Abstract
Herein we describe the first application of direct linear analysis (DLA) to the mapping of a bacterial artificial chromosome (BAC), specifically the 185.1 kb-long BAC 12M9. DLA is a single molecule mapping technology, based on microfluidic elongation and interrogation of individual DNA molecules, sequence-specifically tagged with bisPNAs. A DNA map with S/N ratio sufficiently high to detect all major binding sites was obtained using only 200 molecule traces. A new method was developed to extract an oriented map from an averaged map that included a mixture of head-first and tail-first DNA traces. In addition, we applied DLA to study the conformation and tagging of highly stretched DNA. Optimal conditions for promoting sequence-specific binding of bisPNA to an 8 bp target site were elucidated using DLA, which proved superior to electromobility shift assays. DLA was highly reproducible with a hybridized tag position localized with an accuracy of ±0.7 µm or ±2.1 kb demonstrating its utility for rapid mapping of large DNA at the single molecule level. Within this accuracy, DNA molecules, stretched to at least 85% of their contour length, were stretched uniformly, so that the map expressed in relative coordinates, was the same regardless of the molecule extension.
INTRODUCTION
Growing interest in comparative genomics has spurred interest in technologies that provide affordable genomic scale analysis. DNA mapping, though lower resolution than single base sequencing, can offer rapidity and efficiency and, in many cases, still allow discernment of genomic differences among species or among individuals within a given species (1). The techniques that have single molecule sensitivity and are capable of analyzing long DNA fragments deserve special attention. Single-molecule sensitivity eliminates the need for DNA amplification and could be particularly valuable for discerning haplotypes (2,3). The ability to analyze long DNA molecules comprised of hundreds of kilobases preserves the higher-order information in the genome such as the sequence of linkage disequilibrium blocks (4,5).
Optical mapping developed by Schwartz and colleagues (6) is an example of how very long strands of DNA can be physically mapped by imaging individual stretched DNA molecules attached to a surface and digested with a restriction enzyme. Contrary to DNA fixation on a surface, single molecule mapping of DNA stretched in microfluidic devices and continuously flowing through a reader (7) promised higher throughput. Recently, we utilized a model system of 48.5 kb λ phage DNA to provide a proof-of-concept for a new mapping technology analyzing DNA molecules ‘on the fly’ and known as direct linear analysis (DLA). Compared to optical mapping, DLA has similar resolution, sensitivity and higher throughput (8). The two key components of DLA are: (i) a microfluidic system for flowing and stretching single DNA molecules so that they can be read rapidly in a linear fashion and (ii) fluorescence-based detection that allows the reading of fluorescent tags on single molecules of long DNA. Its combination of features, namely single molecule sensitivity, analysis of very long DNA molecules, and high throughput, makes DLA an attractive approach for many genomic applications. To date, however, DLA has not been shown suitable for practical applications.
The purpose of the present study was to develop and characterize DLA as a technology suitable for practical applications such as mapping very long DNA isolated from cells. In so doing we also demonstrated its utility as a tool for characterizing DNA conformation and tagging. Using bisPNA tags, we demonstrated that DLA provides high precision of determining the hybridized tag positions on a bacterial artificial chromosome (BAC) and uncovered the ability of bisPNA tags to hybridize to both perfectly matching target sequences and to a pair of adjacent mismatching sites, the extended P-loops. Finally, a new method for orienting molecular maps was developed further enhancing the applicability of DLA technology.
MATERIALS AND METHODS
DNA sample
As a long DNA sample, we used 185.1 kb BAC 12M9 (accession no. AL080243) (9). This BAC contains a 177 kb region of human chromosome 22 cloned into the EcoRI sites of the pBACe3.6 vector (10). We obtained Escherichia coli 12M9 clone from P. J. de Jong at CHORI, Oakland CA. Supercoiled BAC was isolated by a slight modification of alkaline lysis protocol (11), linearized with homing endonuclease PI-SceI (NEB, Beverly MA), and evaluated for admixtures of circular and degraded DNA. See Supplemenatry Data for the protocols.
Tagging procedure
For sequence-specific tagging, we used fluorescent bisPNA, which is capable of invading double-stranded DNA under mild non-denaturing conditions by displacing one of the strands (12). The tag complementary to 5′-GAG AAG AA-3′ DNA sequence was synthesized by Applied Biosystems (Bedford, MA): (N)TMR-OO-Lys-Lys-TTC-TTC-TC-OOO-JTJ-TTJ-TT-Lys-Lys(C)
The tag includes tetramethylrhodamine fluorophore (TMR), which is conjugated to N-terminal amino group. The fluorophore, the Watson–Crick (T + C) strand, and the Hoogsteen (T + J) strand are connected with –O– linkers (8-amino-3,6-dioxaoctanoic acid), which form flexible hydrophilic tethers. T, C, and J are thymine, cytosine, and pseudoisocytosine, respectively. The tagging and controls were performed as described in (8) with some modifications. See Supplementary Data for details including tagging mechanism.
Staining of DNA backbone
For DLA, DNA samples were stained with TOTO-3 intercalating dye (Molecular Probes, Eugene, OR) at a concentration of 500 pg/µl, corresponding to 0.76 µM of base pairs. The dye was added to produce base pair to dye molar ratios between 10:1 and 2.5:1. Solutions were incubated for ≥2 h at room temperature or for ≥1 h at 37°C. DNA was stained as close to saturation as possible to have the dye evenly spaced along the DNA backbone to ensure accurate determination of the apparent center of molecule (CM) (8). However, the final DNA sample was not saturated with TOTO-3, in part as a consequence of a competition between TOTO-3 and residual EtdBr, that remains bound to DNA even after multiple isopropanol extractions (data not shown), and also in part due to possible adsorption of the intercalator onto chip surfaces during measurement.
DLA technology
BAC 12M9 was mapped by DLA technology (8). In short, DNA was tagged with fluorescent bisPNA tags and intercalated with TOTO-3, then stretched in a microfluidic chip, and finally ‘read’ by multiple-color epi-illumination confocal microscopy. Intercalator fluorescence revealed both the conformation of individual DNA molecules and also the velocity using the two-spot velocimetry system. Typical velocities were 10–20 µm/ms, and these values were used on an individual molecule basis to convert measurements taken in time into distances. Fluorescence bursts from the DNA-bound tags revealed the underlying sequence motif map.
We used a published analysis (13) and our study of DNA stretching in elongational flow (J. W. Larson et al., manuscript submitted) to design and create a microchip configuration and operating conditions that are adequate for stretching 200 kb-long DNA. The chip design was similar to that presented in Figure 1A of Chan et al (8) and included a 1 µm-deep cavity with a funnel with a 83:1 taper reduction ratio, a taper shape providing W(x) ∼1/x2 profile (W is the channel width, and x is the coordinate along the flow direction), and a 0.6 µm-wide interrogation channel. In some experiments, chips included a post field before the taper (8) that increased the proportion of stretched molecules by 5–10%.
Figure 1.
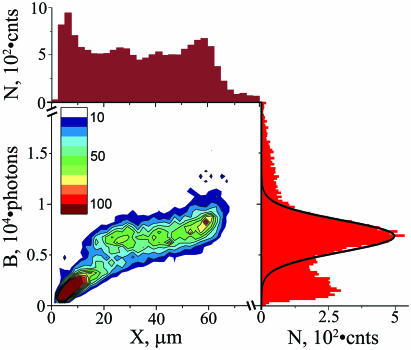
Characterization of DNA stretching. A two-dimensional histogram of burst size (B is the total number of photons per molecule) and length (X is the DNA extension in the direction of flow) for individual DNA molecules is shown as a heat map: 500 photons by 2 µm binning, respectively, and histogram occupancy as indicated in the inset legend. The adjacent histograms show distributions of DNA lengths (maroon at the top) and burst sizes (red at the right) with an interpolated Gaussian curve (black line) corresponding to full size BAC DNA. N is number of events. Data include 15 283 molecules.
Detection channels were synchronized for alignment of different color signals. Every data bin captured the number of photons detected during a 0.1 ms interval (10 kHz frequency). For data processing, we used our own software which selects signals whose fluorescence exceeds a threshold. The presence of a DNA molecule in the detection zone was operationally defined as 5 or more contiguous bins with over-threshold signals. Shorter events (i.e. <5 bins) were discarded; as these spurious flickers are generally from dust particles, very short DNA and other detectable detritus. The apparent CM was determined using the backbone emission profile and used as the main geometrical reference point along each DNA (the origin of coordinates). To compare the theoretical and experimental maps, their origins were aligned and theoretical intensity per single tag was adjusted to match single-target peaks of experimental map. Then the theoretical map was shifted and stretched to obtain the best fit. This procedure accounted for the offset between green and red excitation spots and for the uncertainty of DNA length determination due to thresholding.
RESULTS
Characterization of DNA stretching
Only intact and adequately stretched DNA molecules were selected for analysis. For every DNA molecule, we determined its extension in the direction of flow (X, or ‘length’ for simplicity) and burst size [B is the total number of photons emitted by the stained DNA (8)] and a typical scatter plot of these two parameters is presented in Figure 1. The individual histograms are adjacent to the top and right sides of the scatter plot, respectively (Figure 1). Burst size is proportional to DNA size (14), and the burst size distribution (right histogram in Figure 1) exhibited a single sharp peak centered at 6800 photons consistent with a pure homogenous DNA sample. The width of this peak was the result of two effects. First, different DNA molecules flowed through different sections of the focused light spot and therefore were excited at different intensities. Second, the number of dye molecules varied for different DNA molecules of the same length. We used the area of a Gaussian fit of the peak (black line in the histogram) to estimate the proportion of intact BACs to be 60 ± 2%. The proportion of intact BACs in the sample before injection into a chip was determined independently using pulsed-field gel electrophoresis (PFGE) to be 75 ± 8%. The small difference of 15 ± 10% most likely reflects DNA losses inside the microfluidic chip such as adsorption to walls and possibly some DNA fragmentation.
The distribution of measured DNA lengths (top histogram in Figure 1) had an asymmetric peak centered at 60 µm, showing that the BAC molecules were considerably extended from their relaxed random coil conformations of 2.5 µm [average end-to-end distance, (15)]. This peak asymmetry, with an abrupt border at larger sizes, would be expected for a population of stretched molecules, in which a large portion approached complete stretching. In this case, the position of the abrupt border corresponds to the known DNA contour length. The full size DNA molecules were readily evident in the two-dimensional histogram (Figure 1) as the large cluster centered broadly about the dominant peak in the burst size histogram and ranging in length from 20 to 65 µm, indicating variable stretching efficiency. In most cases, at least 16% of the molecules were stretched to lengths exceeding 54 µm.
The contour length of non-intercalated BAC 12M9 is expected to be 62.9 µm [185.1 kb at 0.34 µm/kb (16)]. However, intercalation can increase the contour length (17). Therefore, our data include molecules as long as 70 µm in some experiments, even though the DNA molecules were not intercalated to saturation. Total numbers of molecules longer than 54 and 64 µm in Figure 1 are 1427 and 124, respectively. This corresponds to 16 and 1.4% of the intact BAC molecules.
It is possible to overstretch DNA beyond its contour length if the applied force exceeds 70 pN (18,19). We avoided overstretching through proper choice of channel geometry and flow velocity (J. W. Larson et al., manuscript submitted). In Figure 1, there is infrequent evidence of DNA molecules longer than 70 µm which also tend to have larger burst sizes outside of the Gaussian distribution (>12 000 photons), but were otherwise relatively scattered in the two-dimensional histogram (hence not visible with the heat map in Figure 1). Most likely, these events were artifacts of two overlapping molecules that were not distinguished by our analysis algorithm.
Obtaining unoriented maps
To obtain a bisPNA binding site map (Figure 2), we selected DNA molecules that were stretched within a narrow size range (the selection interval varied between 1 and 5 µm depending on the number of detected molecules and is stated in every Figure legend), aligned them along their CMs as determined from backbone emission profiles, and calculated the average bisPNA tag fluorescence signal. After averaging, multiple Gaussian peak fitting was used to map tag positions along the DNA. Based on peak width (full width at half maximum height), map resolution was estimated at 1.7 µm or 5 kb. DNA molecules traveled through our chip in both head-first and tail-first orientations, and consequently signal averaging superimposed the two mirror image maps into a single ‘unoriented’ symmetrical map. Maps obtained at different tagging specificities are presented in Figure 2; their analysis is discussed step by step below.
Figure 2.
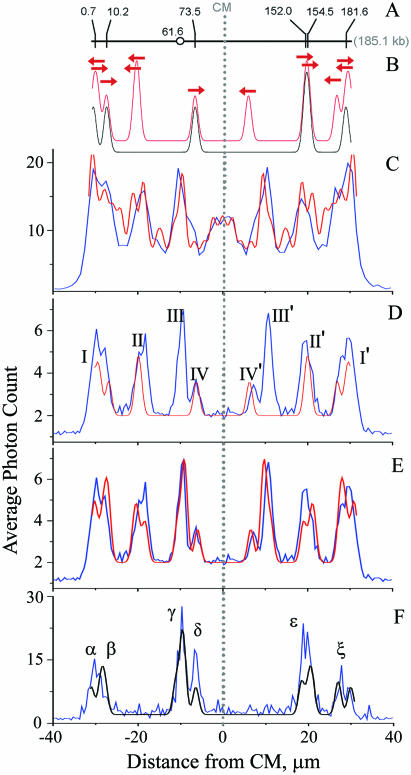
Comparison of theoretical and experimental maps. (A) Positions of 6 GAG AAG AA target sites on BAC 12M9. Circle indicates position of the site responsible for peak III. (B) Theoretical maps for equal mixture of unoriented molecules (red) and for head-first, oriented molecules (black). Sites represented by 5 kb-wide Gaussian. Arrows, one for every target site, show peak positions for head-first or reverse orientations. (C) Unoriented map (blue) observed under low specificity conditions. (D and E) The same unoriented map (blue) observed under high stringency conditions. (F) Oriented map (blue) obtained by selecting head-first DNA molecules having tags at sites IV and/or II′. Each experimental map shows average intensity of the bound fluorescent tags as a function of distance from CM, at 0.5 µm steps. Maps include molecules 62–65 µm long, aligned at their CMs, which are denoted as the zero coordinate. Maps C, D, and F include 1035, 1993 and 87 molecules, respectively. Theoretical oriented (black) and unoriented (red) maps include only target sites (D), plus extended P-loops (E and F), plus SEMM sites (C). See text for details.
Both the width of the selection interval and the extent of stretching of the selected molecules influenced the quality of the maps, i.e. their S/N ratio and resolution. These effects are discussed further.
The accuracy of the observed map was assessed by comparison to a theoretical model (black curve in Figure 2B) consisting of six Gaussian peaks centered on the target site locations (0.7, 10.2, 73.5, 152.0, 154.5 and 181.6 kb as shown in Figure 2A) and sized 5 kb wide (FWHM). An unoriented model was created superimposing the head-first theoretical map with its mirror image (red curve in Figure 2B).
Tagging specificity
bisPNA tags can bind to perfect match target sites as well as single end mismatch (SEMM) sites, whereas other types of mismatch sites do not significantly hybridize and thus were disregarded in our analysis (see Supplementary Data for more discussion). There are 84 SEMM sites in BAC 12M9, many of which hybridized with tags during the incubation. We devised a protocol to enhance tagging specificity. To remove incorrectly bound tags, we incubated the sample at elevated temperature (8,20). Temperature and incubation time were adjusted carefully to preserve maximal binding to perfect target sites, while minimizing incorrect binding. Optimization using both electrophoresis band shift assay (see Supplementary Data for details) and DLA showed 2 h incubation at 68°C to be ideal. The DLA approach to optimize the incubation conditions relied on a cluster of 13 closely positioned SEMM sites between 92.8 and 108.0 kb in BAC 12M9 that does not overlap with any target site. If a sample included residual SEMM-bound tags, they formed an easily detectable peak in the unoriented map between −5.4 and 5.4 µm. As seen in Figure 2C, 1 h incubation at 58°C was not sufficient to remove SEMM-bound tags whereas a 2 h incubation at 68°C showed no trace of the central peak in an unoriented map (Figure 2D and E) signaling that all SEMM-bound tags were gone. Examination of Figure 2D reveals that, overall, there was excellent agreement between the theoretical map and the observed map with the exception of peaks III and III′ (Figure 2C). The origin of these peaks are discussed below. After the inclusion of corresponding binding sites, the theoretical map shows all peaks observed in the actual map (Figure 2E). See more on the construction of theoretical maps in the last section of Results.
Generating an oriented map
Oriented maps can be more informative, so we developed an informatics approach to extract oriented maps using peaks that are uniquely formed by DNA molecules oriented in the same direction. For example, peaks IV and II′ are formed by the sites at 73.5 and 152.0/154.5 kb, respectively, that belong to DNA molecules moving head-first (see similar directed arrows in Figure 2B). Therefore, if we selected only DNA molecules containing peak IV (interval −8 to −5 µm) and peak II′ (interval 17–21 µm) and averaged their maps, we obtained an oriented map (Figure 2F).
Although in this example the oriented map was extracted using a priori knowledge of the tag binding pattern, the approach can be generalized to obtain oriented maps of unknown molecules (see Discussion).
Number of molecules needed to create a map
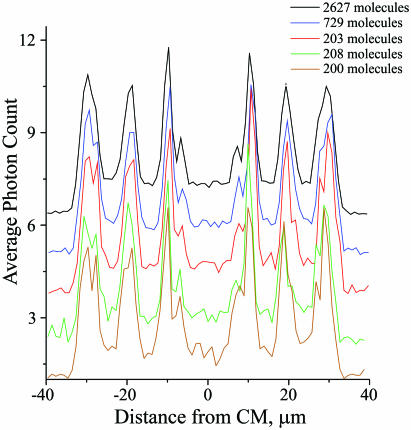
To evaluate how many individual DNA molecules were needed to obtain a map with a good S/N ratio, we compared a typical map obtained by averaging 2627 molecules with maps using a subset of those molecules (Figure 3). Although noise was increased, using only 200–208 molecules was sufficient to observe all peaks.
Figure 3.
Unoriented maps obtained with different numbers of molecules. Upper black map includes 2627 DNA molecules with sizes of 62–66 µm. Blue map includes a subset of 729 DNA molecules within narrower size interval of 63.5–64.5 µm. Three lower maps red, green and brown are obtained with different subsets of the narrow size selection (63.5–64.5 µm), which include 203, 208 and 200 DNA molecules, respectively. Maps were obtained with 1 µm step and displaced along the ordinate to simplify comparison. See Figure 2 legend for other details.
Inclusion of more molecules in averaging, generally speaking, should produce maps with better signal-to-noise ratios. However, selection of wider range of lengths of DNA molecules results in widening of the peaks because their positions depend on the degree of DNA stretching (see below); therefore, even at smaller noise these maps have worse resolution. This trade-off between S/N ratio and resolution is especially visible for the outermost bands I/I′; compare the black map wider selection with others in Figure 3. The rule of thumb is to select the molecules from the narrowest size interval that includes a sufficiently big number of molecules to detect the lowest peak over the noise.
Accuracy of defining tagged sites
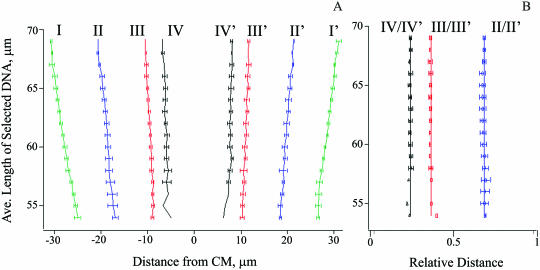
To evaluate the reproducibility of DNA mapping by DLA, we compared unoriented maps obtained from different experiments and generated by averaging the tag signals from DNA that stretched to similar lengths, allowing meaningful comparison when the DNA stretching varied. The positions of the major peaks in maps from nine different experiments, different samples and microchips were determined by multiple Gaussian peak fitting. Figure 4A shows the cumulative results for different DNA length selections; Roman numerals refer to the peak notation in Figure 2D. The bound tag positions were located precisely to within ±0.7 µm, based on the standard deviation, for molecules stretched beyond 59 µm, (Figure 4A), demonstrating the high reproducibility of the procedure. The accuracy was limited by the 0.5 µm step used to calculate the maps and by a crude selection of the peaks that was needed to cover a wide range of DNA elongations. For example, a single Gaussian approximation was adequate for peaks I/I′ and II/II′ for DNA molecules stretched to 60 µm or less. However, the mapping resolution increased at higher degrees of stretching where each of these peaks exhibits two resolved components (see Figure 2D). Similarly, peaks IV or IV′ were difficult to resolve from peaks III or III′, respectively, for DNA maps shorter than 57 µm.
Figure 4.
Analysis of DNA conformation under different degrees of stretching. (A) Measured positions of major peaks in unoriented maps. Roman numbers defined in Figure 2D. (B) Relative distances between peaks calculated as the ratio of distances between symmetric peaks and the outermost peaks LI–I′. Symbols II/II′, III/III′ and IV/IV′ denote the relative distances LII–II′/LI–I′, LIII–III′/LI–I′ and LIV–IV′/LI-I′, respectively. Traces of DNA molecules at 5 µm intervals were averaged, with the ordinate centered on the selection interval. Lines in panel A are drawn by eye; straight vertical lines in panel B are drawn at the positions corresponding to the averages of II/II′, III/III′ and IV/IV′ datasets at 0.68, 0.36 and 0.24, respectively.
Conformation of stretched DNA
Previous work in single DNA molecule dynamics has relied heavily on end-to-end measurements of DNA conformations resulting in relatively little knowledge about stretching dynamics along a molecule. Perkins et al. (17) resolved large internal morphological changes in tethered DNA by fluorescence imaging, but otherwise the tagging strategy employed in the present study for mapping revealed stretching dynamics across the entire molecule with unprecedented spatial resolution (Figure 4). Detailed analysis of the DNA stretching behavior exceeds the scope of this article, but of greatest note, we observed that the tag positions scaled linearly with increasing DNA length, indicative of uniform stretching at all points along the molecule.
Measurements in the current study were performed after a sudden elongational flow, and consequently the highly extended DNA conformations were transient and possibly somewhat relaxed at the time of measurement. For instance, the tension profile along the polymer might have redistributed in the few milliseconds while the polymer emerged from elongational flow. The nature of polymer relaxation from high extension under non-uniform and time varying strain such as in our system is relatively poorly understood. In previous work, it was shown that DNA adopts a steady-state ‘dumbbell’ orientation in elongational flow at lower strain rates above the critical value for the onset of stretching (13), a phenomenon arising from the variation in polymer tension from a maximum in the molecule center to zero at the ends, and also from the propensity of polymers to relax from the ends inwards (21). Therefore, direct extrapolation of DNA stretching behavior to the higher strain rates used in this study suggests that the central tag positions should be relatively insensitive to the overall DNA extension, and that improved stretching should affect the tag positions on the ends most significantly.
To uncover the conformational differences between the central and terminal portions of DNA molecules stretched to varying extents, the peak positions were expressed in relative distances (Figure 4B), normalized against the distance LI–I′ between the outmost peaks I and I′, which are formed by the tags bound very close to the termini. Normalization against the outer peaks was preferred over normalization against the measured DNA length as a more accurate measure of the distance between DNA ends. We also used the distances between symmetric peaks instead of the positions of individual peaks relative to CM to exclude any systematic error due to misalignment of the red and green detection channels. The distances between the peaks II and II′ (LII–II′), III and III′ (LIII–III′), and IV and IV′ (LIV–IV′) were divided by LI–I′ to calculate the relative interpeak distances, which are plotted in Figure 4B as curves II/II′, III/III′, and IV/IV′, respectively, to evaluate uniformity of DNA stretching. Clearly, the maps were identical within experimental uncertainty after the linear transformation despite significant differences in the observed DNA length. Average relative distances between peaks II and II′, III and III′, and IV and IV′ were 0.68 ± 0.02, 0.36 ± 0.01 and 0.24 ± 0.01, respectively. Therefore, BAC molecules longer than 54 µm were stretched uniformly in DLA.
Although the relative distances between the peaks or their positions expressed in relative coordinates did not depend on the extent of DNA stretching, the maps measured for more fully stretched molecules provided better genomic resolution. In other words, our geometric resolution between tags was 1.7 µm which represented 5.0 kb for completely stretched DNA [at 0.34 kb/µm for B-form DNA (16)] or 5.9 kb for DNA stretched to 54 µm.
Characterization of peaks III and III′
As mentioned above, based on the knowledge of the BAC sequence in GenBank, we observed an unexpected tag binding site, visible as peaks III and III′ in the unoriented map (Figure 2C) or peak γ in the oriented map (Figure 2E). Using the position of the unexpected peak in the unoriented map and a conversion factor for µm into kb, determined from the positions of known matching targets, the location of the unknown tag-hybridization site(s) was determined to be at 61.6 ± 2.3 kb. By using electrophoretic mobility shift assays (EMSAs) to examine binding of radioactive bisPNA tags to restriction fragments of BAC 12M9 (see Supplementary Data), two tag binding sites were observed: at 59.2–60.6 kb and 63.5–65.9 kb, independently confirming that these were indeed true binding sites. We resequenced BAC 12M9 in these regions and found that it matched the GenBank deposited sequence and contained no additional perfect match tag sites. Close examination of the DNA sequence revealed the presence of two loci at 59.7 (P1) and 64.4 (P2) kb with potential to form extended P-loops when bound to bisPNA tags. Independent experiments (see Supplementary Data for details) demonstrated that the tags in fact bound to these extended P-loops with stability comparable to that seen for hybridization to perfect match sites, although they included mismatching bases.
Inclusion of extended P-loops in the analysis
Because two extended P-loops clearly produced detectable peaks in unoriented maps under our high stringency hybridization conditions, we also included other similar sites in the analysis. We selected the sequences that included at least two sites with single mismatches that were separated by 1–6 bp and of the two mismatching sites at least one was a SEMM site. These tentative selection rules are in good agreement with our data (see Supplementary Data for details) but by no means are explicit and need further investigation. Sites satisfying these demands were found at 7.4, 147.1 and 172.7 kb and were added to the theoretical map together with P1 and P2. Intensities of the extended P-loop peaks were determined using the measured intensity of III/III′ peak and the independently determined ratio of bisPNA bound to the P1 and P2 sites, which was 1.2:3.0 (see Supplementary Data). The latter site includes three binding sites in proximity and therefore has anomalously high intensity. For all other extended P-loops we used the same intensity as determined for P1 site. The resulting unoriented and oriented theoretical maps are presented in Figure 2E and F, respectively. Clearly, every peak in the theoretical maps now has a matching experimental counterpart and there are no peaks in the experimental maps that are not present in theoretical maps. The peak positions in the experimental and theoretical maps (see Table 1 for the peak positions in Figure 2F) coincide with ±0.5 µm accuracy; the intensities do not always match perfectly because our approximation did not account for the structure variations and, hence, differing binding stabilities in the different extended P-loop sites. Also, unlike the unoriented map, intensities of the peaks in the oriented experimental map (Figure 2F) are distorted because of the way the map was generated with a biased selection of traces that contained a peak δ and/or ɛ but not necessarily other peaks.
Table 1.
The peak positions in the experimental and theoretical oriented maps presented in Figure 2F
| Peak identifiersa | Peak position (µm) | Error (µm)b | |
|---|---|---|---|
| Experimental | Theoretical | ||
| α | −30.25 | −30.91 | 0.66 |
| β | −27.75 | −28.40 | 0.65 |
| γ | −9.75 | −9.66 | −0.09 |
| δ | −6.50 | −6.46 | −0.04 |
| ɛ1 | 18.75 | 18.50 | 0.25 |
| ɛ2 | 19.75 | 20.40 | −0.65 |
| ξ1 | 27.75 | 27.00 | 0.75 |
| ξ2 | 29.50 | 29.90 | −0.40 |
aThe peak notation is the same as in Figure 2F.
bThe error is positive when the experimental peak is to the right from the theoretical one.
DISCUSSION
Genomic mapping is a well-establish and powerful strategy for species identification, for identification of overlapping genomic fragments in sequencing projects, and for other large scale genomic analyses. However, traditional mapping methods are relatively slow, tedious, expensive and require significant amounts of DNA. This study demonstrated that a single molecule mapping technology, namely DLA, is an alternative method for mapping large genomic DNA. Once isolated from bacteria and tagged, 185 kb BAC 12M9 DNA was mapped using DLA in minutes. Multiple sequence-specific tags could be localized along the DNA molecule by observing as few as 200 molecules. The map was highly reproducible and allowed us to discern tag positions with a mapping precision of ±0.7 µm or about ±1.2% of the molecule length. In addition, DLA was found to be very powerful for characterization of the conformation of stretched DNA, and evaluation of binding of sequence-specific tags to DNA.
DNA stretching and sizing
Multiple tag sites could be resolved within the 185 kb BAC 12M9 making it possible to gain insight into stretching behavior along the DNA molecule. Highly extended DNA was stretched uniformly in DLA and hence the relative coordinates of hybridized tags were the same for all elongated DNA molecules (Figure 4). For certain applications such as genome or species identification, the pattern of tag distribution, which can be expressed in relative coordinates, is sufficient; therefore, all the BAC molecules longer than 54 µm could be used for analysis. This relaxed selection rule allows inclusion of a significant portion (>16%) of all the detected BAC molecules for identification. The length of 54 µm constitutes only 85% of the contour length of unintercalated BAC and an even smaller extension of intercalated molecules. However, the maps obtained at longer extensions were more informative because of better resolution.
For some applications, determining DNA contour length between tag sites and between termini (i.e. the size of DNA fragment) is important. Only in this case can geometric length in micrometers be transformed into genetic length in base pairs using a scaling factor that can be determined independently (8). The contour length of a DNA fragment can be evaluated from its burst size. To select completely stretched DNA molecules for analysis and therefore ensure that the measured distance is the contour length, a chart, similar to Figure 1, can be used. Burst size B is proportional to contour length X0 for uniform intercalation (14,22) and therefore the function B/X0 = constant is a straight line going through the coordinates origin; the constant can be determined through measurement of a known DNA sample. The lengths and burst sizes of the traces found in the vicinity of this line belong to completely stretched molecules. This selection approach can be used for analysis of unknown DNA.
DNA tagging
Mapping with DLA requires highly specific tagging. To achieve satisfactory specificity with bisPNA fluorescent tags, the temperature and time of the final incubation were carefully adjusted. Of note, DLA technology itself proved to be a better method than the more traditional EMSA approach for characterizing DNA binding properties of bisPNA. DLA was more rapid, highly sensitive and amenable for analysis of large DNA.
bisPNA tags, in addition to hybridizing with perfect matching target sequences, could form equally stable complexes with a pair of mismatching sites if they were in close proximity i.e. extended P-loops. Extended P-loops were formed previously using a pair of carefully designed bisPNAs (23). In our case, they were formed by pairs of sites in close proximity that had imperfect target sequences. Five extended P-loops were identified in BAC 12M9 for the tag we used. Whereas bisPNAs can discriminate individual perfect match from single mismatch sites, we found that bisPNA tags did not dissociate from extended P-loops sites containing mismatched bases, even after high temperature treatment. If either temperature or incubation time were increased, tags from both target sites and extended P-loops dissociated simultaneously. Therefore, target and extended P-loop sites tagged with bisPNA are indistinguishable; both types should be taken into account when mapping DNA.
Producing oriented maps
In addition to provide a symmetric unoriented map of tag sites along the DNA, DLA was shown capable of generating an oriented map when an asymmetric tagging pattern exists. Although we used a priori knowledge of the tagging pattern to extract an oriented map, this approach can be generalized to orient unknown molecules and analyze mixtures of molecules. The basic principle is to choose any peak in an unoriented map, select all molecules with this particular peak, average their traces and compare this new average sub-map with the original unoriented map. If the new map resembles the original one, includes all the same peaks, then the chosen peak of an unoriented map resulted from both orientations of DNA molecules. In this case, you proceed to another peak of an unoriented map and repeat the process. Once the new sub-map is (i) different from the unoriented map and (ii) reconstructs the unoriented map being superimposed with its own mirror image, then the extracted sub-map is the oriented map. This process does not work if the oriented map is completely symmetric; however, in this case it does not differ from unoriented map anyway. An actual algorithm is more complicated and would involve an iterative analysis of the generated sub-maps and their mirror images to identify asymmetric peaks for obtaining oriented maps. This algorithm would work not only with the peaks that are formed by single DNA orientation, but also with the peaks that are formed by both orientations if one of the orientations inputs more tags than the other. A similar approach, peeling away one after another sub-maps from an averaged unoriented map, can also be used for analysis of mixtures of different DNA fragments. The development of appropriate analytical algorithms is currently in progress.
Specifics of DLA mapping
Some features of DLA distinguish it from other mapping techniques. First, DLA requires a small amount of DNA, a general advantage of single molecule approaches, potentially eliminating the need to create a BAC library to map a genome. This advantage, however, can only be exploited when a technology is available to isolate long DNA fragments directly from genomic DNA. In this regard, we are currently developing an automated sample preparation technology based on concept of digesting a genome with rare-cutting restriction enzyme and then isolating the resulting long DNA fragments for subsequent mapping with DLA.
A second advantage of DLA is its ability to map long fragments, i.e. hundreds of kilobases, which preserves some higher-order information in the genome such as the sequence of linkage disequilibrium blocks (4,5). Assembly of large scale or whole genome maps is also simpler from longer size mapped fragments (24). While about a hundred of 50 kb-long mapped fragments are typically needed to assemble the map of a 0.5 Mb segment (25), the same segment can be covered with only 3–6 overlapping fragments, if they are 100–200 kb-long, using DLA, or the segment can even be mapped as a whole by optical mapping (26). The ability to measure long DNA is especially helpful in analysis of DNA fragments with repeats. The accuracy of map reconstruction depends on the precision of determining the positions of the specific-sequences, which are restriction endonuclease recognition sites in classic and optical mapping and tag-hybridization sites in DLA mapping. The classic restriction mapping precision is limited by PFGE and varies with the fragment length ranging from ±0.5 kb for 10 kb fragments up to ±3.0 kb for 100 kb fragments (27). In comparison, the precision is ±2.06 kb for optical mapping (6) and ±2.1 kb for DLA [±0.7 µm at 0.34 kb/µm for B-form DNA (16)].
A third advantage of DLA is the ability to measure contiguous maps. It allows analysis of a mixture of different DNA fragments in the same experiment. The map of a sufficiently long genome fragment permits identification of a microorganism (28). Therefore, the ability of DLA to measure contiguous maps of long fragments provides the potential for identifying different microorganisms in a mixture of DNA.
DLA shares all the above-mentioned advantages with optical mapping (29,30). However, it has some unique features as well. For example, different tags that hybridize to different target sequences and fluoresce in different spectral regions can be used simultaneously with the same DNA fragment. This way several different maps of the fragment are obtained at once using DLA. Optical mapping also has intrinsic difficulties hampering its implementation: a need to optimize surface treatment in correspondence with the length of analyzed fragments, poor reproducibility of the surface treatment procedure, short shelf life of the derivatized glass, non-specific DNA photolysis catalyzed by DNA-bound intercalators and potentially incomplete cleavage of immobilized DNA by restriction endonucleases (29–31). Contrary to optical mapping, DLA uses microfluidic chips for DNA stretching that can be produced in bulk and have an unlimited shelf life. Fluidic stretching of DNA enables high throughput of molecules and samples because it occurs on the fly and does not involve manual manipulations. Photodamage of DNA does not influence DLA measurements because DNA relaxation time (32) exceeds the measurement time and the DNA backbone trace is uninterrupted regardless of photo-induced nicks. Finally, we perform tagging in solution and, differently from immobilized DNA, in optical mapping all target sites are equally accessible in our case.
Potential applications
In looking ahead, we foresee practical application of DLA for identification of species such as pathogens and other microbes. In fact, microbial identification through genotyping is now the preferred approach (28). In this context, DLA has a number of advantages. It is very sensitive requiring detection and analysis of just hundreds of molecules. In addition, DNA fragments measured by DLA are so long that they provide unique maps sufficient to identify the organism source by comparison to a database of known or informatically deduced patterns. This ability facilitates analysis of mixtures of genomic samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We gratefully acknowledge P. J. de Jong (CHORI, Oakland CA) for providing the E.coli clone with BAC 12M9. We thank Robert S. Langer and Alexander Rich for their comments on the manuscript. This research was partially funded by NSF grants DMI-0213876 and DMI-0320449. Funding to pay the Open Access publication charges for this article was provided by U.S. Genomics, Inc.
Conflict of interest statement. Most authors hold stock and/or stock options in US Genomics, Inc., the makers of the Trilogy 2020 single molecule analyzer. R. Gilmanshin is currently employed by U.S. Genomics and is involved in research on its behalf.
REFERENCES
- 1.Norwood D.A., Sands J.A. Physical map of the Clostridium difficile chromosome. Gene. 1997;201:159–168. doi: 10.1016/s0378-1119(97)00443-5. [DOI] [PubMed] [Google Scholar]
- 2.Ding C., Cantor C.R. Direct molecular haplotyping of long-range genomic DNA with M1-PCR. Proc. Natl Acad. Sci. USA. 2003;100:7449–7453. doi: 10.1073/pnas.1232475100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitra R.D., Butty V.L., Shendure J., Williams B.R., Housman D.E., Church G.M. Digital genotyping and haplotyping with polymerase colonies. Proc. Natl Acad. Sci. USA. 2003;100:5926–5931. doi: 10.1073/pnas.0936399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 5.Weiss K.M., Clark A.G. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 2002;18:19–24. doi: 10.1016/s0168-9525(01)02550-1. [DOI] [PubMed] [Google Scholar]
- 6.Lim A., Dimalanta E.T., Potamousis K.D., Yen G., Apodoca J., Tao C., Lin J., Qi R., Skiadas J., Ramanathan A., et al. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 2001;11:1584–1593. doi: 10.1101/gr.172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tegenfeldt J.O., Bakajin O., Chou C.-F., Chan S.S., Austin R., Fann W., Liou L., Chan E., Duke T., Cox E.C. Near-field scanner for moving molecules. Phys. Rev. Lett. 2001;86:1378–1381. doi: 10.1103/PhysRevLett.86.1378. [DOI] [PubMed] [Google Scholar]
- 8.Chan E.Y., Goncalves N.M., Haeusler R.A., Hatch A.J., Larson J.W., Maletta A.M., Yantz G.R., Carstea E.D., Fuchs M., Wong G.W., et al. DNA mapping using microfluidic stretching and single-molecule detection of fluorescent site-specific tags. Genome Res. 2004;14:1137–1146. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osoegawa K., Mamoser A., Wu C., Frengen E., Seng C., Catanese J., de Jong P.J. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frengen E., Weichenhan D., Zhao B., Osoegawa K., van Geel M., de Jong P.J. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics. 1999;58:250–253. doi: 10.1006/geno.1998.5693. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 12.Nielsen P.E., Egholm M. An introduction to PNA. In: Nielsen P.E., Egholm M., editors. Peptide Nucleic Acids. Protocols and Applications. Norfolk: Horizon Scientific Press; 1999. pp. 1–19. [Google Scholar]
- 13.Perkins T.T., Smith D.E., Chu S. Single polymer dynamics in an elongational flow. Science. 1997;276:2016–2021. doi: 10.1126/science.276.5321.2016. [DOI] [PubMed] [Google Scholar]
- 14.Chou H.-P., Spence C., Scherer A., Quake S.R. A microfabricated device for sizing and sorting DNA molecules. Proc. Natl Acad. Sci. USA. 1999;96:11–13. doi: 10.1073/pnas.96.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strobl G. The Physics of Polymers, 2nd edn. Berlin: Springer; 1997. [Google Scholar]
- 16.Sinden R.R. DNA Structure and Function. San Diego: Academic Press; 1994. [Google Scholar]
- 17.Perkins T.T., Smith D.E., Larson R.G., Chu S. Stretching of a single tethered polymer in a uniform flow. Science. 1995;268:83–87. doi: 10.1126/science.7701345. [DOI] [PubMed] [Google Scholar]
- 18.Bustamante C., Smith S.B., Liphardt J., Smith D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 19.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 20.Demidov V.V., Frank-Kamenetskii M.D. Sequence-specific targeting of duplex DNA by peptide nucleic acids via triplex strand invasion. Methods. 2001;23:108–122. doi: 10.1006/meth.2000.1112. [DOI] [PubMed] [Google Scholar]
- 21.Manneville S., Cluzel P., Viovy J.-L., Chatenay D., Caron F. Evidence for the universal scaling behaviour of a freely relaxing DNA molecule. Europhys. Lett. 1996;36:413–418. [Google Scholar]
- 22.Foquet M., Korlach J., Zipfel W., Webb W.W., Craighead H.G. DNA fragment sizing by single molecule detection in submicrometer-sized closed fluidic channels. Anal. Chem. 2002;74:1415–1422. doi: 10.1021/ac011076w. [DOI] [PubMed] [Google Scholar]
- 23.Demidov V.V., Kuhn H., Lavrentieva-Smolina I.V., Frank-Kamenetskii M.D. Peptide nucleic acid-assisted topological labeling of duplex DNA. Methods. 2001;23:123–131. doi: 10.1006/meth.2000.1113. [DOI] [PubMed] [Google Scholar]
- 24.Shizuya H., Birren B., Kim U.-J., Mancino V., Slepak T., Tachiiri Y., Simon M. Cloning and stable maintenance of 300 kb fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thayer E.C., Olson M.V., Karp R.M. Error checking and graphical representation of multiple-complete-digest (MCD) restriction-fragment maps. Genome Res. 1999;9:79–90. [PMC free article] [PubMed] [Google Scholar]
- 26.Dimalanta E.T., Lim A., Runnheim R., Lamers C., Churas C., Forrest D.K., De Pablo J.J., Graham M.D., Coppersmith S.N., Goldstein S., et al. A microfluidic system for large DNA molecule arrays. Anal. Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 27.Birren B., Lai E. Pulsed-Field Gel Electrophoresis: A Practical Guide. San Diego: Academic Press, Inc.; 1993. [Google Scholar]
- 28.Olive D.M., Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aston C., Hiort C., Schwartz D.C. Optical mapping: an approach for fine mapping. Methods Enzymol. 1999;303:55–73. doi: 10.1016/s0076-6879(99)03006-2. [DOI] [PubMed] [Google Scholar]
- 30.Aston C., Mishra B., Schwartz D.C. Optical mapping and its potential for large-scale sequencing projects. Trends Biotechnol. 1999;17:297–302. doi: 10.1016/s0167-7799(99)01326-8. [DOI] [PubMed] [Google Scholar]
- 31.Taylor J.R., Fang M.M., Nie S. Probing specific sequences on single DNA molecules with bioconjugated fluorescent nanoparticles. Anal. Chem. 2000;72:1979–1986. doi: 10.1021/ac9913311. [DOI] [PubMed] [Google Scholar]
- 32.Ladoux B., Doyle P.S. Stretching tethered DNA chains in shear flow. Europhys. Lett. 2000;52:511–517. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.