Abstract
Fpg is a DNA glycosylase that recognizes and excises the mutagenic 8-oxoguanine (8-oxoG) and the potentially lethal formamidopyrimidic residues (Fapy). Fpg is also associated with an AP lyase activity which successively cleaves the abasic (AP) site at the 3′ and 5′ sides by βδ-elimination. Here, we present the high-resolution crystal structures of the wild-type and the P1G defective mutant of Fpg from Lactococcus lactis bound to 14mer DNA duplexes containing either a tetrahydrofuran (THF) or 1,3-propanediol (Pr) AP site analogues. Structures show that THF is less extrahelical than Pr and its backbone C5′–C4′–C3′ diverges significantly from those of Pr, rAP, 8-oxodG and FapydG. Clearly, the heterocyclic oxygen of THF is pushed back by the carboxylate of the strictly conserved E2 residue. We can propose that the ring-opened form of the damaged deoxyribose is the structure active form of the sugar for Fpg catalysis process. Both structural and functional data suggest that the first step of catalysis mediated by Fpg involves the expulsion of the O4′ leaving group facilitated by general acid catalysis (involving E2), rather than the immediate cleavage of the N-glycosic bond of the damaged nucleoside.
INTRODUCTION
Cellular DNA is continuously subjected to damaging agents from endogenous or exogenous origin (1). To prevent the deleterious effect of such damage (mutagenesis, diseases and cell death), all organisms studied to date have evolved a variety of DNA repair processes specific of the type of DNA damage (2,3). The modifications of the heterocyclic DNA base that are caused by agents involved in normal cellular metabolism resulting in oxidation, alkylation, deamination or base loss [formation of an abasic site, (AP)] are principally addressed by the base excision repair pathway (BER) (4). The DNA glycosylases, the key enzymes of the BER pathway, specifically recognize and excise the damaged base (DNA glycosylase activity) leading to the formation of an AP site in DNA. Among these enzymes, some of them are bifunctional and associated with an AP lyase activity able to cleave at the 3′ side the AP site by a β-elimination mechanism. The 3′-pre-incised AP site is then eliminated by a cleavage at its 5′ side by an AP endonuclease resulting in the formation of a 1 nt gap in DNA. Finally, DNA repair is achieved by the successive action of a DNA polymerase to fill in the gap and a DNA ligase to close up the repaired DNA strand.
Fpg (formamidopyrimidine–DNA glycosylase) is a bacterial DNA glycosylase/AP lyase enzyme involved in the repair of oxidized purines, such as 8-oxoguanine (8-oxoG) and imidazole-ring opened purines (FapyG, FapyA) (5–9). The enzyme eliminates the AP site by successive cleavage at its 3′ and 5′ sides by a βδ-elimination reaction (10,11). Fpg achieves its catalytic process using its P1 N-terminal amino group to perform a nucleophilic attack at the C1′ of the damaged nucleoside. During this process, the enzyme forms an imino enzyme–DNA intermediate which can be stabilized irreversibly by sodium borohydryde (12). Through its activities, Fpg counteracts the mutagenic effects of 8-oxoG and the AP site all together associated with G/T transversions and of the lethal potential of the Fapy residues which constitute DNA polymerase blocks, in vitro (8,9,13–15). As revealed by X-ray crystallographic studies of Fpg bound to a damage-containing DNA, Fpg gains access to its substrate nucleoside by extruding it from the DNA helix (16–20). The resulting extrahelical damaged nucleoside is then inserted into a substrate binding pocket of the enzyme and exposes its C1′ to the nucleophilic attack of the amino group of the P1 N-terminal residue.
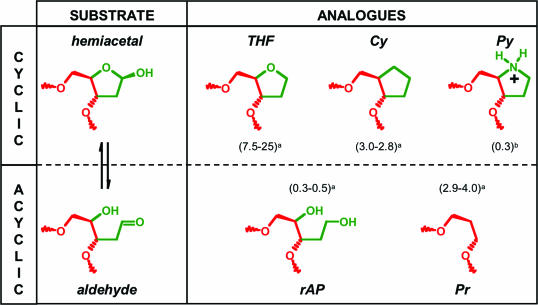
During the past decade, our understanding of the damaged DNA recognition by Fpg has advanced greatly. Structural, chemical and biochemical studies of Fpg interacting with DNA have led to depict the mechanisms underlying of the damaged nucleoside recognition. Two general approaches have been used so far to generate stable abortive Fpg/DNA complexes enabling to study at the molecular level the interaction of this enzyme with its substrates, reaction intermediate and products. One of these relies on site-directed mutagenesis of the active site residues to abolish catalysis, independently from the Fpg specific DNA binding (19). An alternative approach relies on the design and synthesis of uncleavable substrate analogues or inhibitors. One class of inhibitors recently consists of nucleobase analogues with a stabilized N-glycosydic bond, that are not processed by Fpg (21,22). The second class of inhibitors more extensively used for Fpg and other AP lyases contains cyclic and non cyclic AP site analogues that mimic the AP site structure in the catalytic transition state, preventing the Fpg end products synthesis (Figure 1) (23–25). We have employed both strategies to investigate the structural and/or functional determinants of the damaged nucleoside for Fpg specific recognition (16,20,26). In this work, we propose a comparative structural analysis of wild-type and mutant Fpg proteins bound to cyclic and non-cyclic AP site analogues containing DNA. The present four new crystal structures of Fpg/DNA complexes unfavour the recognition of the hemiacetal form of the AP site (Figure 1).
Figure 1.
Structure of abasic site substrate and analogues. Numbers in brackets correspond to the apparent dissociation constants (KD in nM) of EcFpg bound to analogue-containing DNA measured by electrophoresis mobility shift assay. When indicated, the two numbers in brackets indicate the binding constants for the EcFpg and LlFpg enzymes, respectively [a(24); b(25)]. AP site, abasic site; Pr, 1,3-propanediol; THF, tetrahydrofuran; rAP, reduced abasic site; Pyr, pyrolidine; Cy, cyclopentane.
MATERIALS AND METHODS
DNA and proteins
Self-complementary 14mer single-stranded oligonucleotides were purchased from Eurogentec (Belgium). After purification according to the procedure described by Pereira et al. (26), they were annealed with their complementary strand to generate blunt- or sticky-ended DNA duplexes containing an AP site analogue, 1,3-propanediol (Pr) or tetrahydrofuran (THF), at position 7 or 8. The 12 DNA duplexes (listed in Table 1) used in this study have nucleotide sequences on each side of the target site similar to those previously used (16,26). Wild-type LlFpg (W) and mutant P1G-LlFpg (M) (in which the catalytic P1 of the enzyme was replaced by G1) were purified as described previously. Each protein (W or M) was mixed with a 1.3 molar excess of DNA duplex to generate 24 protein/DNA complexes which were tested for systematic crystallization.
Table 1.
DNA duplexes used in this study
| DNA sequences | DNA nomenclature |
|---|---|
| GCTCTTTXTTTCTC | 8(X) |
| CGAGAAACAAAGAG | |
| GCTCTTTXTTTCTC | 8(X)3′ |
| GCGAGAAACAAAGA | |
| GCTCTTTXTTTCTC | 8(X)5′ |
| GAGAAACAAAGAGC | |
| CTCTTTXTTTCTCG | 7(X) |
| GAGAAACAAAGAGC | |
| CTCTTTXTTTCTCG | 7(X)3′ |
| CGAGAAACAAAGAG | |
| CTCTTTXTTTCTCG | 7(X)5′ |
| AGAAACAAAGAGCG |
The DNA nomenclature convention is as followed: the first number indicated the position from the 5′ end of the AP site analogue in the damaged strand and the letter (X) indicated the AP site analogue used (X = Pr or THF). For duplexes with sticky ends, the polarity of the overhang base is indicated by 3′ or 5′.
Crystallization
The 12 P1G-LlFpg (M)/DNA complexes were crystallized at room temperature by vapour diffusion with the hanging drop method, using Crystal Screen 1 (Hampton Research). For the initial crystallization trials, a 3 µl drop was prepared by mixing 1.5 µl of complex (5 mg/ml) with an equal volume of the reservoir solution. Numerous crystals appeared within a few days in several conditions for most of the complexes. The most promising results were obtained with the complexes M/7(Pr)5′ and M/7(THF)5′ with 1.4 M Sodium Citrate, 0.1 M Sodium HEPES, pH 7.5. After optimization of these conditions (1.2–1.6 M Sodium Citrate, 0.1 M Sodium HEPES, pH 7.5–8.5, complex concentration: 2 mg/ml, complex/reservoir solution ratio in the drop: 1/1 and 1/2), crystals of M/7(Pr)5′ and M/7(THF)5′ complexes with dimensions 100 × 100 × 700 µm were obtained. Similar conditions were used to crystallize the identical complexes named W/7(Pr)5′ and W/7(THF)5′ with the wild-type enzyme (W). All these complexes crystallize in the tetragonal space group P41212, with cell dimensions of a = b = 91 Å and c = 142 Å.
Data collection
Crystals selected for cryo-crystallographic experiments were soaked in their crystallization buffer supplemented with 15% glycerol as a cryo-protectant, before being flash-cooled at 100K in a nitrogen gas stream. Diffraction data were collected at the ID14 and BM30 beamlines at the ESRF (Grenoble, France) to 1.8 and 1.9 Å resolution for M/7(Pr)5′ and M/7(THF)5′ complexes, respectively, and to 1.9 Å resolution for both W/7(Pr)5′ and M/7(THF)5′ complexes. Each dataset was integrated with MOSFLM (27) and reduced by SCALA (28). Conversion of I to F was performed using TRUNCATE. Other calculations were carried out with the CCP4 program suite (29). Detailed statistics are presented in Table 2.
Table 2.
Diffraction data
| Enzyme | P1G-LlFpg (M) | Wild-type LlFpg (W) | ||
| Complex name | M/7(Pr)5′ | M/7(THF)5′ | W/7(Pr)5′ | W/7(THF)5′ |
| Wave lengths (Å) | 0.933 | 0.976 | ||
| Beamline | ID14 | BM30 | ||
| Parameters (Å) | a = b = 92.1 c = 142.5 | a = b = 91.9 c = 142.4 | a = b = 91.9 c = 142.1 | a = b = 90.8 c = 141.8 |
| Space group | P41212 | |||
| Resolution (Å) | 1.80 | 1.90 | 1.90 | 1.90 |
| Unique reflections | 56 858 | 48 703 | 48 727 | 43 717 |
| Redundancy | 11.6 | 9.6 | 2.5 | 6.7 |
| Completeness (%) | 97.6 | 100.0 | 99.2 | 97.5 |
| 〈I/σ(I)〉 | 6.1 | 5.2 | 16.5 | 10.2 |
| B-factor (Å2) (Wilson plot) | 28.3 | 27.7 | 30.7 | 29.4 |
| Rsym all data (last shell | 0.074 (0.627) | 0.073 (0.488) | 0.062 (0.53) | 0.045 (0.147) |
Phasing and refinement
The structures of both complexes formed with P1G-LlFpg (M/7(Pr)5′ and M/7(THF)5′) were solved by the molecular replacement method with the AMoRe program (30) using the atomic coordinates of P1G-LlFpg enzyme (protein atoms only) as the research model, [PDB code 1kfv, (16)] and diffraction data comprised between 20 and 4 Å resolution. The best solution obtained after translation function search corresponds to R-factors of 46.0 and 43.1% and correlation coefficients of 50.0 and 48.4%, respectively.
The whole refinement was performed with the CNS program (31). Prior to the refinement process, 5% of the reflection data were selected randomly and excluded from the refinement procedure (32). For each complex, the protein model was submitted to a cycle of simulated annealing at 3000K followed by some energy minimization cycles. At this stage, the electron density map indicated unambiguously the location of the whole DNA molecule, which was added manually in the protein model using the O program (33). The atomic models were then refined further by energy minimization and atomic B-factor refinement. Zinc ion, glycerol, water molecules and polypeptide regions that were missing in the initial model were added progressively to the models during the refinement procedure. Further refinement statistics are shown in Table 3.
Table 3.
Refinement
| Complex name | M/7(Pr)5′ | M/7(THF)5′ | W/7(Pr)5′ | W/7(THF)5′ |
|---|---|---|---|---|
| PDB code | 1NNJ | 1PJJ | 1PJI | 1PM5 |
| R.m.s.d. bond lengths | 0.0050 Å | 0.0049 Å | 0.0055 Å | 0.0046 Å |
| R.m.s.d. bond angles | 1.21° | 1.24° | 1.19° | |
| 〈B-factor〉(Å2) (DNA only) | 31.0 (38.4) | 31.0 (38.4) | 34.5 (43.1) | 30.9 (40.8) |
| Rfactor | 0.215 | 0.202 | 0.210 | 0.196 |
| Rfree (32) | 0.236 | 0.220 | 0.237 | 0.216 |
The structures of the complexes formed with wt-LlFpg (W/7(Pr)5′ and W/7(THF)5′) were built in the electron density map calculated after a step of rigid body refinement of these former models (REFMAC, (34)). The refinement was then conducted according to the same protocol.
Accession numbers
Atomic coordinates of these four models have been deposited in the Protein Data Bank under the accession codes 1NNJ [M/7(Pr)5′], 1PJJ [M/7(THF)5′], 1PJI [W/7(Pr)5′] and 1PM5 [W/7(THF)5′].
RESULTS
General overview of the enzyme–DNA complexes
This work presents the crystal structures of the wild-type (W) and the catalytic defective P1G-LlFpg mutant (M) bound to a DNA duplex containing either a 1,3-propanediol (Pr) or a THF (Figure 1) AP site analogue named as M/7(Pr)5′, M/7(THF)5′, W/7(Pr)5′ and W/7(THF)5′ (see the oligonucleotide nomenclature, Table 1).
Enzyme structure
These models were solved by the molecular replacement method based on the previous model of P1G-Ll-Fpg complexed to a 13mer DNA duplex containing a 1,3 propanediol nucleotide (16). The root-mean-square deviation (r.m.s.d.) values calculated on the backbone Cα atoms of the enzyme (∼0.15–0.5 Å) indicate that the enzyme Fpg adopts a similar conformation in all five models independently of the crystal packing and of the lesion type. These models are also equivalent to the ones of Fpgs purified from other organisms (Bacillus or Escherichia coli) and crystallized in different conditions with other damaged bases and different oligonucleotide sequences (17–19). The superposition with these latter models leads to small r.m.s.d. as well (∼1.1 Å). Similar to the other crystal structures of Fpg containing an AP site (17,18), the αF-β9 loop connecting the H2TH motif and the zinc finger is also partially disordered in all these four models, independently of the lesion, of the oligonucleotide sequence and length, and of the nature of the enzymes (W or M).
DNA structure
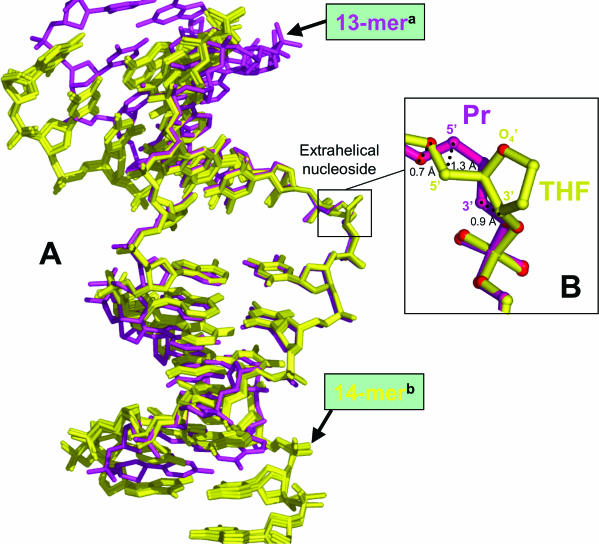
The presented models of DNA–Fpg complexes were crystallized with DNA duplexes harbouring 5′ overhanging complementary bases (Table 1). Structural differences between these models and the previous structural work result from the extremities of the DNA used in the crystallization. 14mer duplexes with cohesive ends were considered in this present work while the first model of LlFpg complex was based on a 13mer duplex with blunt ends (16). As previously described for the crystal structures of Fpg-damaged DNA complexes, the oligonucleotide duplex, globally structured as a B-DNA helix, is kinked towards the major groove at the lesion site (16–20) (Figure 2). Extensive contacts between Fpg residues and the damaged strand of the duplex are responsible for this distortion of the DNA helix axis (16–20). The DNA torsion is centred on the extruded damaged nucleoside and stabilized by the intercalation in the minor groove of three Fpg residues (M75, R109 and F111 in Lactococcus lactis numbering) in the hole resulting from the expulsion of the damage. Superimposition of the four new structures with the P1G-LlFpg-13mer DNA complex shows that the DNA torsion is similar for the 14mer DNA duplexes (∼70°) and for the 13mer DNA duplex [∼60°; CURVES, (35)] (Figure 3). This fits well with the bend observed in 16mer DNA duplex containing a reduced AP site (rAP) bound by Fpg from Bacillus stearothermophilus (18). The superposition of all DNA molecules used for these structural studies shows that divergence on atomic positions is larger at the oligonucleotides extremities than at the five central nucleotides, centred on the damaged base (Figure 3). In the vicinity of the extrahelical AP site where the enzyme contacts the DNA backbone, the structure of the 13 and 14mer bound duplexes superimpose perfectly independently of the lesion type and/or the enzyme nature. Residues at the interface between DNA and the enzyme have identical geometries with respect to previous structural data. Therefore, the crystal packing, the nucleotide sequence and the type of lesion have little impact on specific protein–DNA interactions and lesion recognition.
Figure 2.
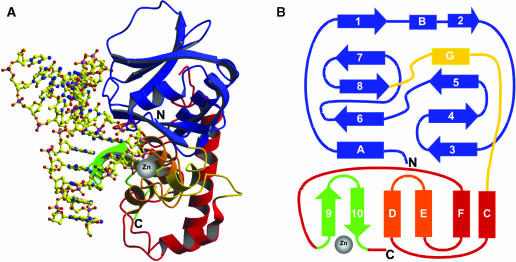
Overview of the Fpg/DNA complex. (A) Ribbon diagram and (B) topology of LlFpg bound to DNA. The ribbon representation corresponds to the wild-type LlFpg bound to the Pr site-containing 14mer DNA duplex [the complex named M/7(Pr)5′, Tables 2 and 3]. LlFpg is constituted of an N-terminal domain (blue and yellow), a C-terminal domain (red, orange and green) and two long loops (αG-αC and αF-β9). The N-terminal domain consists of a two-layered β-sandwich including the α-helices A, B and G. The C-terminal domain contains the four α-helices C, D, E and F (red and orange) and the β-hairpin loop β9–β10 of the zinc finger (green). P1 which serves as a nucleophile in the glycosylase/lyase process points out on the protein surface toward the extrahelical damaged nucleoside at the interphase between the two Fpg globular domains and DNA. β-strands and α-helices are represented by arrows and rectangles, respectively. The zinc atom coordinated by four cysteines of the Fpg zinc finger is indicated by a grey sphere. The colour code follows the previous definition of the enzyme domains (16). DNA atoms are represented by yellow ball-and-sticks. Figure generated by Molscript (41).
Figure 3.
Structure of AP site analogue-containing DNA bound to Fpg. (A) Superposition of the DNA crystal structures. In magenta, Pr-containing the blunt ended 13mer DNA bound to the M enzyme a(16) and in yellow, Pr- and THF-containing 14mer DNA duplexes harbouring sticky ends bound to the M or W enzyme b(this work). (B) Zoom around the DNA lesion for Pr- and THF-containing 14mer DNA duplexes bound to the wild-type (W) LlFpg enzyme. The extrahelical Pr and THF sites are painted in magenta and yellow, respectively. Largest atomic deviations between the two models are indicated. Figure generated by Pymol (DeLano.http://www.pymol.org).
Fine structures of Pr- and THF-containing DNA bound by mutant and wild-type Fpg
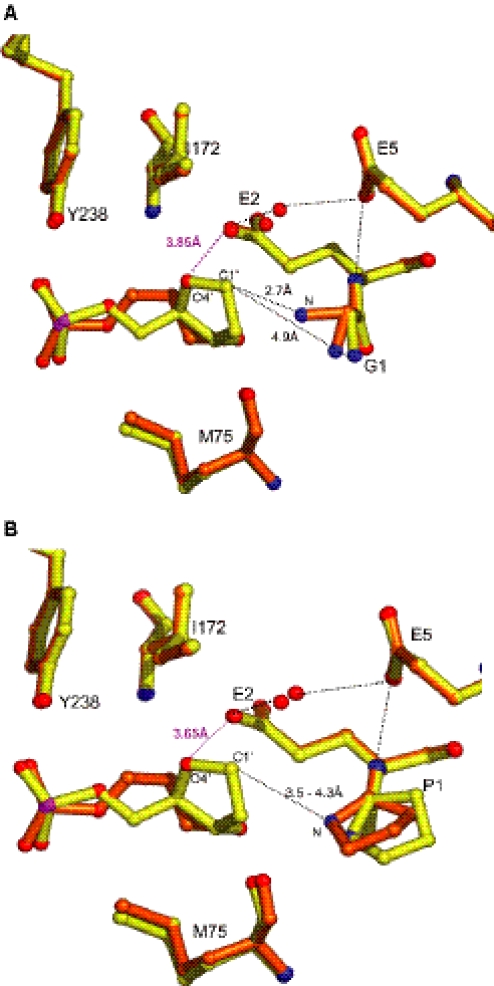
For both AP site analogues, the damaged nucleoside is flipped out off the DNA helix. At the lesion site, the B-DNA helical parameters are strongly affected by the flip-out and the local compression of the phosphate-phosphate distance (Table 4). In all structures, the AP site analogue is not directly contacted by Fpg residues (Figure 4). The residue G1 adopts two alternative conformations when the lesion is Pr but only one with THF. In the THF/M complex, the amino group of G1 is 4.9 Å away of the C1′ atom of the damage (Figure 4A). In the THF/W and Pr/M complexes, the variation in position is restricted for the catalytic residue P1, and the amino group of P1 and the THF C1′ atom are at VDW distances (3.5–4.3Å) (Figure 4B). Superposition of the four models shows that the mutation of the N-terminal residue or the lesion type does not affect the positions of the surrounding residues although THF is bigger than Pr. The additional atoms of THF (C1′, C2′ and O4′) are kept at VDW distances from the closest enzyme atoms.
Table 4.
DNA backbone torsion angles α, γ and ζ of AP site-containing DNA bound by the Fpg protein Pr, THF and redAP are for 1,3-propanediol, tetrahydrofuran and reduced AP site, respectively
| Protein–DNA complex | Backbone torsion angles (°) | PDB ID code | References | ||||
|---|---|---|---|---|---|---|---|
| AP site | DNA size (bp) | Protein | α (p0–O′5) | γ (C′4–C′5) | ζ (O′3–p−1) | ||
| Pr | 13 | P1G-LlFpg | −69.78 | −51.63 | −62.08 | 1KFV | (16,26) |
| Pr | 14 | P1G-LlFpg | −62.63 | −52.59 | −72.37 | 1NNJ | This work |
| Pr | 14 | wt LlFpg | −66.66 | −49.59 | −68.15 | 1PJI | This work |
| THF | 14 | P1G-LlFpg | 63.70 | −171.65 | −55.84 | 1PJJ | This work |
| THF | 14 | wt LlFpg | 63.71 | −174.63 | −56.76 | 1PM5 | This work |
| rAP | 16 | wt BstFpg | −157.90 | −65.71 | −62.27 | 1L1T | (18) |
Bold numbers highlight the torsion angle values of the THF site, which were significantly different from those of the other AP site analogues, Pr and rAP.
Figure 4.
Superposition of the AP sites (THF and Pr). AP site-containing DNA bound to (A) the P1G mutant enzyme and (B) to the wild-type enzyme. Figure generated by Pymol (DeLano.http://www.pymol.org). Structures with Pr and THF were painted in orange and in yellow, respectively.
In addition, the structures of the Pr and THF sites exhibit conspicuous conformational differences, especially at the phosphodiester backbone level of the target site (–C5′–C4′–C3′–) (Table 4 and Figure 3B). The THF nucleotide is therefore less extrahelical than the Pr site. The larger deviation between these two AP sites concerns the C5′ atoms (1.3 Å displacement, Figure 3B). Superposition of the lesions bound to the wild-type enzyme (W) and the P1G catalytic defective mutant (M) does not exhibit major differences.
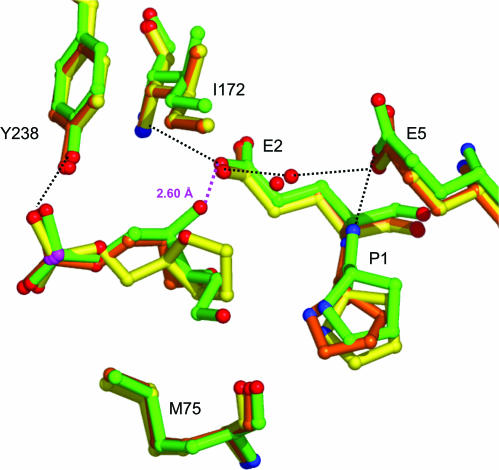
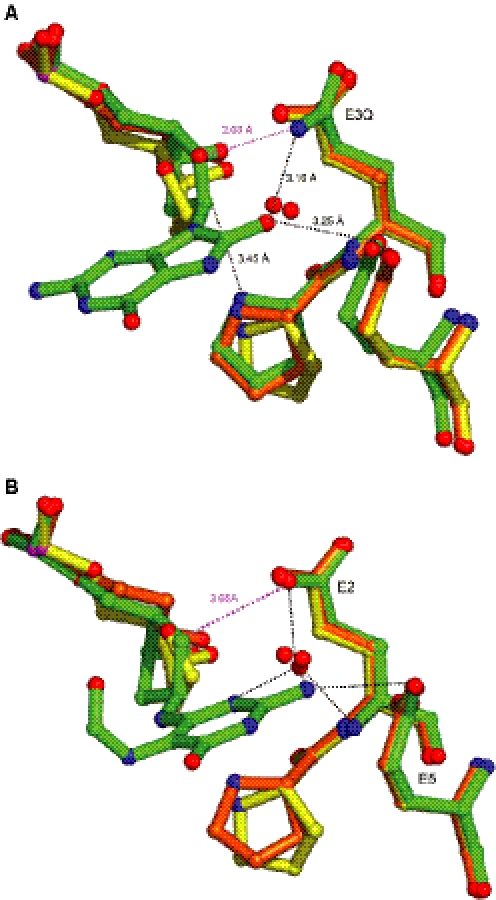
Superposition of our wild-type enzyme/THF- (or) Pr-DNA complexes and with that of the reduced AP (rAP) site bound to the B.stearothermophilus wild-type Fpg shows that the Pr lesion gives the best fit with rAP (18) (Figure 5). The extrahelical position of rAP compares well with the one of the Pr site. In the three complexes, the surrounding protein residues adopt the same conformation except P1 that lies closer to the rAP site. A major difference between these models consists in the H-bond formed between the carboxylate group of E2 and the C4′-OH group of rAP. Figure 6A also indicates that Pr fits better than THF to the 8-oxoG lesion in its complex with the B.stearothermophilus E2Q-Fpg defective mutant (19). The O8 atom of the 8-oxoG-containing DNA is replaced by a solvent molecule conserved in all complexes of Fpg bound to an AP site (Figure 6A). Position of the P1 residue is also similar in the Fpg/Pr–DNA and Fpg/8–oxoG–DNA complexes. With the FapyG containing structure (20), the FapyG carbacycle occupies a position intermediate between Pr and THF. FapyG is, however, less extrahelical. Distance between the C6′ atom of FapyG and the carboxylate group of E2 is comparable with the equivalent distance in the THF containing structures (Figure 6B).
Figure 5.
Superposition of the THF and Pr binding site with the deoxyribose-ring opened rAP site (17). THF in yellow; Pr in orange and rAP in green. Figure generated by Pymol (DeLano.http://www.pymol.org).
Figure 6.
Superposition of AP sites with damaged nucleobase. (A) with the 8-oxoG (19) and (B) with the cFapyG (20). Figure generated by Pymol (DeLano.http://www.pymol.org).
DISCUSSION
The 3D structures of bound AP site analogues correlate with functional studies
As compared with previous published crystal structures, the four models we present in this study show that the interaction of Fpg with a damaged nucleoside is independent of the crystal packing and of the oligonucleotide sequence that contains the lesion (16–20). All the structures of Fpg solved with an abasic site-containing DNA exhibit similar protein:DNA interactions and DNA torsions (16–18). The fine differences concern the geometry of the lesions (Pr or THF) and may explain the difference in affinity measured for each of these damaged nucleosides. Indeed in previous studies, we have investigated the AP site structural determinants required for a good recognition (23,24) (Figure 1). We showed that among the AP site analogues, THF was the least efficiently recognized by Fpg and we proposed that this lower binding constant was due to its heterocyclic oxygen which is negatively discriminated by the enzyme. This hypothesis was supported by the fact that the replacement of this oxygen atom (present in THF) by a non-polar group (like a CH2-group in the carbocyclic cyclopentanol AP site analogue, Cy, Figure 1) enhanced Fpg binding 3- to 8-fold. Similar binding enhancement has also been observed with several DNA glycosylases for the positively charged pyrrolidine AP site analogue, Py, as compared with THF (36,37) (Figure 1). All these functional data have suggested that the hemiacetal form of the AP site was better recognized by Fpg than the ring-closed forms and that a negatively charged Fpg residue repulses the electronegative heterocycle oxygen of THF. Our present structures confirm that the closer electronegative Fpg atoms belong to the strictly conserved E2 residue (Figure 4). The E2 carboxyl side chain is invariable whatever the lesion site or the source in the crystallographic models (16–20). Interestingly, the OE2 carboxyl group of E2 was shown to hydrogen bond the C4′-OH group of the rAP site analogue and of the Schiff base intermediate (17,18) (Figure 5). Site-directed mutagenesis of E2 has also shown that this residue is essential for Fpg activity (38). As a matter of fact, the E2Q Fpg mutant was 2-fold less active than the wild-type enzyme on an AP site-containing DNA used as substrate. The hydrogen bond between E2 and the C4′-OH of the AP site ring-opened form would stabilize the sugar in its acyclic form maintaining it in an optimal orientation for catalysis. We had already anticipated this interaction comparing the Fpg binding to rAP with other cyclic and non-cyclic AP site analogues and suggested that the ring-opened aldehyde tautomer of the AP site is the substrate active form for the Fpg AP lyase activity (24). From the comparison of our structures with those of rAP bound by Fpg and reduced Schiff base, we can propose that the electronegative charge of the THF heterocyclic oxygen is repulsed up to 3.8 Å towards the DNA major groove by the carboxyl group of E2, leading to significant structural changes of the THF phosphodiester backbone (Figure 5 and Table 4). These new structural data are consistent with the less efficient recognition of THF by Fpg compared with the other designated AP site analogues (24,25). This structural peculiarity of THF would also be related to its mutagenic effects in vivo. Indeed, recent data suggested that a very small structural difference in the AP site model (THF or true AP site) influences the AP site mutagenic spectrum greatly (39,40). Cytosine was most frequently incorporated opposite a natural AP site (‘C-rule’), followed by thymine. In the case of THF, adenine, not cytosine, was most favourably incorporated. These biochemical and functional studies have also shown that THF blocks replication less than a natural AP site. All these data suggest that THF and the true AP site are not recognized in the same way by DNA polymerases. This is also consistent with their recognition by Fpg.
Comparison between the recognition of the extruded damaged base and the extruded AP site
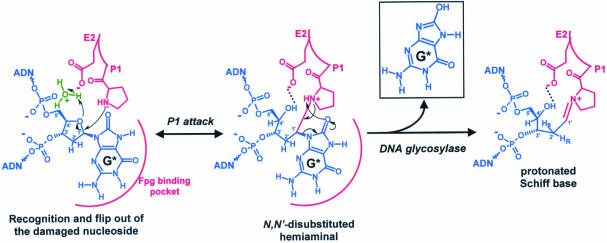
If the cycle of THF is supposed to mimic the flipped out damaged base sugar after Fpg base excision (DNA glycosylase activity), its C5′ and C4′ would not superpose with the Pr ones (Figure 3A). Similar observations can be made comparing the DNA backbone of THF with the one of the reduced abasic site (rAP), the AP site analogue which is proposed to mimic better the structure of the AP site immediately after the cleavage of the damaged base N-glycosidic bond (18,24). The position of the heterocyclic oxygen of THF was also significantly different from that of the C4′-OH of the rAP deoxyribose-ring opened which was hydrogen bound by the E2 residue (Figure 5). The restrained cyclic form of THF seems to prevent this site from adopting a perfect extrahelical conformation. THF is less extruded from the DNA helix than Pr and rAP sites (Figure 5). Contrary to THF, the Pr site considered as the minimal AP site structure for Fpg-specific binding displays a phosphodiester backbone which fits nicely with that of rAP (Figure 5). Recently, we have solved the structure of LlFpg bound to a formamidopyrimidic residue analogue (cFapydG) containing the same 14mer DNA duplex (Figure 6B) (20). Similar to 8-oxodG, FapydG is a high efficiency Fpg substrate (6). The orientation of the cyclopentane ring of cFapydG is significantly different from that of THF. Here again, THF appears to be less extrahelical than cFapydG. The replacement of the heterocyclic oxygen (present in the natural damaged nucleoside and in THF) by a methyl group would explain this observation. Indeed, the neutral group of the cyclopentane of cFapydG is less repulsed by the OE2 carboxyl group of E2 than the electronegative 4′-heterocyclic oxygen of THF. This assumption may be supported by the crystal structure of Fpg from B.thermophilus [the E2Q defective mutant in DNA glycosylase activity; (38)] bound to an 8-oxodG-containing 16mer DNA duplex (Figure 6A) (19). Similarly, the non-electronegativity feature of Q2 abolishes the repulsion of the heterocyclic oxygen of the 8-oxodG. Interestingly, the structure of cyclopentane moiety of cFapydG fits perfectly with that of the deoxyribose of 8-oxodG. In addition, the DNA backbone (C5′–C3′) at the lesion sites Pr, rAP, cFapydG and 8-oxodG are very similar, while that of THF diverges significantly. As we proposed previously, the E2 residue would collaborate with the nucleophile P1 residue to form the imino enzyme–DNA intermediate (the transient N, N′-disubstituted hemiaminal) in which the opening of the deoxyribose ring associated to the damaged nucleoside occurs prior to the cleavage of the N-glycosidic bond of the damaged base (DNA glycosylase activity) (20,24). In the present and previous models of Fpg bound to damaged DNA, the observed angle of attack of the P1 nucleophile is highly unfavourable for the N-glycosidic bond cleavage, suggesting that initial step of catalysis would rather consist in the expulsion of the O4′ leaving group, than in the cleavage of the N-glycosidic bond (Figure 7). In this mechanistic model, E2 acts as a general acid catalyst for the DNA glycosylase process, establishing a hydrogen bond with the C4′-OH of the deoxyribose-opened form. Finally, the hydrogen bond stabilizes the cationic Schiff base intermediate and sets the extrahelical nucleosidic substrate in an optimal orientation for the AP lyase process. This mechanistic model explains why the rAP site is the very best Fpg high affinity ligand (24) and why the mutation E2Q is strongly deleterious for both Fpg glycosylase and lyase activities (38).
Figure 7.
The ring-opened form of the damaged base deoxyribose is the sugar active form for Fpg catalysis. After the recognition and the flip out of the damaged nucleoside inner the Fpg substrate binding pocket, the P1 nucleophile attacks the anomeric C1′ carbon to form a transient aminal intermediate in which E2 acts as general acid catalyst. Thus, expulsion of the damaged base leaving group occurs in the subsequent step of forming the protonated Schiff base intermediate. The catalytic E2 could now stabilize the cationic Schiff base intermediate through a favourable electrostatic interaction, providing the thermodynamic driving force for expulsion of the damaged base. G* indicates 8-oxoG. A similar molecular mechanism can be drawn for the Fapy residue as substrate.
Acknowledgments
The authors are greatly indebted to the staff of BM30 and ID14 for its help in data collection (ESRF-Grenoble, France). The authors would like to thank P. Amara for her critical reading. K.P. was supported by a doctoral fellowship from the Ministère de la recherche et de la technologie (France). This work was supported by the Ligue contre le cancer (Région Centre, France), by Electricité de France (EDF) and by the Association pour la recherche contre le cancer (ARC, France). Funding to pay the Open Access publication charges for this article was provided by the Centre National de la Recherche Scientifique (CNRS, France).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T., Wood R.D. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg E.C., Meira L.B. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage. DNA Repair. 2004;3:1617–1638. doi: 10.1016/j.dnarep.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Dogliotti E., Fortini P., Pascucci B., Parlanti E. The mechanism of switching among multiple BER pathways. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:3–27. doi: 10.1016/s0079-6603(01)68086-3. [DOI] [PubMed] [Google Scholar]
- 5.Chetsanga C.J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3683. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiteux S., O'Connor T.R., Laval J. Formamidopyrimidine–DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czeczot H., Tudek B., Lambert B., Laval J., Boiteux S. Escherichia coli Fpg protein and UvrABC endonuclease repair DNA damage induced by methylene blue plus visible light in vivo and in vitro. J. Bacteriol. 1991;173:3419–3424. doi: 10.1128/jb.173.11.3419-3424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tchou J., Kasai H., Shibutani S., Chung M.H., Laval J., Grollman A.P., Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl Acad. Sci. USA. 1991;88:4690–4696. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaels M.L., Miller J.H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J. Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailly V., Verly W.G., O'Connor T., Laval J. Mechanism of DNA strand nicking at apurinic/apyrimidinic sites by Escherichia coli [formamidopyrimidine]DNA glycosylase. Biochem. J. 1989;262:581–589. doi: 10.1042/bj2620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connor T.R., Laval J. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc. Natl Acad. Sci. USA. 1989;86:5222–5226. doi: 10.1073/pnas.86.14.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zharkov D.O., Rieger R.A., Iden C.R., Grollman A.P. NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J. Biol. Chem. 1997;272:5335–5341. doi: 10.1074/jbc.272.8.5335. [DOI] [PubMed] [Google Scholar]
- 13.Thomas D., Scot A.D., Barbey R., Padula M., Boiteux S. Inactivation of OGG1 increases the incidence of G. C→T. A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet. 1997;254:171–178. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- 14.Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 2003;31:517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tudek B. Imidazole ring-opened DNA purines and their biological significance. J. Biochem. Mol. Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 16.Serre L., Pereira de Jesus K., Boiteux S., Zelwer C., Castaing B. Crystal Structure of the Lactococcus lactis formamidopyrimidine–DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21:2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilboa R., Zharkov D.O., Golan G., Fernandes A.S., Gerchman S.E., Matz E., Kycia J.H., Grollman A.P., Shoham G. Structure of formamidopyrimidine–DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- 18.Fromme J.C., Verdine G.L. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nature Struct. Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 19.Fromme J.C., Verdine G.L. DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 20.Coste F., Ober M., Carell T., Boiteux S., Zelwer C., Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by the Fpg DNA glycosylase. J. Biol. Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 21.Wiederholt C.J., Delaney M.O., Greenberg M.M. Interaction of DNA containing Fapy.dA or its C-nucleoside analogues with base excision repair enzymes. Implications for mutagenesis and enzyme inhibition. Biochemistry. 2002;41:15838–15844. doi: 10.1021/bi025903e. [DOI] [PubMed] [Google Scholar]
- 22.Ober M., Linne U., Gierlich J., Carell T. The two main DNA lesions 8-Oxo-7,8-dihydroguanine and 2,6-diamino-5-formamido-4-hydroxypyrimidine exhibit strongly different pairing properties. Angew. Chem. Int Ed Engl. 2003;42:4947–4951. doi: 10.1002/anie.200351287. [DOI] [PubMed] [Google Scholar]
- 23.Castaing B., Boiteux S., Zelwer C. DNA containing a chemically reduced apurinic site is a high affinity ligand for the E.coli formamidopyrimidine-DNA glycosylase. Nucleic Acid Res. 1992;20:389–394. doi: 10.1093/nar/20.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castaing B., Fourrey J.L., Hervouet N., Thomas M., Boiteux S., Zelwer C. AP site structural determinants for Fpg specific recognition. Nucleic Acids Res. 1999;27:608–615. doi: 10.1093/nar/27.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schärer O.D., Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 26.Pereira de Jesus K., Serre L., Hervouet N., Bouckson-Castaing V., Zelwer C., Castaing B. Crystallization and preliminary X-ray crystallographic studies of a complex between the Lactococcus lactis Fpg DNA-repair enzyme and an abasic site containing DNA. Acta Crystallogr. 2002;58D:679–682. doi: 10.1107/s0907444902001397. [DOI] [PubMed] [Google Scholar]
- 27.Leslie A.W.G. Integration of macromolecular diffraction data. Acta Crystallogr. 1999;55D:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 28.Evans P.R. Data reduction. Proceedings of CCP4 Study Weekend on Data Collection and Processing; Warrington, UK: Danesbury Laboratory; 1993. pp. 114–122. [Google Scholar]
- 29.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;50D:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Navazza J. On the computation of the fast rotation function. Acta Crystallogr. 1993;49D:588–591. doi: 10.1107/S0907444993005141. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A.T., Adams P.D., Clore G.M., Delano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges N., Pannu N.S., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54D:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Brünger A. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;403:859–866. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 33.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47A:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov G., Vagin A.A., Dodson E.J. Refinement of molecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53D:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Lavery R., Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 36.McCullough A.K., Scharer O., Verdine G.L., Lloyd R.S. Structural determinants for specific recognition by T4 endonuclease V. J. Biol. Chem. 1996;271:32147–32152. doi: 10.1074/jbc.271.50.32147. [DOI] [PubMed] [Google Scholar]
- 37.Schärer O.D., Nash H.M., Jiricny J., Laval J., Verdine G.L. Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases. J. Biol. Chem. 1998;273:8592–8597. doi: 10.1074/jbc.273.15.8592. [DOI] [PubMed] [Google Scholar]
- 38.Lavrukhin O.V., Lloyd R.S. Involvement of phylogenetically conserved acidic amino acid residues in catalysis by an oxidative DNA damage enzyme formamidopyrimidine glycosylase. Biochemistry. 2000;39:15266–15271. doi: 10.1021/bi001587x. [DOI] [PubMed] [Google Scholar]
- 39.Avkin S., Adar S., Blander G., Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otsuka C., Sanadai S., Hata Y., Okuto H., Noskov V.N., Loakes D., Negishi K. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 2002;30:5129–5135. doi: 10.1093/nar/gkf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraulis P.J. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]