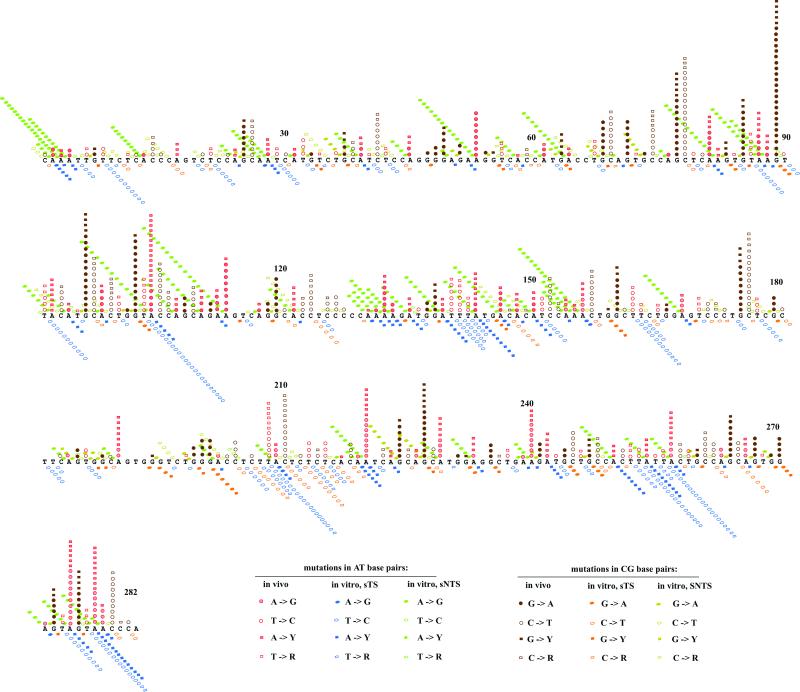
Figure 2.
Three-dimensional representation of SHM and mouse pol η substitution spectra. The in vivo spectrum is shown by vertical columns of red and brown symbols, each representing a single occurrence. The spectrum of mutations produced by pol η during synthesis of the nontranscribed strand is shown by green and khaki symbols extending from the mutable position to the left and behind the sequence. The spectrum of mutations produced by pol η during synthesis of the transcribed strand is shown by blue and yellow symbols extending from the mutable position to the right and in front of the sequence.
Comparison of in Vivo and in Vitro Substitution Specificity at G-C Pairs.
To compare the types of substitutions generated during SHM in the VκOx1 gene in mice to those generated by pol η during synthesis of each DNA strand, all three spectra are displayed in Fig. 2 by using the sequence of the non-transcribed strand. The in vivo hotspots in the database used likely reflect intrinsic properties of SHM (6, 7). For substitutions at G-C base pairs, two substantial differences between the in vivo and in vitro spectra were apparent. First, substitutions at G-C comprise a very different proportion of the spectra. Fifty-four percent (496 of 916, see Table 2) of the mutations in vivo were at G-C pairs (brown symbols in Fig. 2). However, only about 26% of the mouse pol η error spectra in vitro were at G-C pairs (khaki and yellow symbols for synthesis of the non-transcribed strand and transcribed strand, respectively). Secondly, the distribution of mutations at G-C pairs in the in vivo spectrum is very different from the distribution in the two in vitro spectra (Fig. 2). Five or more G to A transitions were observed in vivo at base pairs 25, 68, 77, 85, 96, 119, 160, 175, 224, 264, 272, and 275 (brown circles, 87 total substitutions). However, not a single G to A was generated by mouse pol η at any of these positions during synthesis of either strand (Fig. 2). These obvious differences are supported by statistical analysis revealing no significant correlation between SHM of the VκOx1 gene in mice and errors at G-C base pairs generated by mouse pol η during synthesis of either strand. This result was the case when all errors at G-C pairs were considered (Table 3, line 1), or when the RGYW and WRCY hotspots for SHM were analyzed (Table 4).
Table 3.
Correlation between in vivo and in vitro spectra
| Site (no.) | Spectra compared
|
||
|---|---|---|---|
| VκOx1 in vivo and mp2-(IG-sNTS) | VκOx1 in vivo and mp2-IG-(sTS) | mp2-IG-(sTS) and mp2-IG-(sNTS) | |
| G-C (146) | −0.05 | 0.05 | 0.01 |
| A-T (136) | 0.33 (<0.001) | 0.12 (0.03) | −0.07 |
Table 4.
Mutations in different mutable motifs
| Spectrum | Sequence motif
|
|||
|---|---|---|---|---|
| WA | TW | RGYW | WRCY | |
| VκOx1 | 3.1 | 1.5 | 4.2 | 4.0 |
| mp2-IG-sNTS | 4.4 | 1.5 | 0.7 | 0.2 |
| mp2-IG-sTS | 1.9 | 6.8 | 0.6 | 0.9 |
The values listed represent the fold increase in occurrence of mutations at a mutable site above the average occurrence of mutations at non-hotspot sites. Underlined values represent a statistically significant correlation (P < 0.001) between a mutable motif and the distribution of mutations, as revealed by using a Monte Carlo procedure (5, 27).
Comparison of SHM to Errors at A-T Pairs During Synthesis of the Non-Transcribed Strand.
Several features of somatic mutations at A-T base pairs in mice correlated well with the spectrum of substitutions at A-T base pairs generated by mouse pol η (Tables 3 and 4). The correlation was particularly strong for errors during synthesis of the non-transcribed DNA strand, with Pcc values of <0.001 (Table 3; see definition of Pcc). Over 50% of the substitutions at A-T base pairs in all three spectra were transitions (Table 2). Among these, the in vivo spectrum contained two times as many A to G transitions (Table 2 and Fig. 2, filled red circles) as T to C transitions (open red circles). The bias for A to G substitutions during SHM in mice (Table 2, line 1) most closely matches transition errors made by pol η during synthesis of the non-transcribed strand (Table 2, line 3, mostly A to G). For substitutions in specific sequence contexts, errors by mouse pol η were overrepresented at the WA sequence motifs (TW in the sTS spectrum) by factors of 4- to 7-fold (Table 4). This result is similar to the preference for substitutions in the WA motif during SHM at the same sequence in the mouse (Table 4). A number of matches were also seen when individual A-T hotspots were considered. Table 5 lists five examples in the VκOx1 transgene (taken from Fig. 2) where mutations at A-T pairs were frequently generated during SHM in vivo and during synthesis of the non-transcribed strand by pol η in vitro. Collectively, these observations support the hypothesis that errors made by pol η during synthesis of the non-transcribed strand of the VκOx1 transgene contribute to SHM in mice.
Table 5.
Substitution hotspots at A-T pairs
| Position | Sequence | No. of transitions/ transversions observed
|
||
|---|---|---|---|---|
| In vivo | sNTS | sTS | ||
| Correlation of SHM with the sNTS spectrum | ||||
| 53 | GAG AA GGT | 8/0 | 15/0 | 0/0 |
| 87 | GTG TA AGT | 4/4 | 11/0 | 0/1 |
| 113 | CAG AA GTC | 8/1 | 18/0 | 0/0 |
| 132 | CAA AA GAT | 5/2 | 8/0 | 0/2 |
| 240 | CTG AA GAT | 7/3 | 4/0 | 0/1 |
| 274 | GAG TA GTA | 10/6 | 6/0 | 0/1 |
| Correlation of SHM with the sTS spectrum | ||||
| 103 | TGG TA CCA | 4/2 | 1/0 | 17/7 |
| 140 | GGA TT TAT | 5/1 | 1/0 | 9/0 |
| 141 | GAT TT ATG | 4/2 | 0/0 | 6/1 |
| 163 | GGC TT CTG | 4/1 | 1/0 | 10/3 |
| 208 | TCT TA CTC | 5/6 | 0/0 | 18/1 |
| Correlation of SHM with both in vitro spectra | ||||
| 104 | TGG TA CCA | 4/15 | 14/0 | 0/5 |
| 143 | ATT TA TGA | 2/7 | 7/0 | 0/9 |
| 220 | CAC AA TCA | 8/6 | 8/0 | 1/2 |
| 257 | TAT TA CTG | 3/6 | 5/0 | 1/2 |
| 277 | TAG TA ACC | 6/9 | 7/0 | 0/13 |
The clusterm program (27) was used for hotspot prediction. The mutated base is underlined.
Comparison of SHM to Errors at A-T Pairs During Synthesis of the Transcribed Strand.
Several observations suggest that pol η may also generate mutations in vivo during synthesis of the transcribed DNA strand. First, numerous T to C transitions are observed in vivo, and they are also characteristic of the sTS spectrum (Table 2). Second, a similar ratio of transversions at A-T pairs is observed in vivo and in the sTS spectrum (Table 2). Third, mutations at A-T pairs are frequently generated during SHM in vivo at locations matching errors made by pol η during synthesis of the transcribed strand (Fig. 2. and see examples in Table 5). There were significant correlations between somatic mutations at A-T base pairs and the spectrum of substitutions generated when mouse pol η synthesized the transcribed DNA strand (Table 3, Pcc = 0.03), and between in vivo substitutions and the TW motif [Table 4, P(W ≤ Wrandom) < 0.001].
Comparison of SHM to Errors at A-T Pairs During Synthesis of Both Strands.
Table 5 shows five examples of in vivo hotspots that match errors by pol η during synthesis of both strands. When mutations at A and T bases were analyzed separately, significant correlations were observed between somatic mutations and errors generated by pol η during synthesis the both strands (Table 6). Both results indicate that pol η may operate in vivo on both strands. This finding prompted multiple linear regression analysis of the relationship between all three mutational spectra simultaneously. A highly significant positive correlation was found between the distributions of somatic mutations and pol η errors at A-T pairs (Table 7). A larger regression coefficient B (which represents the independent contributions of each of in vitro spectrum to the prediction of the SHM spectrum) was found for the sNTS comparison, once again suggesting that errors during synthesis of the non-transcribed strand make a larger contribution to SHM.
Table 6.
Correlations between spectra for substitutions at either adenine or thymine
| Base (no.) | Spectra compared
|
||
|---|---|---|---|
| VκOx1 and mp2-IG-sNTS | VκOx1 and mp2-IG-sTS | mp2-IG-sTS and mp2-IG-sNTS | |
| A sites (71) | 0.28 (<0.001) | 0.19 (0.02) | 0.00 |
| T sites (65) | 0.19 (0.03) | 0.41 (<0.001) | 0.09 |
Nos. in parentheses are Pcc values (see note to Table 3).
Table 7.
Multiple regression analysis of the correlation between somatic mutation and pol η errors at A-T pairs
| Spectrum | B value | β value | P level |
|---|---|---|---|
| mp2-IG-sNTS | 0.35 | 0.45 | 0.000001 |
| mp2-IG-sTS | 0.20 | 0.27 | 0.0006 |
The spectrum of somatic mutations was used as the dependent variable. Multiple regression procedures estimate a linear equation of the form Y = A + B1⋅X1 + B2⋅X2 + … + Bn⋅Xn where A is a constant and B are regression coefficients that represent the independent contributions of each independent variable to the prediction of the dependent variable. This type of correlation is also referred to as a partial correlation. Partial correlation coefficients β are closely related to B coefficients, such that a test of significance of β (P level) helps to evaluate the importance of each independent variable separately.
Discussion
The similarities reported here between the specificity of SHM in mouse and the error specificity of DNA polymerase η in vitro are consistent with the hypothesis that mammalian DNA polymerase η contributes to a subset of mutations during SHM. This finding supports the interpretations of our earlier study (8). In the present study, using the same VκOx1 target sequence, in vivo–in vitro correlations are seen for substitutions at A-T pairs but not G-C pairs, for errors during synthesis of the non-transcribed strand and to a lesser extent the transcribed strand, for transitions because of misincorporation of dGMP opposite template T, for transversions when in vitro synthesis of the transcribed strand is considered, and for errors in the WA sequence motif. Each of these specificity features represents a preference, not an exclusive characteristic. Collectively, they suggest that pol η (or an undiscovered polymerase with similar error specificity) conducts inaccurate DNA synthesis of both DNA strands of Ig genes, with some preference used for synthesis of the non-transcribed strand.
In principle, errors by pol η could be made during continuous DNA synthesis of the whole undamaged non-transcribed strand (frequently) or the whole transcribed strand (rarely). Continuous synthesis by using unmodified templates is unlikely based on earlier studies (7, 30, 31), especially including the fact that pol η rarely produces mutations at G-C pairs in vitro whereas these predominate in SHM spectra and are often found in the same lineage with A-T mutations (7, 30). Alternatively, inaccurate DNA synthesis by pol η could occur in short patches, as suggested here by the non-random substitution pattern at A-T pairs in the in vivo spectrum (red symbols in Fig. 2) and discontinuous correlation of hotspots with two in vitro spectra. Indeed, 16 SHM hotspots at A-T pairs in the VκOx1 gene correlate with errors in the sNTS spectrum, the sTS spectrum, and both spectra (Fig. 2 and Table 5), and these hotspots are interspersed throughout the 282-bp sequence. Short patch synthesis is also consistent with the intrinsically low processivity of pol η (refs. 32 and 33; and F.H., unpublished results). Therefore, our data are consistent with the possibility that pol η performs inaccurate short-patch DNA synthesis after being recruited to specific locations on the transcribed strand to perform synthesis of the non-transcribed strand, and less frequently, to locations on the opposite strand. This finding suggests that specific signals may attract pol η to specific regions on the nontranscribed or transcribed DNA strand.
How might pol η recruitment occur? SHM depends on transcription and the distance from the transcription initiation site (34), and it may be initiated at nicks in DNA (35). Excision repair of certain types of DNA damage involves nicking the DNA, is coupled to transcription, and requires short patch DNA synthesis. Pol η is clearly one polymerase involved in cellular response to DNA damage. These facts suggest a model wherein an IG gene is somehow targeted for damage that initiates a process of incision, excision, and inaccurate short-patch gap-filling that leads to SHM at A-T pairs. Although the nature of the putative damage is unknown, SHM does require the AID gene (36, 37), which is a cytidine deaminase that can use deoxycytidine as a substrate (38, 39). Cytosine deamination produces uracil, which codes like thymine and can yield C to T transitions without the need for an error-prone polymerase. However, uracil removal from DNA by base excision repair involves synthesis of short patches of DNA (40). A number of mammalian DNA polymerases have already been implicated in gap-filling during various excision repair reactions, including DNA polymerases β, δ, ɛ, ι, and λ. Thus, it is reasonable to hypothesize that inaccurate pol η might be recruited to contribute to somatic hypermutation via a short gap-filling reaction. It is also possible that SHM could be triggered by an abasic site that stalls replication, thus recruiting an inaccurate polymerase for extension by several nucleotides before accurate replication resumes (see ref. 41 for discussion of the role of abasic sites in SHM). Thus, damage to G-C base pairs might lead to mutations at G-C pairs and concomitantly recruit enzymes for SHM at A-T base pairs. This possibility is consistent with the observation that five of six WA SHM hotspots correlating with the pol η sNTS spectrum are flanked by a 5′-template G (Table 5). The idea can also account for the fact that the average number of in vivo mutations in all WA motifs fused with RGYW motifs is greater than in WA sites not fused with RGYW (11, 42). The strand bias in mutations in A-T sites would then partly depend on the local concentration and orientation of modified G-C pairs and the distance of putative A-T hotspots from these cytosines in each strand and should vary from gene to gene, which is again what is seen in a larger number of target sequences (30). Because the signal (damaged nucleotide) and the patch synthesized with low fidelity may be on opposite strands, their simultaneous repair could result in double strand breaks, another hallmark of SHM (43). Finally, if a modified nucleotide were to somehow persist, it could give rise to mutations in several cell divisions, as observed in vivo (30).
The lack of a correlation between SHM at G-C pairs and the error specificity of pol η is consistent with the participation of one or more other polymerases in SHM. Possibilities include pol ι (14–16), pol μ (ref. 19, but see refs. 44 and 45) and pol ζ (17, 18). Pol ζ is expressed in germinal centers, and evidence suggests that it participates in SHM, possibly by extending mismatches during the transition from pol η to accurate replicative DNA polymerases (17, 18). If pol ι was somehow recruited to copy deaminated cytosine, it could produce both transitions and transversions because of its low fidelity when copying template T or U (16, 46).
In models involving chemical modification of the template, preferential mutagenesis at G-C pairs in RGYW motifs would be determined not by the DNA polymerase but by the specificity of a DNA modifying enzyme (for example, AID, ref. 39, or an enzyme regulated by AID, ref. 47). In this circumstance, little or no correlation of error specificity of DNA polymerases with RGYW motifs would be expected, as observed here and earlier (8, 15, 16). Whatever role other polymerases may have, the contribution of pol η to SHM, when triggered, may be substantial. This result is suggested by the fact that 54% of somatic mutations in normal individuals occur at A-T sites, whereas only 18% of mutations at A-T sites were found in XP-V patients lacking pol η (12). This result and our earlier specificity analysis (11) led us to estimate that perhaps as much as 30% of SHM might be due to synthesis errors by pol η.
Acknowledgments
We are grateful to Drs. K. Bebenek and V. Poltoratsky for comments on the manuscript. This study was supported in part by an American Institute for Cancer Research (AICR) grant to C. Milstein (MRC Laboratory of Molecular Biology, Cambridge, U.K.) and in part by Russian Foundation of Fundamental Research (RFFR) Grant 02-04-48342.
Abbreviations
- SHM
somatic hypermutation
- IG
Ig gene
- Pcc, the probability that an observed correlation is due to random fluctuation.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tonegawa S. Nature (London) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Allen D, Cumano A, Dildrop R, Kocks C, Rajewsky K, Rajewsky N, Roes J, Sablitzky F, Siekevitz M. Immunol Rev. 1987;96:5–22. doi: 10.1111/j.1600-065x.1987.tb00506.x. [DOI] [PubMed] [Google Scholar]
- 3.Neuberger M S, Milstein C. Curr Opin Immunol. 1995;7:248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 4.Storb U. Immunol Rev. 1998;162:5–11. doi: 10.1111/j.1600-065x.1998.tb01424.x. [DOI] [PubMed] [Google Scholar]
- 5.Rogozin I B, Kolchanov N A. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 6.Betz A G, Rada C, Pannell R, Milstein C, Neuberger M S. Proc Natl Acad Sci USA. 1993;90:2385–2388. doi: 10.1073/pnas.90.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milstein C, Neuberger M S, Staden R. Proc Natl Acad Sci USA. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogozin I B, Pavlov Y I, Bebenek K, Matsuda T, Kunkel T A. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 9.Spencer J, Dunn M, Dunn-Walters D K. J Immunol. 1999;162:6596–6601. [PubMed] [Google Scholar]
- 10.Oprea M, Cowell L G, Kepler T B. J Immunol. 2001;166:892–899. doi: 10.4049/jimmunol.166.2.892. [DOI] [PubMed] [Google Scholar]
- 11.Rogozin I B, Pavlov Y I, Kunkel T A. Nat Immunol. 2001;2:983–984. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 12.Zeng X, Winter D B, Kasmer C, Kraemer K H, Lehmann A R, Gearhart P J. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 13.Rada C, Ehrenstein M R, Neuberger M S, Milstein C. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 14.Poltoratsky V, Goodman M F, Scharff M D. J Exp Med. 2000;192:F27–F30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poltoratsky V, Woo C J, Tippin B, Martin A, Goodman M F, Scharff M D. Proc Natl Acad Sci USA. 2001;98:7976–7981. doi: 10.1073/pnas.141222198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank E G, Tissier A, McDonald J P, Rapic-Otrin V, Zeng X, Gearhart P J, Woodgate R. EMBO J. 2001;20:2914–2922. doi: 10.1093/emboj/20.11.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz M, Verkoczy L K, Flajnik M F, Klinman N R. J Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- 18.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik M F, Diaz M, Casali P. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz J F, Dominguez O, Lain de Lera T, Garcia-Diaz M, Bernad A, Blanco L. Philos Trans R Soc Lond B Biol Sci. 2001;356:99–109. doi: 10.1098/rstb.2000.0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada A, Masutani C, Iwai S, Hanaoka F. Nucleic Acids Res. 2000;28:2473–2480. doi: 10.1093/nar/28.13.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda T, Bebenek K, Masutani C, Rogozin I B, Hanaoka F, Kunkel T A. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 22.Bebenek K, Kunkel T A. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 23.Sharpe M J, Milstein C, Jarvis J M, Neuberger M S. EMBO J. 1991;10:2139–2145. doi: 10.1002/j.1460-2075.1991.tb07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel T A. Nature (London) 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Fernandez A, Milstein C. Proc Natl Acad Sci USA. 1993;90:9862–9866. doi: 10.1073/pnas.90.21.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams W T, Skopek T R. J Mol Biol. 1987;194:391–396. doi: 10.1016/0022-2836(87)90669-3. [DOI] [PubMed] [Google Scholar]
- 27.Babenko V N, Rogozin I B. Biofizika. 1999;44:632–638. [PubMed] [Google Scholar]
- 28.Rogozin I B, Kondrashov F A, Glazko G V. Hum Mutat. 2001;17:83–102. doi: 10.1002/1098-1004(200102)17:2<83::AID-HUMU1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.Cariello N F. Mutat Res. 1994;312:173–185. doi: 10.1016/0165-1161(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 30.Michael N, Martin T E, Nicolae D, Kim N, Padjen K, Zhan P, Nguyen H, Pinkert C, Storb U. Immunity. 2002;16:123–134. doi: 10.1016/s1074-7613(02)00261-3. [DOI] [PubMed] [Google Scholar]
- 31.Bertocci B, Quint L, Delbos F, Garcia C, Reynaud C A, Weill J C. Immunity. 1998;9:257–265. doi: 10.1016/s1074-7613(00)80608-1. [DOI] [PubMed] [Google Scholar]
- 32.Masutani C, Kusumoto R, Iwai S, Hanaoka F. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bebenek K, Matsuda T, Masutani C, Hanaoka F, Kunkel T A. J Biol Chem. 2001;276:2317–2320. doi: 10.1074/jbc.C000690200. [DOI] [PubMed] [Google Scholar]
- 34.Rada C, Milstein C. EMBO J. 2001;20:4570–4576. doi: 10.1093/emboj/20.16.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong Q, Maizels N. Genetics. 2001;158:369–378. doi: 10.1093/genetics/158.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 37.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 38.Longacre A, Storb U. Cell. 2000;102:541–544. doi: 10.1016/s0092-8674(00)00075-1. [DOI] [PubMed] [Google Scholar]
- 39.Martin A, Bardwell P D, Woo C J, Fan M, Shulman M J, Scharff M D. Nature (London) 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 40.Wilson S H, Sobol R W, Beard W A, Horton J K, Prasad R, Vande Berg B J. Cold Spring Harbor Symposia on Quantitative Biology. Vol. 65. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 143–155. [DOI] [PubMed] [Google Scholar]
- 41.Storb U, Peters A, Klotz E, Kim N, Shen H M, Kage K, Rogerson B, Martin T E. Curr Top Microbiol Immunol. 1998;229:11–19. doi: 10.1007/978-3-642-71984-4_2. [DOI] [PubMed] [Google Scholar]
- 42.Dorner T, Lipsky P E. Nat Immunol. 2001;2:982–984. doi: 10.1038/ni1101-982. [DOI] [PubMed] [Google Scholar]
- 43.Rogozin I B, Kunkel T A, Pavlov Y I. Trends Immunol. 2002;23:12–13. doi: 10.1016/s1471-4906(01)02112-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Wu X, Yuan F, Xie Z, Wang Z. Mol Cell Biol. 2001;21:7995–8006. doi: 10.1128/MCB.21.23.7995-8006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertocci B, De Smet A, Flatter E, Dahan A, Bories J C, Landreau C, Weill J C, Reynaud C A. J Immunol. 2002;168:3702–3706. doi: 10.4049/jimmunol.168.8.3702. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Yuan F, Wu X, Wang Z. Mol Cell Biol. 2000;20:7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. J Exp Med. 2002;195:529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]