Abstract
Sequencing pathogen genomes is costly, demanding careful allocation of limited sequencing resources. We built a computational Sequencing Analysis Pipeline (SAP) to guide decisions regarding the amount of genomic sequencing necessary to develop high-quality diagnostic DNA and protein signatures. SAP uses simulations to estimate the number of target genomes and close phylogenetic relatives (near neighbors or NNs) to sequence. We use SAP to assess whether draft data are sufficient or finished sequencing is required using Marburg and variola virus sequences. Simulations indicate that intermediate to high-quality draft with error rates of 10−3–10−5 (∼8× coverage) of target organisms is suitable for DNA signature prediction. Low-quality draft with error rates of ∼1% (3× to 6× coverage) of target isolates is inadequate for DNA signature prediction, although low-quality draft of NNs is sufficient, as long as the target genomes are of high quality. For protein signature prediction, sequencing errors in target genomes substantially reduce the detection of amino acid sequence conservation, even if the draft is of high quality. In summary, high-quality draft of target and low-quality draft of NNs appears to be a cost-effective investment for DNA signature prediction, but may lead to underestimation of predicted protein signatures.
INTRODUCTION
Draft sequencing requires that the order of base pairs in cloned fragments of a genome be determined usually at least four times (4× depth of coverage) at each position for a minimum degree of draft accuracy. This information is assembled into contigs, or fragments of the genome that cannot be joined further due to lack of sequence information across gaps between the contigs. To generate high-quality draft, usually ∼8× coverage is optimal (1). Finished sequence, without gaps or ambiguous base calls, usually requires 8× to 10× coverage, along with additional analyses, often manual, to orient the contigs relative to one another and to close the gaps between them in a process called finishing. In fact, it has been stated that ‘the defining distinction of draft sequencing is the avoidance of significant human intervention’ (1), although there are computational tools that may also be capable of automated finishing in some circumstances (2).
While some tabulate the cost differential between high-quality draft versus finished sequences to be 3- to 4-fold, and the speed differential to be >10-fold (1), others state that the cost differential is a more modest 1.3- to 1.5-fold (3). In either case, draft sequencing is cheaper and faster. Experts have debated whether finished sequencing is always necessary, considering the higher costs (1,3,4).
Thus, here we set out to determine whether draft sequence data are adequate for the computational prediction of DNA and protein diagnostic signatures. By a ‘signature’ we mean a short region of sequence that is sufficient to uniquely identify an organism down to the species level, without false negatives due to strain variation or false positives due to cross reaction with close phylogenetic relatives. In addition, for DNA signatures, we require that the signature be suitable for a TaqMan reaction (e.g. composed of two primers and a probe of the desired Tms). Limited funds and facilities in which to sequence biothreat pathogens mean that decision makers must choose wisely which and how many organisms to sequence. Money and time saved as a result of draft rather than finished sequencing enables more target organisms, more isolates of the target and more near neighbors (NNs) of the target to be sequenced. However, if draft data do not facilitate the generation of high-quality signatures for detection, the tradeoff of quantity over quality will not be worth it.
We used the Sequencing Analysis Pipeline (SAP) (5,6) to compare the value of finished sequence, real draft sequence and simulated draft sequence of different qualities for the computational prediction of DNA and protein signatures for pathogen detection/diagnostics. Marburg and variola viruses were used as model organisms for these analyses, due to the availability of multiple genomes for these organisms. We hope that variola may serve as a guide for making predictions about bacteria, in which the genomes are substantially larger, and thus the cost of sequencing is much higher than for viruses. Variola was selected as the best available surrogate for bacteria at the time we began these analyses because:
it is double-stranded DNA;
it has a relatively low mutation rate, more like bacteria than like the RNA or shorter DNA viruses that have higher mutation rates and thus higher levels of variation;
it is very long for a virus, albeit shorter than a bacterial genome;
we have access to many genomes, which were sequenced by our collaborators at the US Centers for Disease Control and Prevention in Atlanta, GA;
there are finished genomes available, so we can compare actual finished data with simulated draft data.
Only recently have a fairly large number of Bacillus anthracis genomes become available to us. However, since only some of these are finished, currently we cannot compare finished with draft results for this bacterial genome.
MATERIALS AND METHODS
The sequencing analysis pipeline uses the DNA and protein signature pipelines
The draft SAP simulations are nearly identical to those using finished genomes, described previously (5,6). The SAP (Figure 1) performs stochastic (Monte Carlo) simulations and includes our DNA and Protein Signature Pipelines as components, which will be summarized briefly below. It is necessary to describe what the signature pipelines do before the SAP can be clearly described, so signature pipelines will be discussed first. As a step within the DNA and Protein Signature Pipelines, DNA sequence alignments of multiple draft genomes are required. For this we use the WGASA software, also summarized below. Once each of these components of the SAP has been presented, the SAP itself will be described.
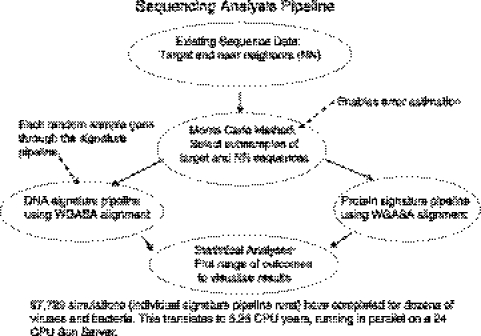
Figure 1.
Diagram of the SAP. For an SAP run, first a pool of target genome and a pool of NN genomes are collected. Then many random subsamples of target and NN genomes are selected from the pool, and each subsample is run through either the DNA signature pipeline or the protein signature pipeline, which identify regions conserved among target genomes and unique relative to non-target genomes, where unique regions are evaluated by comparing to a large sequence database of all currently available bacterial and viral complete genomes or the non-redundant protein database, excluding NNs from the NN pool that are not in that random subsample. Thus, each run of the SAP requires many runs of the DNA or protein signature pipelines with different random samples, generating a range of outcomes that are plotted on range plots.
DNA signature pipeline
The DNA Signature Prediction Pipeline, described in detail elsewhere (7–10), finds sequence regions that are conserved among target genomes by creating a consensus based on a multiple sequence alignment. WGASA is the software used in the analyses here to create an alignment and will be discussed below. Next, the DNA Signature Pipeline identifies regions that are unique in the target sequence consensus relative to all other non-target bacterial and viral sequences that we have in a >1 Gb database, which is frequently updated from the NCBI GenBank sequence database (11) and other sources (e.g. our collaborators at the CDC, USDA and other public sources, such as TIGR, Sanger Institute and the Joint Genome Institute). From the conserved, unique regions, signatures are selected based on the requirements of a particular technology, in this case, TaqMan PCR. These signature candidates may then proceed for further in silico screening (BLAST analyses to look for undesired inexact matches) before undergoing laboratory screening.
Protein signature pipeline
Protein signature prediction and SAP methods have previously been described in detail (6). The following briefly describes the procedure. First, target genomes are aligned using WGASA. A set of gene (start, end) pairs for both the plus and minus strands relative to the reference genome is required. This implies that coding frames for the translation of nucleic acid codons into amino acids for each protein of the target organism's genome have been correctly determined. From the aligned genomes, nucleotide codons are translated into amino acid sequence based on the gene locations, and conserved strings of six or more amino acids among all the target genomes are recorded. These conserved fragments are then compared with the NCBI GenBank non-redundant (nr) database of amino acid sequences, unveiling peptides that are unique to the target species. For our computations, we require that if a peptide signature is longer than six amino acids, then every sub-string of length six amino acids is also conserved and unique. There may be many conserved and unique peptide signatures on the same and on different proteins. The resulting conserved, unique peptides that are at least six amino acids long from open reading frames are considered to be protein signature candidates.
Signature peptides may be used as targets for antibody or ligand binding and may be developed for use in detection, therapeutics or vaccines (12,13). Since the signature regions are highly conserved within a species, it is likely that they are functionally important to the organism's survival or reproduction. Those signatures that land on or near protein active sites may be developed into therapeutics, since antibody or ligand binding may interfere with protein function. Signature regions may even be considered as vaccine targets, since these unique peptides may evoke a specific response in the host (14,15).
WGASA, a new sequence alignment tool
For draft genomes, WGASA, or Whole Genome Analysis through Scalable Algorithms, is used to align multiple sequences. This is the only available tool that enables multiple sequence alignment of draft genomes and that is capable of aligning large or many genomes. WGASA requires at least one finished reference genome and the others may be draft.
Only recently has it become possible to use the DNA Signature Pipeline to predict signature candidates for draft genomes. This capability is due to the invention of software for multiple sequence alignment of draft genomes with at least one completed full genome. WGASA was developed by David Hysom, Chuck Baldwin and Scott Kohn in the Computations directorate at Lawrence Livermore National Laboratory. They designed the software in close communication with members of our bioinformatics team, and it is tailored for our needs of generating diagnostic and forensic pathogen signatures.
WGASA can efficiently align large (e.g. bacterial) genomes. In addition, the developers have created a parallel version that runs in minutes, allowing the SAP simulations, involving thousands of calls to WGASA, to complete in a feasible time frame. In addition to the SAP analyses, this tool has enabled us to revisit signature predictions for several important organisms, such as the food-borne pathogen Listeria, that were previously problematic because some of the sequences were available only in draft format.
The tool requires that there be one or more complete, finished genome, and any number of draft sequences. It is based on suffix-tree algorithms (16). It requires that anchors, identical sequence fragments of user-specified length, be found in each of the genomes to be aligned. Thus, there must be some level of sequence conservation among the genomes in order to discover anchors of sufficient length (e.g. 35–60 bp) that are present in all the genomes. Then the regions between the anchors are aligned using a tool, such as clustalw or HMMer. The algorithm functions most efficiently if anchors are frequent and dispersed across the genomes to provide even coverage. If substitutions, deletions, insertions or gaps in sequence information (e.g. between contigs) result in an anchor's absence in one or more of the genomes, then those regions must be aligned using clustalw, which is slower and more memory intensive for large amounts of sequence data. Similar to all anchor-based alignment algorithms, WGASA is dependent upon a high degree of co-linearity across all input genomes.
SAP
DNA analyses
The SAP for DNA signature analyses operates as follows. First, all available complete genomes of target were gathered into a pool with the total genome count called T. A second pool was created of all available NN complete genomes, with the total count of sequences called N. Next, we selected 10 random samples of size t targets and n NNs, for all t ranging from 1 to T and all n ranging between 1 and minimum(10,N). We ran the DNA Signature Prediction Pipeline for each sample, with signature prediction based on conservation among the t target strains and uniqueness relative to a >1 Gb database minus those NNs in the NN pool that were not chosen in that sample. Thus, for each sample, signature candidates were predicted as though we had only t target and n NN sequences, as well as the rest of the less-closely related organisms in our database that are not considered NNs. In addition to the number of TaqMan signature candidates, the fraction of the genome that is conserved among the t target sequences was also calculated. Based on the combined results of the many signature pipeline runs using random target samples of size t and n, we assessed how much sequence data, that is, the values of t and n, was required to approximate the number of signature candidates c that were predicted when the full data set (all target and NN sequences, t = T and n = N) was analyzed with the signature pipeline. Using the full data set will yield the fewest signatures, because lack of conservation or uniqueness will winnow away all unsuitable candidates.
Thus, the SAP performs Monte Carlo sampling from the target and NN genomes, runs each sample through the signature pipeline and summarizes the results of the hundreds of signature pipeline runs in a single plot. On our 24 CPU Sun server, up to seven signature pipeline simulations may be run in parallel, each requiring ∼15–22 min for viral genomes. All of the SAP analyses of dozens of bacteria and viruses to date have used a total run time of 6.26 years (operating in parallel), with an average pipeline run time of 0.522 h, and a process time span of 2.32 years.
The span of predictions generated by different random samples of genomes is illustrated using range plots (Figures 2–8). Along the y-axis, whole numbers represent the number of target strains t and the incremental values between the integers represent the number n of NN genomes. Only Figures 3, 5 and 6 have the incremental n values, because for the other plots of target sequence conservation only, the number of NNs was not relevant, and for the protein analyses NN comparisons were not made (described below). Outcomes of the number of signature candidates or the fraction of the target genome that is conserved are plotted along the x-axis as horizontal lines spanning the range (of predicted numbers of signatures or fraction conserved) for the s random samples of size (t,n) with the median and quantiles of the range indicated by colored, short vertical lines. If a random sample of t target strains and n NN strains were sequenced, there would be a 90% chance that the number of signature candidates for that sample would be less than or equal to the 90% quantile mark. The expected outcome is a reduction in the number of signature candidates or the fraction of the genome that is conserved as the number of target and NN sequences used in the simulations increases, due to a reduction in conservation from additional targets and a reduction in uniqueness from additional NNs.
Figure 2.
Range plots of the conserved fraction of the target genome for Marburg virus (A) finished sequences and (B) draft sequences. The range of values from different random samples for a given sample size (number of target sequences) is drawn as a horizontal line. The 75th quantile of each range is marked with a short vertical tick. The conserved fraction using all of the target sequences is given in the box labeled ANS (for ‘answer’) and marked with a vertical line along this value on the x-axis.
Figure 8.
Range plots for the number of protein signature candidates for variola virus (A) finished sequence, (B) simulated draft target sequence with a low error rate, and (C) simulated draft target sequence with an intermediate error rate and (D) simulated draft target sequence with a high error rate.
Figure 3.
Range plots as described in the methods for the number of TaqMan signature candidates for Marburg virus for (A) finished and (B) draft sequences. To discriminate samples in which zero NNs were used, the range is drawn as a horizontal gray line, and when n > 0, the range is drawn as a black line. The best estimate of the true value is the quality measure determined using the entire target and NN pools, and is represented by a vertical black line. This best estimate plus a constant c = 20 is at the location of the vertical dashed line and was selected to indicate a reasonable distance from the true answer. The 75% quantile for each range is shown with a black, vertical tick mark.
Figure 5.
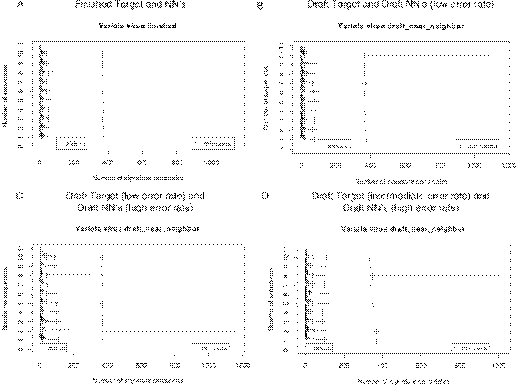
Range plots for the number of TaqMan signature candidates for variola virus (A) finished target and NN sequences, (B) simulated draft target and NN sequences with a low error rate, (C) simulated draft target sequences with a low error rate and draft NN sequences with a high error rate and (D) simulated draft target sequences with an intermediate error rate and draft NN sequences with a high error rate.
Figure 6.
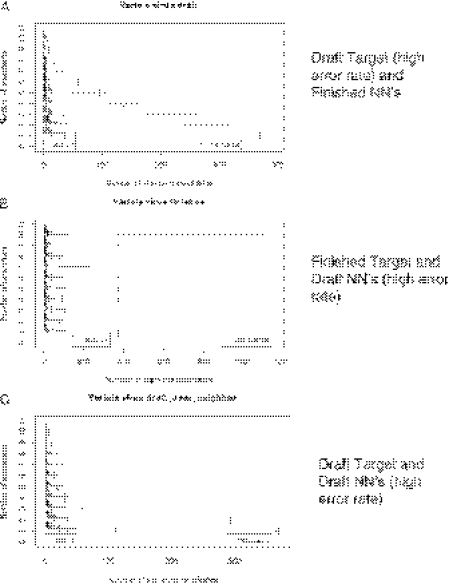
Range plots for the number of TaqMan signature candidates for variola virus (A) simulated draft target with a high error rate and finished NNs, (B) finished target and draft NNs with a high error rate and (C) draft target and draft NNs with a high error rate.
Protein analyses
The SAP analyses for proteins proceed much like that for DNA signatures. Random samples of size t target sequences are generated, where t ranges from 1 to T, the total number of target sequences in the pool. Either finished data, actual draft data, or draft data simulated as described below are aligned using WGASA. The protein signature prediction pipeline is run on each random sample, and the range, median, 75th and 90th quantiles of the number of protein signature candidates for the samples of a given target size t is plotted in range plots as described above.
Our DNA SAP analyses examined the effects of both the number of target as well as the number of NN sequences, but our protein SAP analyses investigated the effects of only the number of target sequences. This is because composing the lists of NN proteins for random, temporary exclusion from the protein nr database (to estimate the value of that NN sequence data) would be difficult to automate for rapid, high-throughput computations. Thus, we compared the target proteins with all the proteins in nr, regardless of their phylogenetic relationship to the target. This was comparable with DNA SAP results using all available NN data.
THE SEQUENCE DATA
Actual draft: Marburg virus
We had sequence data for four strains of Marburg virus, both the actual draft and the finished versions of those same isolates, provided for these analyses by a colleague working at Lawrence Livermore National Laboratory. The draft sequence was of ∼3× to 6× coverage, which enabled us to compare SAP results using the same strains in finished form. The identities of these sequences are provided in the Appendix. For the draft Marburg analyses, we selected one finished strain, the reference strain from GenBank (gi|13489275|ref|NC_001608.2| Marburg virus, complete genome), as the WGASA reference genome, and then used random sub-samples from the four draft genomes. Marburg was the only organism for which we could obtain a sufficient number of draft genomes for the SAP Monte Carlo simulations. A total of 814 simulations for DNA signatures, i.e. individual runs of the DNA signature pipeline, and 48 simulations for protein signatures were performed using Marburg finished and draft data, requiring an average of 15 min per simulation.
Simulated draft: variola virus
We used finished sequence data generously provided by collaborators at the US CDC for 28 variola major genomes and 22 NN genomes from the Orthopox family. The sequence identities are provided in the Appendix. Since we did not have real draft data available, we developed a program to simulate draft sequence from finished sequence, based on guidance from two colleagues who have been involved in sequencing efforts and the finishing process in the Biology and Biotechnology Research Program at Lawrence Livermore National Laboratory. In outline, the draft simulator program randomly cuts a genome into contigs of a size randomly selected from an exponential distribution. Stochastic simulation also determines whether there are gaps or overlaps between contigs, as well as the size of the gap or overlap. Sequencing errors are also simulated.
The following paragraphs describe the draft simulation process in greater detail. First, the 5′ end of the sequence is simulated as missing or present according to a random (Bernoulli) trial based on the probability of there being a gap in the sequence data. If simulations randomly determine that the first part of the sequence is missing, then the size of the missing segment is selected randomly from a uniform distribution ranging from the minimum gap size to the maximum gap size. The length of the first contig is selected randomly from an exponential distribution with a non-zero minimum contig size and a maximum contig size that is a fraction of the mean genome length for the species. The mean of this exponential distribution is also specified as a fraction of the mean genome length.
Next, a random Bernoulli trial again determines whether there is a gap or overlap between the first and second contigs, and the size of the gap or overlap is chosen from the appropriate uniform distribution (range for gap size = 1–2000 bases, range for overlap size = 20–40 bases). The size of the contig is selected from the exponential distribution as described above. Additional contigs are simulated in a similar manner.
Within each contig, sequencing errors are simulated based on the size of the contig, and whether the base position is at an end (first or last 100 bases) or in the middle of the contig. For long, double-stranded DNA viruses (e.g. variola) and bacteria, the sequencing error rates are larger at the beginning and end of a contig than in the middle, and small contigs are more likely to contain sequencing errors than are large contigs. In contrast, due to differences in generating the products for Sanger sequencing that are employed for smaller RNA and DNA viruses, there are often more sequencing errors in the middle of contigs for such smaller viral draft genomes. Although we did not specifically simulate draft for RNA and short DNA viruses, our simulator should work with minor modification to a few parameters. Thus, there are four parameters that must be specified for simulating sequencing errors: (i) the size cutoff for small versus large contigs, (ii) the probability of errors in the middle portion of small versus large contigs, (iii) the length of the contig ends where sequencing is either less accurate (bacteria and long double-stranded DNA viruses) or more accurate (small viruses, RNA viruses) and (iv) the probability of sequencing errors at the contig ends. If there is a sequencing error at a particular base, we assumed that that base is randomly changed to one of the other three bases with equal probability. Although additional features could be added to the draft simulation tool, the stochastic features that we have incorporated capture the main features of draft sequence and produce data that are suitable for SAP analyses.
We performed six sets of analyses using simulated variola draft. Three sets of simulated variola draft runs of the SAP used the following parameters:
probability of a gap between contigs = 0.95;
probability of overlap between contigs = 0.05;
minimum gap size if there is a gap (uniform distribution) = 1 bp;
maximum gap size = 2000 bp;
minimum overlap if there is overlap (uniform distribution) = 20 bp;
maximum overlap = 40 bp;
minimum contig size (exponential distribution) = 2000 bp;
maximum contig size = 0.5 × (mean genome length) bp
mean contig size = 0.05 × (mean genome length) bp
cutoff size for small versus large contigs = 10 000 bp;
probability of sequence errors inside large contigs = 0.01;
probability of sequence errors inside small contigs = 0.05;
length of contig ends = 100;
probability of sequence errors in the contig ends = 0.20;
We will refer to the above set of simulations as those with a high probability of sequencing errors, or low-quality draft. The other three simulated variola draft runs used all the same parameters as above, except that the sequencing error rates were dramatically lower, more in line with error rates of 10−5/base that the US Centers for Disease Control and Prevention (CDC) has indicated for their draft variola genomes:
probability of sequence errors inside large contigs = 10−5;
probability of sequence errors inside small contigs = 10−4;
probability of sequence errors in the contig ends = 10−3;
These runs were referred to as low error rate, or high-quality draft. Finally, we performed SAP runs using high error rate (low quality) simulated draft of the NN sequences and intermediate quality simulated draft of target genomes, using the following probabilities of sequencing errors:
probability of sequence errors inside large contigs = 10−3;
probability of sequence errors inside small contigs = 10−3;
probability of sequence errors in the contig ends = 10−3;
The intermediate quality simulated draft is consistent with error rates for draft sequencing cited in the literature (1,3).
SAP experiments using simulated draft variola
For the parameter values specified above, three SAP experiments were simulated. In the first, only the target sequences were simulated into draft, and the NN sequences remained as finished sequences. In the second, the NN sequences were converted to simulated draft and the target sequences remained as finished. In the third, both target and NN sequences were simulated into draft. In the second and third cases, all the NNs were run through the draft simulator each time they were chosen, so that the draft sequences (i.e. location and extent of gaps and sequence errors) differ for the same genome among samples. In the first and third cases, the target sequences must be aligned, and WGASA requires that one of the sequences be a finished genome for reference. Thus, for each random sample from the pool of target genomes, one genome was randomly selected to be the finished genome, and so was left as finished sequence, and the other genomes in the sample were replaced with simulated draft sequence (by running through the draft simulator) before alignment. As with NNs, target draft sequences differ for the same genome among samples due to the randomness of the draft simulation each time it is run. In addition, the target genome that is chosen to be the finished, reference genome differs between samples, and the other target genomes in the sample simulation are replaced with simulated draft versions of the actual finished sequences. Then these sequences were aligned using WGASA and the SAP process was run as described above. A total of 1101 stochastic simulations per ‘experiment’ were performed, requiring ∼18 min per simulation. Each simulation involved randomly selecting the subset of target and NN sequences to be included, simulating the draft data based on the finished genomes, aligning the target sample, and finally running the DNA Signature Pipeline. There were four combinations examined: (i) finished variola and finished NN, (ii) draft variola and finished NN, (iii) finished variola and draft NN and (iv) draft variola and draft NN, with each of the draft runs repeated for both low and high sequencing error rates. The combination (iv) was also run with intermediate quality simulated draft variola and low-quality simulated draft NNs. In total, there were eight computational experiments for the finished and simulated draft variola data.
Sequencing cost estimates
We have used the following function to estimate viral sequencing costs, based on discussion with our laboratory colleagues involved in sequencing and finishing.
| 1 |
This is merely a rough estimate, and the actual costs of sequencing any given organism may differ substantially from this rule-of-thumb calculation. In Equation 1, it is assumed that the cost of sequencing viruses does not decline for sequencing second and subsequent isolates. While this may be a false assumption in cases where isolates are similar to one another, in other cases where the new sequences are divergent, as isolates from different outbreaks or for viruses with rapid mutation rates, the cost is especially unlikely to decline. In addition, the $0.40/bp figure for draft of 6× to 8× coverage could range from $0.30 to $0.50/bp using shotgun sequencing, but may be as low as $0.10–$0.20/bp if primer walking works well (i.e. known primer sites are found in new isolates). Finishing could be 1–3 times more than the cost of draft, so we used a factor of 2 times more (draft $0.4/bp, finished $0.4 + 0.8/bp) in the equation above as a reasonable estimate. With rapidly evolving sequencing technologies and costs, these are only rough guides that may quickly become outdated.
RESULTS
Marburg virus
It may be substantially less expensive, on the order of 3-fold, to generate draft compared with finished sequence data for an organism like Marburg virus, according to estimates using Equation 1. For example, for $45K, one could sequence either two finished genomes or one finished and three draft. However, draft sequencing of this low quality (3× to 6×) for Marburg causes a dramatic decline in the ability to computationally eliminate regions of poor conservation, and thus to exclude poor signature regions (Figure 2). This occurs because gaps in the draft data of some of the sequences mask sequence variation among strains. Using the best available data, all six finished genomes, there is 75.2% sequence conservation. The deficiencies of draft data give a false impression that there is 92.6% sequence conservation (Table 1). Each additional finished genome reduces the conserved fraction by ∼5%, compared with a reduction of only 2% per genome for the draft data.
Table 1.
Summary of results using 28 variola genomes (finished or simulated draft as indicated) and 22 NN genomes from the Orthopox family, as well as the finished and draft Marburg results
| Species | Simulated draft or finished target | Draft or finished NNs | Percent conserved sequence (%) | Percent conserved and unique sequence (%) | Number TaqMan DNA signature candidates | Number conserved and unique regions | Longest conserved and unique region |
|---|---|---|---|---|---|---|---|
| Variola major virus | Simulated draft, high error rate | Finished | 58.30 | 57.79 | 0 | 4 | 23 |
| Variola major virus | Simulated draft, high error rate | Simulated draft, high error rate | 58.68 | 58.36 | 0 | 8 | 23 |
| Variola major virus | Finished | Simulated draft, high error rate | 98.90 | 3.91 | 1 | 71 | 49 |
| Variola major virus | Simulated draft, low error rate | Finished | 98.61 | 4.60 | 1 | 89 | 49 |
| Variola major virus | Simulated draft, low error rate | Simulated draft, low error rate | 98.65 | 4.43 | 0 | 86 | 49 |
| Variola major virus | Simulated draft, low error rate | Simulated draft, high error rate | 98.76 | 4.42 | 0 | 88 | 49 |
| Variola major virus | Simulated draft, intermediate error rate | Simulated draft, high error rate | 96.67 | 14.06 | 0 | 191 | 52 |
| Variola major virus | Finished | Simulated draft, low error rate | 98.84 | 4.05 | 1 | 80 | 49 |
| Variola major virus | Finished | Finished | 98.90 | 3.99 | 1 | 76 | 49 |
| Marburg virus | Real draft, 3× to 6× coverage | Finished | 92.60 | 83.31 | 43 | 250 | 198 |
| Marburg virus | Finished | Finished | 75.19 | 74.36 | 0 | 38 | 41 |
The percent of the target genome that is conserved varies slightly among the runs using finished target sequences because different genomes were randomly selected to be the reference strain in each multiple sequence alignment.
The overestimation of conservation using draft Marburg data also results in overestimation of the number of signature candidates (Figure 3). Samples of four draft targets plus one finished reference yield 43 signature candidates. A smaller sample size of only two draft targets and one finished reference generate upwards of 80 candidates. These results differ from those using finished genomes, where the lack of sequence conservation is more evident and there are zero TaqMan signatures conserved among all strains. Most combinations of four finished genomes are sufficient to eliminate non-conserved signatures (Figure 3A). Although predictions that there are 0 signature candidates shared among all finished strains may seem to argue against TaqMan methods, in fact this information provides important guidance for the development of TaqMan signatures with degenerate bases or a set of signatures that will, in combination, pick up all sequenced strains. Other analyses indicate that there are TaqMan signatures conserved among five of the six strains, so that two signatures would form a minimal set that could detect both the one divergent and the other five strains.
Variola virus simulated draft
Estimated sequencing costs of draft variola and draft NNs indicate that draft may require only one-quarter to one-half the costs of finished sequencing. Simulations of high-quality draft data indicated that it is as good as finished data for diagnostic signature prediction. The conservation range plots (Figure 4A and B) are virtually identical for finished and high-quality draft, and indicate that ∼98% of the genome is conserved among sequenced isolates. For intermediate quality draft (Figure 4C) the conservation range plot is also similar to that for finished sequence, showing that ∼97% of the genome appears to be conserved. The range plots for the number of TaqMan signature candidates are very similar for finished sequence data, high or intermediate quality draft target, and high or low-quality draft NNs (Figure 5A–D).
Figure 4.
Range plots for the conserved fraction of the target genome for variola virus for (A) finished sequence, (B) simulated draft sequence with a low error rate, (C) simulated draft with an intermediate error rate and (D) simulated draft with a high error rate.
In contrast to the results using high-quality simulated draft or actual Marburg draft, simulations of low-quality variola draft target illustrate that sequence conservation may be underestimated compared with results with finished sequence data, due to sequencing errors (Table 1 and Figure 4D). With low-quality draft target, it appears that only 58% of the genome is conserved among isolates.
Low-quality (high error rate) draft NN data, however, yield results that are very similar to those with high-quality draft or finished NN data, as long as the target sequence information is of intermediate to high quality (Figure 5C–D and Figure 6): At least four NN sequences are necessary to ensure that signature regions are unique, whether the NN data are low- or high-quality draft or finished. That is, our simulations indicate that low-quality draft NN data are adequate for predicting DNA signatures, as long as there is good quality target sequence data. This results because errors in the NN sequences occur at random locations that differ in each NN sequence. As long as at least one of the NNs has enough correct sequence to eliminate each of the non-unique target regions, then the unique regions of the target can be determined.
The results illustrated in the figures are emphasized by the data in Table 1. This table shows the fraction of the target genome that is conserved and conserved+unique, the number of conserved+unique regions that are at least 18 contiguous base pairs long and the number of base pairs in the largest of these regions, since these are the sections that are of sufficient length for one or possibly more primers to be located. The number of these regions is similar for finished data and for draft with a low error rate. Low-quality draft (with a high rate of sequencing errors) for the target data, however, gives the false impression that there are fewer and shorter regions that are conserved and suitable as signature regions than is actually the case.
There is an artifact in some of our results that is a consequence of the order in which we calculate conservation and then uniqueness, although this does not affect the signatures that are predicted. First, a conservation gestalt is generated from the sequence alignment, in which non-conserved bases are replaced by a dot (‘.’). Then uniqueness is calculated based upon perfect matches of at least 18 bp long between the conservation gestalt and a large sequence database of non-target sequences. Non-conserved bases in the conservation gestalt may break up a region into conserved fragments of <18 bases long, and as a result these short fragments are not tested for uniqueness. Consequently, if there is a low level of conservation, then we may overestimate the fraction of the genome that is unique. For example, in Table 1 the conserved+unique fraction is 4% with finished variola target data, but is overestimated at 58% with low-quality draft. This artifact does not, however, affect TaqMan signature prediction, since the regions suitable for primers and probes must have at least 18 contiguous, conserved bases, and all of these are tested for uniqueness, i.e. there is no underestimation of uniqueness in conserved fragments that are at least 18 bp in length, and thus no underestimate of uniqueness in the predicted signatures. We are working to eliminate this issue in future versions of the software.
Protein SAP results
Protein results show a large disparity between finished and draft data. There are 113 protein signature candidates for finished Marburg data compared with only two protein signature candidates for Marburg draft (Figure 7). For variola, using all available target data, 97, 14, 6 and 0 protein signatures are predicted using finished, low error, intermediate error and high error draft target data, respectively (Figure 8). Thus, sequencing errors substantially reduce the detection of amino acid sequence conservation, even if sequencing errors occur at the low rate of 10−4–10−5 across most of the genome.
Figure 7.
Range plots for the number of protein signature candidates for Marburg virus (A) finished and (B) draft sequence data.
The pattern of how additional sequences reduce the number of protein signature candidates also differs for draft compared with finished sequence data. With finished data, there is a large range in the number of peptide signature candidates predicted with 17 or fewer variola genomes, and this range narrows around the lower bound with >17 genomes. With 16 genomes, the 75% quantile mark approaches the final predicted number of 97 signatures (Figure 8A). This pattern indicates that there is a set of 97 peptides that are highly conserved among all currently sequenced variolas, which are unlikely to be eroded even as more sequence data are obtained. In other words, additional sequence data are probably not needed at this time in order to computationally predict good peptide signature targets, and as few as 16 finished target sequences would most likely have been adequate to generate this same list of ∼100 peptide signatures.
Draft data, in contrast, whether they are of low or high quality, mask the above pattern (Figure 8B and C): the range and 75th quantile of the number of peptide signatures gradually decline with each additional target sequence (rather than a sudden, sharp drop as is seen with the finished data), suggesting that additional target sequences would continue to erode the number of peptide signatures. This occurs since sequencing errors occur at random, in different locations in each of the draft target genomes, and obscure the truly conserved peptides. One might falsely infer from peptide SAP results based on the draft data that additional sequencing (beyond the 28 variola major genomes used here) would be useful in generating peptide signature candidates. In actuality, however, SAP analyses using the finished sequence data indicate that there are already ample sequence data for peptide signature prediction.
DISCUSSION AND CONCLUSIONS
The failure of draft sequencing for Marburg at 3× to 6× coverage or of simulated variola draft with a high error rate to facilitate the prediction of detection signatures highlights a need for finished viral sequences, or at least for draft of high quality such as 8×. Otherwise, a large number of signature candidates either will fail in screening because they are incorrectly designated as conserved among strains (as observed with the Marburg results), or too few regions will be classified as conserved (as observed with variola), and thus not be considered for signatures.
The variola simulations with intermediate to high-quality draft (that is, a low error rate, approximating what one might observe with 8× coverage) target and/or NN genomes deliver virtually the same results as finished genomes. Considering that it costs approximately three times as much to generate finished sequence as it does draft, our analyses indicate that investing in more high-quality draft target genomes is better than investing in fewer finished genomes. For our analyses, only one target strain must be finished, and the remaining target sequences and all the NNs may be provided as draft. Our results indicate that NN sequencing may be of low coverage, and thus of low quality, without serious detriment to signature prediction, as long as there are at least four NN draft genome sequences.
If high-quality draft sequence is used, and it appears that there is too little sequence conservation among target strains, one might relax specifications for 100% conservation among strains for diagnostic signature prediction. Calculations indicate that it is often possible to generate signatures if one allows a base to be considered ‘conserved’ if it is present in only a fraction of the genomes (e.g. 75%) rather than the standard requirement for 100% conservation when finished sequence data are used. We have used this ‘ratio-to-win’ option to generate signature candidates for some highly divergent RNA viruses (for which we have finished sequence), although usually our preference is to include degenerate bases, especially when there are only a few bases with heterogeneity among strains in a given signature candidate. Using a ratio-to-win approach may be particularly important for the generation of protein signature candidates, since draft target data severely compromises the ability to detect conserved strings of amino acids.
In summary, intermediate to high-quality draft sequencing of target genomes, combined with low-quality draft sequencing of close phylogenetic relatives, is sufficient for the prediction of DNA diagnostic signatures. Prediction of peptide/protein signature candidates, in contrast, requires finished sequencing to avoid substantial underestimation of conserved peptide regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
This work was performed under the auspices of the US Department of Energy by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48. This work was supported by the Intelligence Technology Innovation Center. Sequence data from the CDC and colleagues at Lawrence Livermore National Laboratory, which we have used in our analyses, are gratefully acknowledged: P. Chain at LLNL provided draft Marburg sequence data; Variola and Orthopox genome sequences were generously shared by our CDC colleagues J. Esposito, I. Damon, S. Sammons, M. Frace, J. Osborne, R. Kline, and M. Olsen-Rasmussen. Although some of the sequence data used in these analyses is not yet in the public domain, it is currently submitted for publication. We also thank D. Hysom, C. Baldwin and S. Kohn for providing us with the WGASA alignment software, and E. Skowronski and P. Chain for guidance in developing a draft simulator. Funding to pay the Open Access publication charges for this article was provided by the Intelligence Technology Innovation Center.
Conflict of interest statement. None declared.
APPENDIX
Marburg finished genomes used in these analyses
| Fasta header | Sequence length | Sequence description |
|---|---|---|
| gi|13489275|ref|NC_001608.2| Marburg virus, complete genome | 19 112 | Marburg virus, complete genome |
| Unpublished strain 1 | 19 113 | Unpublished sequence of Marburg virus |
| PP3 | 19 113 | AY430365 |
| PP4 | 19 112 | AY430366 |
| Ozolin | 19 151 | AY358025 |
| Unpublished strain 2 | 19 083 | Unpublished sequence of Marburg virus |
Draft Marburg sequence data (intermediate versions of the same finished sequences given above): unpublished strain 1, 17 contigs ranging from 859 bases to 29 302 bases; PP3, 15 contigs ranging from 778 bases to 24 884 bases; PP4, 15 contigs ranging from 779 bases to 19 728 bases; unpublished strain 2, 42 contigs ranging from 818 bases to 40 767 bases.
Near neighbors for Marburg virus
| Fasta header | Sequence length |
|---|---|
| gi|23630482|gb|AY142960.1| Zaire Ebola virus strain Mayinga subtype Zaire, complete genome | 18 959 |
| gi|21702647|gb|AF499101.1| Zaire Ebola virus strain Mayinga, complete genome | 18 960 |
| gi|11761745|gb|AF272001.1| Zaire Ebola virus strain Mayinga, complete genome | 18 959 |
| gi|10313991|ref|NC_002549.1| Zaire Ebola virus, complete genome | 18 959 |
| gi|33860540|gb|AY354458.1| Zaire Ebola virus strain Zaire 1995, complete genome | 18 961 |
| Raw sequence of Ebola virus strain Zaire-95 from LLNL on Aug 29 2003 1:13PM | 18 961 |
| gi|15823608|dbj|AB050936.1| Reston Ebola virus genomic RNA, complete genome | 18 890 |
| gi|22789222|ref|NC_004161.1| Reston Ebola virus, complete genome | 18 891 |
Variola major sequence data
| Fasta header | Sequence length | Sequence description |
|---|---|---|
| gi|9627521|ref|NC_001611.1| Variola virus, complete genome | 185 578 | Variola virus, complete genome |
| gi|623595|gb|L22579.1| VARCG Variola major virus (strain Bangladesh-1975) complete genome | 186 103 | Variola major virus (strain Bangladesh-1975) complete genome |
| 26 Unpublished CDC sequences | Various | Variola virus, complete genome |
Near neighbors for variola
| Fasta header | Sequence length | Sequence description |
|---|---|---|
| gi|20152989|gb|AF482758.1| Cowpox virus strain Brighton Red, complete genome | 224 501 | Cowpox virus strain Brighton Red, complete genom |
| gi|30519405|emb|X94355.2| CV41KBPL Cowpox virus strain GRI-90, complete genome | 223 666 | Cowpox virus strain GRI-90, complete genome |
| gi|30844336|ref|NC_003663.2| Cowpox virus, complete genome | 224 499 | Cowpox virus, complete genome |
| 1 cowpox genomes | – | Unpublished CDC sequence |
| gi|17974913|ref|NC_003310.1| Monkeypox virus, complete genome | 196 858 | Monkeypox virus, complete genome |
| 3 Monkeypox genomes | Various | Unpublished sequence from the CDC |
| gi|29692106|gb|AY243312.1| Vaccinia virus strain WR, complete genome | 194 711 | Vaccinia virus strain WR, complete genome |
| gi|9790357|ref|NC_001559.1| Vaccinia virus, complete genome | 191 737 | Vaccinia virus, complete genome |
| gi|47088326|gb|AY603355.1| Vaccinia virus strain Acambis 3000 Modified Virus Ankara (MVA), complete genome | 166 722 | Vaccinia virus strain Acambis 3000 Modified Virus Ankara (MVA), complete genome |
| 3 Vaccinia genomes | Various | Unpublished CDC sequence |
| gi|22164589|ref|NC_004105.1| Ectromelia virus, complete genome | 209 771 | Ectromelia virus, complete genome |
| Ectromelia virus (Naval) | 207 620 | http://athena.bioc.uvic.ca/pbr/ncbi_gb/pocsBrowser.php?ECTV-Nav |
| 1 Taterapox genome | – | Unpublished CDC sequence |
| gi|18640237|ref|NC_003391.1| Camelpox virus, complete genome | 205 719 | Camelpox virus, complete genome |
| gi|19717929|gb|AY009089.1| Camelpox virus CMS, complete genome | 202 205 | Camelpox virus CMS, complete genome |
| 1 Buffalopox genome | – | Unpublished CDC sequence |
| gi|46401901|ref|NC_005858.1| Rabbitpox virus, complete genome | 197 731 | Rabbitpox virus, complete genome |
| RPXV-UTR_forward_1-197731 | 197 731 | Raw sequence of Rabbitpox virus (strain Utrecht) from poxvirus.org on Aug 05 2003 1:59PM |
REFERENCES
- 1.Branscomb E., Predki P. On the high value of low standards. J. Bacteriol. 2002;184:6406–6409. doi: 10.1128/JB.184.23.6406-6409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon D., Desmarais C., Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser C.M., Eisen J.A., Nelson K.E., Paulsen I.T., Salzberg S.L. The value of complete microbial genome sequencing (you get what you pay for) J. Bacteriol. 2002;184:6403–6405. doi: 10.1128/JB.184.23.6403-6405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouck J., Miller W., Gorrell J.H., Muzny D., Gibbs R.A. Analysis of the quality and utility of random shotgun sequencing at low redundancies. Genome Res. 1998;8:1074–1084. doi: 10.1101/gr.8.10.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner S.N., Lam M.W., Mulakken N.J., Torres C.L., Smith J.R., Slezak T.R. Sequencing needs for viral diagnostics. J. Clin. Microbiol. 2004;42:5472–5476. doi: 10.1128/JCM.42.12.5472-5476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner S.N., Kuczmarksi T.A., Zhou C.E., Lam M.W., Slezak T.R. A System to Assess Genome Sequencing Needs for Viral Protein Diagnostics and Therapeutics. J. Clin. Microbiol. 2005;43:1807–1817. doi: 10.1128/JCM.43.4.1807-1817.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slezak T., Kuczmarski T., Ott L., Torres C., Medeiros D., Smith J., Truitt B., Mulakken N., Lam M., Vitalis E., et al. Comparative genomics tools applied to bioterrorism defence. Brief. Bioinform. 2003;4:133–149. doi: 10.1093/bib/4.2.133. [DOI] [PubMed] [Google Scholar]
- 8.Fitch J.P., Gardner S.N., Kuczmarski T.A., Kurtz S., Myers R., Ott L.L., Slezak T.R., Vitalis E.A., Zemla A.T., McCready P.M. Rapid development of nucleic acid diagnostics. Proc. IEEE. 2002;90:1708–1721. [Google Scholar]
- 9.Fitch J.P., Chromy B.A., Forde C.E., Garcia E., Gardner S.N., Gu P., Kuczmarksi T.A., Melius C., McCutchen-Maloney S.L., Milanovich F.M., et al. Biosignatures of pathogen and host. Proceedings of the IEEE Workshop on Genomic Signal Processing and Statistics (GENSIPS); Raleigh, NC. 2002. [Google Scholar]
- 10.Gardner S.N., Kuczmarski T.A., Vitalis E.A., Slezak T.R. Limitations of TaqMan® PCR for detecting divergent viral pathogens illustrated by hepatitis A, B, C, and E viruses and human immunodeficiency virus. J. Clin. Microbiol. 2003;41:2417–2427. doi: 10.1128/JCM.41.6.2417-2427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Rapp B.A., Wheeler D.L. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews T., Salgo M., Greenberg M., Chung J., DeMasi R., Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD 4 lymphocytes. Nature Rev. Drug Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 13.Okkels L.M., Brock I., Follmann F., Agger E.M., Arend S.M., Ottenhoff T.H., Oftung F., Rosenkrands I., Andersen P. PPE protein (Rv3873) from DNA segment RD1 of Mycobacterium tuberculosis: strong recognition of both specific T-cell epitopes and epitopes conserved within the PPE family. Infect. Immun. 2003;71:6116–6123. doi: 10.1128/IAI.71.11.6116-6123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGaughey G., Barbato G., Bianchi E., Freidinger R., Garsky V., Hurni W., Joyce J., Liang X., Miller M., Pessi A., et al. Progress towards the development of a HIV-1 gp41-directed vaccine. Curr. HIV Res. 2004;2:193–204. doi: 10.2174/1570162043484933. [DOI] [PubMed] [Google Scholar]
- 15.Choy W., Lin S., Chan P., Tam J., Lo Y., Chu I., Tsai S., Zhong M., Fung K., Waye M., et al. Synthetic peptide studies on the severe acute respiratory syndrome (SARS) coronavirus spike glycoprotein: perspective for SARS vaccine development. Clin. Chem. 2004;50:1036–1042. doi: 10.1373/clinchem.2003.029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giegerich R., Kurtz S., Stoye J. Efficient implementation of lazy suffix trees. Software-Practice and Experience. 2003;33:1035–1049. [Google Scholar]