Abstract
Ribosomes synthesize proteins according to the information encoded in mRNA. During this process, both the incoming amino acid and the nascent peptide are bound to tRNA molecules. Three binding sites for tRNA in the ribosome are known: the A-site for aminoacyl-tRNA, the P-site for peptidyl-tRNA and the E-site for the deacylated tRNA leaving the ribosome. Here, we present a study of Escherichia coli ribosomes with the E-site binding destabilized by mutation C2394G of the 23S rRNA. Expression of the mutant 23S rRNA in vivo caused increased frameshifting and stop codon readthrough. The progression of these ribosomes through the ribosomal elongation cycle in vitro reveals ejection of deacylated tRNA during the translocation step or shortly after. E-site compromised ribosomes can undergo translocation, although in some cases it is less efficient and results in a frameshift. The mutation affects formation of the P/E hybrid site and leads to a loss of stimulation of the multiple turnover GTPase activity of EF-G by deacylated tRNA bound to the ribosome.
INTRODUCTION
The ribosome is a molecular machine, translating mRNA into a protein. Amino acids, which are to be incorporated into a nascent peptide are delivered to the ribosomes attached to tRNA. Within the ribosome, the tRNA traverses three binding sites. The A-site accepts the incoming aminoacyl-tRNA. The P-site holds the tRNA with the peptide attached, which is to be transferred to the new amino acid residue in the course of the peptidyltransferase reaction. During peptide transfer, the A-site bound tRNA gains a peptide and the P-site bound tRNA becomes deacylated. For the synthesis to be continued, a specific GTPase–elongation factor G (EF-G) has to catalyse the translocation of peptidyl-tRNA to the P-site, thus making the A-site capable of accepting another aminoacyl-tRNA. According to a classical model, the ribosome should be able to function with only the A- and P-sites (1,2). Later, additional site specific for the deacylated tRNA was discovered (3) and afterwards identified in all examined living organisms (4–6). This site was named E-site (for exit of tRNA from the ribosome).
The location of the E-site on the ribosome was studied by cryo-EM (7), and X-ray structure analysis (8,9). The 23S rRNA nucleotides C2394, G2112 and G2116, being in contact with E-site bound tRNA were identified by chemical probing (10). Modification interference experiments had shown that contact with C2394 was essential for the binding (11). Later, direct contact of this nucleotide with the 3′-terminal adenosine residue of the E-site tRNA was confirmed by X-ray crystallography (8). Since deletion of the terminal adenosine residue abolished tRNA binding to the E-site (12,13), both partners of the A76–C2394 interaction appeared to be crucial for the binding of tRNA to the E-site. 2′-hydroxyl groups at the tRNA residues 71 and 76 were also shown to be necessary for the E-site binding (14). According to Gnirke et al. (15), codon–anticodon interactions are necessary for the E-site binding (16).
The functional role of the E-site has been extensively discussed in the literature and still is a matter of controversy (17,18). Nierhaus and colleagues suggested the concept of the ‘allosteric three-site model’ (19). The major postulates of this model are that binding to the E-site is strong and codon-dependent (16), and that the binding to the A- and E-sites is negatively coupled (15,20). The functional role of this latter feature is to prevent binding of a non-cognate aminoacyl-tRNA to the A-site (21) and to maintain the reading frame (22). The properties of the allosteric three-site model had been disputed in the literature (7,17,23). Particularly, the codon–anticodon interactions in the E-site (24) and negative coupling between the A- and E-site occupation (25) were questioned. It was suggested that E-site binding was transient and unstable, so being only necessary for lowering of the activation energy barrier for translocation. Later, however, a contact between codon and anticodon at the E-site was visualized, although for the non-cognate tRNA (9).
The role of the E-site in translocation deserves special attention. Translocation is catalysed by EF-G, which hydrolyses a GTP molecule in the course of this process. The action of elongation factor G is required only after completion of the peptidyl transferase reaction. Thus, the factor, which binds 70 Å away from the peptidyl transferase centre, must ‘sense’ whether the peptide is attached to the CCA-end of the tRNA in the P-site or to that in the A-site. Modulation of the uncoupled GTPase activity of EF-G by P-site bound tRNA was discovered by Chinali and Parmeggiani (26) and studied under various conditions (27–29). Later, Zavialov and Ehrenberg (30) studied this phenomenon in more detail. It was shown that not only GTPase activity, but also the binding of the EF-G to the ribosome was under the control of the tRNA in the P-site. It appears that for the function of EF-G, the most essential parameter is the state of the CCA-end of the tRNA bound to the P-site, rather than that of the tRNA bound to the A-site. In agreement with this, anticodon stem–loop bound to the A-site could be translocated by EF-G, thus indicating that CCA-end of the tRNA, bound to the A-site is not required for translocation (31).
Wintermeyer and colleagues (27) observed that deacylated tRNA bound to the P-site stimulates the activity of EF-G. This stimulation was compromised if peptidyl-tRNA was bound to the P-site, nor was it detected if a tRNA analogue incapable of entering the E-site was located at the P-site. In their search for determinants essential for tRNA translocation, Feinberg and Joseph (14) revealed that 2′-hydroxyl groups necessary for the E-site binding were also crucial for the translocation. Thus, it appears that all determinants, essential for the E-site binding are also necessary for the fast and efficient translocation.
How can EF-G detect that the peptidyl moiety is attached to the tRNA in the P-site? Why does the activity of EF-G correlate with the tRNA affinity to the E-site? In the 1980s, Moazed and Noller (32) in their ‘hybrid model’ postulated that after peptide transfer the deacylated CCA-end of the tRNA spontaneously moves to the E-site on the large ribosomal subunit, while its anticodon remains locked in the P-site of the small subunit until the EF-G-driven translocation occurs. At the same time, the CCA-end of the A-site bound tRNA carrying the peptide group is moved to the P-site. This process has also been disputed in the literature (33). However, it is clear that translocation of the CCA-ends of the tRNAs on the large subunit precedes translocation of the anticodons and mRNA on the small subunit. Although it requires EF-G under certain conditions, the formation of the hybrid states does not need GTP hydrolysis (33,34). The activity of EF-G might depend, not on the absence of a peptide bound to the CCA-end of the tRNA at the P-site, but rather correlates with the ability to form the hybrid P/E-site (27), even if the hybrid sites are forming only as an intermediate in translocation (30,33). It should be noted, however, that not all models of the translocation consider the hybrid sites formation. According to the α–ɛ model (23) based on iodine cleavage studies of ribosome-bound tRNA, deacylated tRNA is translocated to the E-site without significant alterations of the tRNA environment on the ribosome (35). Contacts of the 3′-terminal region of tRNA could not be monitored by the iodine cleavage, so even the α–ɛ model allows the formation of the A76–C2394 interaction as the first step of translocation.
To test the roles of the E- and P/E-sites in the elongation cycle, we created mutant E.coli ribosomes that were severely compromised in their E-site binding as indicated by various assays. The mutant ribosomes were tested for the efficiency of various stages of the elongation cycle in vitro, and for their translation error rates in vivo. Our results give us an increased knowledge of the function of the E-site.
MATERIALS AND METHODS
The C2394G mutation was made by standard site-directed mutagenesis procedures in a SphI–BamHI fragment of the rrnB operon cloned into M13 mp19 phage and subcloned into the plasmid pLK1192U (36) essentially as described (37). Transformation of the AVS69009 strain (38), plasmid substitutions and checking the purity of mutant rRNA in the cells were carried out as described (37). Translational fidelity was measured using a set of reporter strains (39,40). For translation fidelity assays, the cells were harvested at the same stage of growth, namely mid-log phase. Doubling times were measured by monitoring A600 in Luria–Bertani medium at 37°C, using three independent clones to monitor the growth rate of each strain. Doubling times were calculated by a least square linear approximation of the logarithms of the optical density of the cultures in their log phase.
Ribosomes were isolated from the AVS69009 strain, expressing only mutant rDNA. The isolation was carried out using the subunit reassociation technique (41). The wild-type ribosomes, used as a control were purified from the same strain, carrying the same plasmid without the C2394G mutation. All binding experiments were made in a buffer favourable for E-site binding (41) consisting of 20 mM HEPES–K pH 7.5, 6 mM Mg(OAc)2, 150 mM NH4OAc, 2 mM spermidine, 0.05 mM spermine and 4 mM 2-mercaptoethanol. Preparation of recombinant EF-G, EF-Tu, EF-Ts, phenylalanine-, methionine- and lysine-tRNA synthetases and transformylase (42), [14C]Phe-tRNAPhe, [14C]Lys-tRNALys and [3H]AcPhe-tRNAPhe was as described (43), except that aminoacyl-tRNAs were HPLC-purified. Radioactively labelled tRNAPhe with and without the 3′-terminal adenine was prepared by T7 transcription from the tRNAPhe gene, amplified by PCR using 3′-primers with 2′-OMe residues to obtain a transcript with a homogeneous 3′ end (44). To label , natural (Sigma) was 5′-labelled with [γ-32P]ATP by polynucleotide kinase.
Binding studies were performed in 20 µl volume, containing 2 pmol of ribosomes. Each experiment was repeated at least twice with independent ribosome preparations. The natural mRNA analogue was T7-transcribed from a PCR-amplified fragment of the plasmid pET33b (Novagen). Either polyU or the natural mRNA analogue was added to a final concentration of 0.5 µg/µl or in a 2- to 3-fold excess over ribosomes, respectively. Equal amounts of ribosomes and mRNA were used for toe-printing. For binding of tRNAPhe, or AcPhe-tRNAPhe to the P-site, a 2-fold excess of tRNA was incubated with ribosomes for 10 min at 37°C. Deacylated tRNA titrations were performed similarly, except that the amount of tRNA was varied. Binding of AcPhe-tRNAPhe to the A-site was accomplished similarly, by incubation of a 2-fold excess of AcPhe-tRNAPhe with P-site filled ribosomes for 10 min at 37°C. A-site binding of Phe-tRNAPhe or Lys-tRNALys, in 2-fold excess over ribosomes, was facilitated by addition of stoichiometric amounts of recombinant EF-Tu and one-tenth amounts of EF-Ts in the presence of 0.5 mM GTP. The reaction mixtures were incubated for 10 min at 37°C. Translocation was induced by addition of equimolar amounts of recombinant EF-G in the presence of 0.5 mM GTP. Nitrocellulose binding was performed as described (43).
Chemical modification of the ribosomes and their functional complexes was made as described (37), with the exception that the buffer conditions and complex concentrations were exactly as in the binding experiments described above. Toe-printing (45) was performed with the natural mRNA analogue, with the 5′-[32P]-labelled primer pre-hybridized 50 nt downstream from the AUG codon. GTPase assays were made as in Leonov et al. (43). The buffer conditions were 60 mM Tris-Cl, pH 7.8, 80 mM NH4Cl, 6 mM MgCl2, 8 mM β-mercaptoethanol (28). EF-G (0.8–70 nM) was added together with 0.5 mM GTP and 0.01 µCi/µl [γ-32P]GTP (final volume 35 µl) to the ribosomal complex (0.07 µM) containing polyU (1.4 µg/µl) and tRNAPhe. Reactions were running at 20°C and stopped after 40 min incubation by addition of formic acid to 10% concentration. If the time course of reaction is to be monitored, the EF-G concentration was kept at 7 nM, and reaction was stopped at 1–60 min by addition of formic acid. Thin layer chromatography was carried out as described (46).
RESULTS
In our study, the nucleotide C2394 of the 23S rRNA was altered to G. The AVS69009 strain of E.coli in which all the chromosomal rDNA operons were deleted (38) was transformed with a plasmid expressing the mutant rDNA. After substitution of the housekeeping rDNA plasmid, a strain was obtained with all the 23S rRNA molecules carrying the mutation at position 2394. The absence of wild-type 23S rRNA was confirmed by primer extension analysis (data not shown).
The mutation C2394G in the 23S rRNA leads to a slower growth rate at 37°C (doubling time 103 ± 2 min) than that of the wild-type strain (85 ± 2 min). Practically no growth of the strain, carrying the mutation C2394G in the 23S rRNA could be detected at 30°C. To check the ability of the mutant ribosomes to bind tRNA to the E-site, ribosomes carrying the C2394G 23S rRNA mutation were prepared from the corresponding E.coli strains.
Inability of the mutants to accommodate tRNA at the E-site
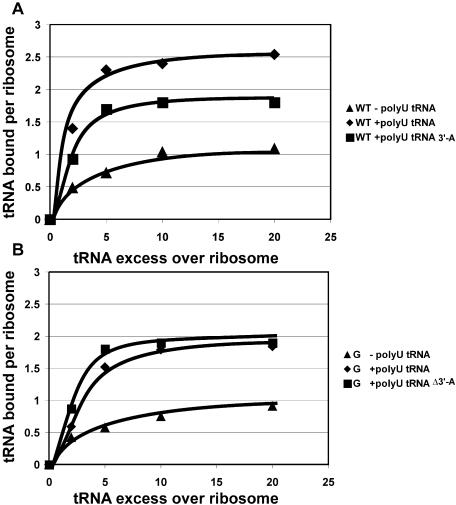
The binding of the deacylated tRNA to ribosomes was studied as in the classical experiments of Rheinberger et al. (4). Since binding to the E-site depends critically on the ionic conditions (7), we used a polyamine-containing buffer favourable for E-site binding in all the experiments. Ribosomes were titrated with deacylated radioactive tRNAPhe, with or without polyU as a template, and the amount of bound tRNA was determined by filtration through nitrocellulose. As an additional control, tRNAPhe lacking the 3′-terminal adenosine was used, as this should not be able to bind to the E-site (12,13). As expected (Figure 1A), wild-type ribosomes in the presence of polyU showed a binding approaching the three molecules of deacylated tRNA per ribosome. The actual value observed was 2.5 tRNA molecules per ribosome, explained by the low affinity of deacylated tRNA for the A-site of the wild-type ribosomes. The absence of the 3′-terminal adenosine of tRNA reduces the binding to two tRNA molecules per wild-type ribosome (bound to the P- and A-sites), the value observed being 1.8, while in the absence of mRNA only a single molecule binds (to the P-site; value observed 1.0). The C2394G mutant ribosomes (Figure 1B) showed a binding approaching only 2 tRNA molecules per ribosome in the presence of the polyU template (i.e. to the P- and A-sites). The binding curves for normal tRNA and tRNA lacking the 3′-terminal adenosine were practically identical for the mutant ribosomes. Since tRNA without the 3′-terminal adenosine is known not to bind to the E-site (12,13), while retaining its binding to the A- and P-sites, the obvious conclusion is that the mutant ribosomes are specifically impaired in their E-site binding.
Figure 1.
Binding of deacylated tRNAPhe to ribosomes. The graph shows the number of tRNA molecules bound to wild-type ribosomes (A), or to ribosomes carrying the C2394G mutation (B), versus tRNA excess over ribosomes. The curves are marked according to the presence or absence of a polyU template and to the tRNA molecule used. Closed triangles indicate ribosomes with presence of tRNA and absence of polyU. Closed diamonds indicate ribosomes with presence of both tRNA and polyU. Closed squares correspond to ribosomes with presence of polyU and tRNA-Δ3′A. ‘tRNA’ corresponds to intact tRNAPhe, ‘tRNA-Δ3′A’ corresponds to tRNAPhe lacking the 3′-terminal adenine.
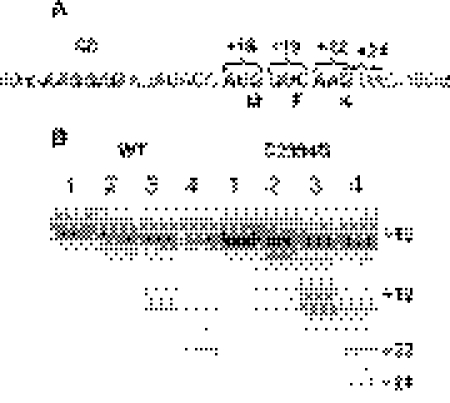
We monitored the direct binding of deacylated to the E-site of ribosomes programmed with a natural mRNA analogue coding for the MF peptide (Figure 2A). For this purpose, the P-site was filled with AcPhe-tRNAPhe, thus positioning the AUG codon at the E-site. Wild-type ribosomal complexes readily bound to the E-site, as revealed by filtration through nitrocellulose. In contrast, ribosomal complexes carrying the C2394G mutation with the P-site occupied in the same way were significantly compromised in their ability to bind (Table 1). The observed 11% binding could, however, be explained by the deacylated tRNA binding to the fraction of ribosomes (∼10%), whose P-site was not occupied during preincubation with AcPhe-tRNAPhe.
Figure 2.
Stepwise translation of the MFK-encoding mRNA analogue. (A) Partial sequence of the mRNA analogue. The Shine–Dalgarno sequence, the codons being translated and the corresponding toe-print stops are marked. The toe-print stop is repeatedly appearing at the nucleotide +16 relative to the 5′-most nucleotide at the P-site. (B) Toe-print analysis of the stepwise translation progress. ‘WT’ and ‘C2394G’ mark the complexes formed by wild-type ribosomes and ribosomes carrying the C2394G mutation, respectively. Lane 1, complex of ribosomes (0.1 µM), MFK-mRNA (0.1 µM) and (0.2 µM). Lane 2, ribosomes (0.1 µM), MFK-mRNA (0.1 µM), (0.2 µM) and AcPhe-tRNAPhe (0.2 µM). Lane 3, same as in lane 2 but after the addition of EF-G*GTP (0.1 µM). Lane 4, corresponds to lane 3 but with Lys-tRNALys*EF-Tu*GTP (0.2 µM) added. Reverse transcriptase stops are marked by numbers starting from the A in the initiation AUG codon. The primer was hybridized 50 nt downstream of the AUG codon.
Table 1.
Direct binding of deacylated to the E-site of MF-programmed ribosomes
| tRNA added | tRNA bound per ribosome | |||
|---|---|---|---|---|
| WT | C2394G | |||
| AcPhe-tRNAPhe (P-site) | (E-site) | AcPhe-tRNAPhe (P-site) | (E-site) | |
| AcPhe-tRNAPhe | 0.77 ± 0.04 | 0.95 ± 0.06 | ||
| AcPhe-tRNAPhe, | 0.76 ± 0.03 | 0.44 ± 0.05 | 0.91 ± 0.06 | 0.11 ± 0.03 |
Elongation cycle abnormalities associated with the mutation C2394G in the 23S rRNA
Ribosomes carrying the C2394G mutation were tested in vitro for the ability to form different functional complexes. For this purpose, we used the mRNA, coding for MFK peptide, the set of transfer RNAs and elongation factors. Each functional complex was characterized by a number of techniques. Filtration through the nitrocellulose was used to determine the amount of tRNA bound to the ribosome (Table 2); toe-printing was used to assign the tRNA to the particular binding site (Figure 2); and chemical modification allowed us not only to monitor tRNA binding by an independent approach, but also to distinguish between the ‘classic’ and ‘hybrid’ tRNA binding sites (Figure 3).
Table 2.
Deacylated , remaining on the ribosome at different steps of the translation in vitro of the MFK-encoding mRNA analogue
| Stage of elongation | bound per ribosome | |
|---|---|---|
| WT | C2394G | |
| binding to the P-site | 0.82 ± 0.01 | 0.83 ± 0.10 |
| AcPhe-tRNAPhe binding to the A-site | 0.76 ± 0.03 | 0.69 ± 0.05 |
| EF-G*GTP, translocation | 0.70 ± 0.10 | 0.45 ± 0.08 |
| Lys-tRNALys*EF-Tu*GTP binding to the A-site | 0.45 ± 0.05 | 0.40 ± 0.04 |
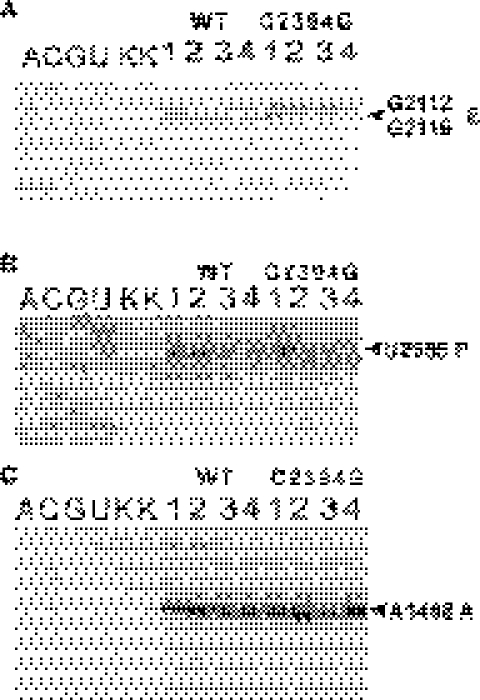
Figure 3.
Footprints of tRNAs in the various functional complexes. Lane numbers correspond to (1) empty ribosomes; (2) ribosomes carrying MFK-mRNA and ; (3) ribosomes carrying MFK-mRNA, and AcPhe-tRNAPhe; (4) ribosomes with MFK-mRNA, , AcPhe-tRNAPhe and EF-G*GTP; (A, C, G, U) correspond to sequencing lanes, (K) to the unmodified rRNA. Modification with kethoxal was used in (A), CMCT in (B) and DMS in (C). Primers complementary to the 23S rRNA positions 2281–2301 (A), 2649–2667 (B) and 16S rRNA positions 1440–1458 (C) were used for the extension reactions. Primer extension stops corresponding to the nucleotides protected by tRNA are indicated, together with their tRNA binding site specificity.
At the first stage, MFK-programmed ribosomes were incubated with radioactive deacylated [32P] . The level of binding was ∼82% (Table 2) for both wild-type and mutant ribosomes. Toe-printing shows that the binding occurs to the P-site, as expected (Figure 2B, lane 1). However, the main difference could be seen in the chemical modification assay (Figure 3). Wild-type ribosomes accommodate deacylated to the hybrid P/E-site. The G2112/G2116 residues, attributed to the E-site became protected (Figure 3A, lane 2); extent of modification being 0.23 ± 0.03, relative to 1 for the free ribosomes. P-site specific residue U2585 remained unprotected (Figure 3B, lane 2); extent of modification being 1.0 ± 0.1 relative to the unprotected ribosomes. The different situation was observed in the case of C2394G mutant. Here, the P-site specific residue was protected from modification (Figure 3B, lane 2); extent of protection being 0.38 ± 0.03, relative to the free ribosomes. The E-site specific nucleotides (Figure 3A, lane 2) were almost unprotected—0.69 ± 0.04 modification relative to the free ribosomes. This result indicated that the P/E hybrid site formation was damaged by the C2394G mutation.
After the filling of the P-site with , the A-site was occupied by AcPhe-tRNAPhe. Approximately 50% of the A-sites became occupied by AcPhe-tRNAPhe, which led to the band doubling on the toe-print (Figure 2B, lane 2). The low magnesium concentration, which caused the low non-enzymatic binding of AcPhe-tRNAPhe could not be avoided here, since those were the conditions for the highest E-site affinity, indispensable for one studying the effect of the C2394G mutation, affecting this very site. The release of deacylated tRNA at that stage was ∼10% for both wild-type and mutant ribosomes (Table 2). Chemical modification revealed AcPhe-tRNAPhe and being in the hybrid A/P- and P/E-sites for the wild-type ribosomes and in the ‘classical’ A- and P-sites for the mutant ribosomes. The P-site specific U2585 was protected in both types of ribosomes (Figure 3B, lane 3). The protection of the A-site specific A1408 residue of the 16S rRNA in both wild-type and mutant ribosomes leads to the conclusion that the A-site on the 30S subunit is occupied to the level of 0.55 ± 0.03 (Figure 3C, lane 3), consistent with the result of the toe-print (Figure 2B, lane 2). However, protection of the G2112/G2116 residues of 23S rRNA was very modest for the C2394G mutant (relative modification 0.60 ± 0.05), but high (relative modification 0.17 ± 0.01) for the wild-type ribosomes (Figure 3A, lane 3), supporting the hybrid sites formation in the latter case. Although we cannot exclude that some tRNA could be found in the hybrid P/E state even in the mutant ribosome, it is clear that the efficiency of the hybrid site formation is significantly decreased.
Translocation was then induced by addition of EF-G and GTP to the ribosomal complexes with AcPhe-tRNAPhe in the A-site (Figure 3B and C, lane 3). Translocation of AcPhe-tRNA to the P-site did not cause the release of the deacylated tRNA from the wild-type ribosomes (Table 2), as in the experiments of Nierhaus and colleagues (15). However, for the mutant ribosomes the loss of deacylated tRNA (Table 2) was equal to the amount of translocation (Figure 2B, lane 3). The deacylated tRNA, remained on the mutant ribosomes was bound to the P-site, as was shown by toe-printing (Figure 2B, lane 3). The same conclusion could be deduced for the chemical modification experiment. After translocation, the wild-type ribosomes holds deacylated tRNA in the E-site, there it protected G2112/G2116 residues (Figure 3A, lane 4); relative modification being 0.18 ± 0.01. A significantly weaker protection could be observed for the C2394G mutant (Figure 3A, lane 4); relative modification being 0.56 ± 0.03. We cannot exclude that deacylated tRNA could be translocated to the E-site of the mutant ribosomes. It is obvious, however, that the E-site of the mutant ribosomes is not able to retain the deacylated tRNA stably.
After the translocation of deacylated tRNA to the E-site of the wild-type ribosomes and, apparently, the release of the major part of the deacylated tRNA to the solution for the C2394G mutant, the A-site was enzymatically filled with Lys-tRNALys with the help of EF-Tu, EF-Ts and GTP. Immediately after A-site occupation and peptidyl transferase reaction, a further round of translocation was catalyzed by the EF-G present in the mixture (Figure 2B, lane 4). The deacylated tRNA, which remained bound to the E-site of the wild-type ribosomes was released to the solution (Table 2). According to the toe-print, the deacylated tRNA fraction, which remained on the ribosomes after that step occupied P-site (Figure 2B, lane 4). No further release of deacylated tRNA from the mutant ribosomes was observed. It means that for pre-translocation complexes formed by the mutant ribosomes, the release of tRNA happened during the translocation, or quickly after.
Several other differences between the wild-type ribosomes and ribosomes, carrying the C2394G mutation could be noted from the step-by-step formation of the functional complexes. It is clear that, although for mutant ribosomes, translocation could be catalysed by EF-G, it is less efficient in the case of translocation of AcPhe-Lys-tRNALys, but not for the AcPhe-tRNAPhe. Whereas for the wild-type ribosomes, both translocation reactions go to the same high extent (66% for the first translocation and 68% for the second translocation), the C2394G mutant ribosomes show similar translocation efficiency (69%) for the first translocation, but only a partial translocation (43%) of AcPhe-Lys-tRNALys (compare Figure 2B, lane 4). Moreover, an additional reproducible toe-print signal, specific for the ribosomes carrying the C2394G mutation appeared after binding of Lys-tRNALys, peptide transfer and translocation (Figure 2, lane 4). This signal corresponds to an incorrect translocation of AcPhe-Lys-tRNALys in the +2 frame. This signal is specific for the translocation and could not be explained by the re-binding of the Lys-tRNALys, since no detectable toe-print could be observed for Lys-tRNALys.
Defects in the E- and P/E-site formation affects EF-G function
It is generally agreed that E-site tRNA binding is important for translocation, since the derivatives of tRNA with diminished ability to bind the E-site slows down translocation (14,27). We decided to check the ability of EF-G to bind either empty ribosomes, carrying C2394G mutation (Figure 4, lanes 4 and 5) or their pre-translocation complex (Figure 4, lanes 2 and 3). The latter complex was created by sequential addition of fMet- and Phe-tRNAPhe*EF-Tu*GTP to the MFK-programmed ribosomes. EF-G binding was stabilized either by addition of GMPPNP (Figure 4, lanes 2 and 4) or by GTP and fusidic acid (Figure 4, lanes 3 and 5). The binding of EF-G was demonstrated for both the empty ribosomes and pre-translocation complex, and for both GMPPNP and fusidic acid stabilized conformations (Figure 4), although binding of EF-G to the mutant C2394G ribosomes was slightly reduced practically in all complexes investigated (Table 3).
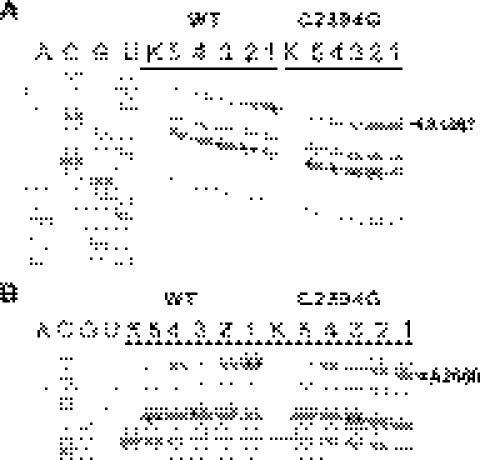
Figure 4.
Interaction of ribosomes carrying the C2394G mutation with EF-G, monitored by footprinting. (A) DMS footprints of EF-G on the GAC of the wild-type (WT) and mutant (C2394G) ribosomes. (A, C, G, U)—correspond to sequencing lanes; (K)—to the unmodified rRNA. Other lanes of the gel are marked according to the ribosomal complex, modified with DMS. (1) empty ribosomes; (2) ribosomes, MFK-mRNA, fMet-, Phe-tRNAPhe*EF-Tu*GTP, EF-G*GMPPNP; (3) ribosomes, MFK-mRNA, fMet-, Phe-tRNAPhe*EF-Tu*GTP, EF-G*GTP, fusidic acid; (4) ribosomes, EF-G*GMPPNP; (5) ribosomes, EF-G*GTP, fusidic acid. The primer extension stop corresponding to the nucleotide A1067 protected from DMS modification by EF-G is marked. (B) DMS footprints of EF-G on the Sarcin–Ricin loop of the wild-type and mutant ribosomes. Lanes are marked as in (A). The primer extension stop corresponding to the nucleotide A2660 protected from DMS modification by EF-G is marked.
Table 3.
EF-G binding to the different ribosomal complexes, as monitored by protection from chemical modification
| Ribosomal complex | WT | C2394G |
|---|---|---|
| Relative modification of the nucleotide 2661 | ||
| Ribosomes, MFK-mRNA, fMet-, Phe-tRNAPhe*EF-Tu*GTP, EF-G*GTP, fusidic acid | 0.79 ± 0.04 | 0.90 ± 0.05 |
| Ribosomes, MFK-mRNA, fMet-, Phe-tRNAPhe*EF-Tu*GTP, EF-G*GMPPNP | 0.31 ± 0.06 | 0.56 ± 0.05 |
| Ribosomes, EF-G*GTP, fusidic acid | 0.59 ± 0.10 | 0.73 ± 0.08 |
| Ribosomes, EF-G*GMPPNP | 0.11 ± 0.04 | 0.37 ± 0.04 |
| Relative modification of the nucleotide 1067 | ||
| Ribosomes, MFK-mRNA, fMet-, Phe-tRNAPhe*EF-Tu*GTP, EF-G*GTP, fusidic acid | 0.86 ± 0.10 | 1.01 ± 0.10 |
| Ribosomes, MFK-mRNA, fMet-, Phe-tRNAPhe*EF-Tu*GTP, EF-G*GMPPNP | 0.62 ± 0.07 | 0.74 ± 0.05 |
| Ribosomes, EF-G*GTP, fusidic acid | 0.73 ± 0.10 | 0.80 ± 0.05 |
| Ribosomes, EF-G*GMPPNP | 0.50 ± 0.05 | 0.63 ± 0.04 |
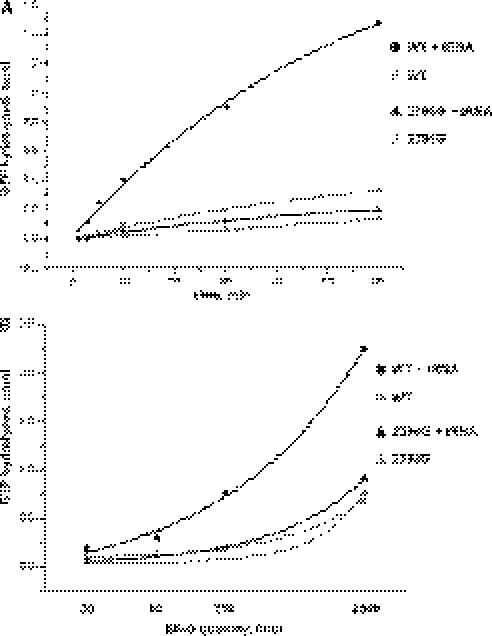
The P/E hybrid state formation was damaged by the C2394G mutation. However, this state is one of the translocation intermediates (33). Its formation precedes GTP hydrolysis (32,34). Since EF-G binding to the mutant ribosomes was reduced only slightly, we decided to investigate the influence of the mutation on the ribosome-induced GTP hydrolysis by EF-G and other steps of the EF-G cycle, which could be measured as the multiple turnover GTPase activity. We monitored the multiple turnover GTPase activity of EF-G induced by empty ribosomes and polyU-programmed ribosomes carrying deacylated tRNAPhe. In agreement with Zavialov and Ehrenberg (30) and Lill et al. (27), we observed increased stimulation of the GTPase reaction of EF-G by the complex of wild-type ribosomes with deacylated tRNAPhe (Figure 5A). Empty ribosomes carrying the C2394G mutation stimulate the GTPase activity of EF-G slightly less efficiently; this effect might be attributed to the minor decrease in the binding of EF-G to the empty ribosomes, carrying C2394G mutation (Figure 5). A dramatic difference between the wild-type and mutant ribosomes was found in the stimulation of EF-G GTPase activity by ribosome-bound deacylated tRNAPhe (Figure 5B). According to the structure probing data, deacylated tRNA strongly prefer binding to the P/P-site of mutant ribosomes, in contrast to the P/E-site in case of the wild type. It is likely that the lack of the multiple turnover GTPase activity stimulation by deacylated tRNA is due to the inefficiency of the hybrid P/E-site formation caused by the C2394G mutation. This result suggests that formation of the P/E hybrid state at least shows a good correlation with the activation of EF-G. Alternatively, the lack of stimulation of the multiple turnover GTP hydrolysis by the mutant ribosomes, carrying deacylated tRNA, could reside in some other stage of the EF-G working cycle, which we have no technical possibility to monitor.
Figure 5.
Stimulation of the EF-G-catalysed GTPase reaction by complexes of wild-type ribosomes (WT), and ribosomes carrying the C2394G mutation (C2394G). (A) The time course of multiple turnover [γ-32P]GTP hydrolysis by EF-G, stimulated by either empty ribosomes or the complex of polyU-programmed ribosomes, carrying deacylated tRNAPhe in the P-site. (B) The dependences of the GTP hydrolysis rates on the concentration of EF-G. The hydrolysis was stimulated by either the empty ribosomes or the complex of polyU-programmed ribosomes with deacylated tRNAPhe. The curves are marked as indicated on the right-hand side of the graphs. Closed circles correspond to the polyU programmed wild-type ribosomes carrying deacylated tRNAPhe; grey circles correspond to the empty ribosomes; closed triangles correspond to the polyU programmed 2394G ribosomes carrying deacylated tRNAPhe; grey triangles correspond to the empty 2394G ribosomes.
Defects in the E- and/or P/E-site binding causes increased frameshifting and stop-codon readthrough in vivo
Using a toe-print assay, we were able to detect aberrant translocation of the correct tRNA substrates in vitro. To check whether this effect could be detected in vivo, we applied a set of lacZ encoding plasmids, constructed by O'Connor and Dahlberg (39). Each plasmid in this set (with the exception of pSG25, which was used as a control) contains a mutation in the lacZ gene, which would lead to the production of a non-functional protein if the translation of the corresponding mRNA were to proceed without mistakes. Thus, the active enzyme could only be synthesized as a result of a translational error, such as frameshifting, stop codon readthrough or misreading, compensating the mutation encoded in the DNA. Both stop codon readthrough and frameshifting levels were elevated in the strains expressing the mutant rDNA (Table 4). In contrast, the mutation C2394G had no influence on translational misreading. The synthesis of the wild-type galactosidase from the control reporter plasmid was also elevated in the C2394G mutant. Some other rRNA mutations, e.g. in the helix 34 of the 16S rRNA, could also cause an increase in the expression of the wild-type lacZ gene (47). The elevated levels of UAA readthrough and −1 frameshifting were sufficiently above the overall increase in translation, caused by the C2394G mutation, thus allowing to attribute them safely to the direct influence on the fidelity. An influence of the E-site occupation on maintaining the translation reading frame was suggested by the experiments reported by Nierhaus and colleagues (22). However, it was not possible in the past to test this idea in an in vivo experiment. Our data provide evidence for the influence of E-site binding on translational accuracy.
Table 4.
Effect of the C2394G mutation on translational fidelity
| Test plasmid | Type of translation error | WT | C2394G |
|---|---|---|---|
| pSG 853 | UAA readthrough | 2.2 ± 0.6 | 7.3 ± 0.6 |
| pSG 12-6 | UAG readthrough | 7 ± 0.4 | 12 ± 0.5 |
| pSG 3/4 | UGA readthrough | 43 ± 2 | 64 ± 3 |
| pSG lac7 | +1 frameshift | 41 ± 2 | 74 ± 4 |
| pSG 12DP | −1 frameshift | 44 ± 2 | 84 ± 5 |
| f'CSH 103 | Glu/Gln substitution | 0.6 ± 0.1 | 0.6 ± 0.1 |
| pSG25 | no error | 4280 ± 70 | 5990 ± 80 |
Values are expressed in Miller units of the β-galactosidase activity. The higher is the activity, the more frequent is the translational error event.
DISCUSSION
The role of the ribosomal E-site in ribosomal function has been a matter of debates for years. Although the results of many experiments were published, they often contradicted each other. Here we have described a mutant, C2394G of the 23S rRNA, which is severely compromised in the binding of deacylated tRNA to the E- and P/E-sites. This was proven by a set of methods, including saturation binding of deacylated tRNA, direct binding to the E-site, stepwise translation and structure probing. The cells expressing only the mutant rDNA were viable, thus indicating that the lack of stable tRNA binding to the E-site could be tolerated, at least at 37°C. Moderate increase in frameshifting and stop-codon readthrough were found associated with the C2394G mutation in vivo. Increased frameshifting frequencies associated with the mutation could be due to the relaxed fixation of the tRNAs in the post-translocational state, when instead of two tRNAs as in the wild-type ribosomes, only a single P-site bound tRNA holds the reading frame. The frameshifting might also be related to the incorrect translocation, as evidenced by the toe-printing experiments in vitro.
The mutation C2394G affects translocation. Why the first translocation is equally efficient for the wild-type ribosomes and the C2394G mutant ribosomes, but the second one is not? The first translocation causes the substitution of at the P-site with pept-tRNAPhe, while the second one leads to the substitution of tRNAPhe with pept-tRNALys. Comparison of the affinities of different deacylated tRNA species to the P-site shows that tRNAPhe binds more tightly than , while tRNALys is one of the weakest P-site binders (15). Moreover, during the first translocation the interaction of a Shine–Dalgarno sequence of mRNA with 16S rRNA could retain, while it should be destroyed during the second translocation.
There are many indications that translocation proceeds through an intermediate formation of the hybrid A/P- and P/E-states (27,30,32,34). The hybrid states cannot be formed before the peptide transfer, so this might be a mechanism to ensure that EF-G binding and action only occur at the proper time. Interestingly, it was shown that binding of tRNA, able to move to the hybrid P/E-site of ribosomes even if A/P-site is empty seems to be sufficient to stimulate the GTPase activity of EF-G (27,30). However, a direct influence of the deacylated 3′ end of the tRNA, bound to the P-site could not be ruled out. Here, we observed no stimulation of the multiple turnover GTPase activity of EF-G by deacylated tRNA bound to ribosomes carrying the C2394G mutation. Chemical footprinting clearly shows that deacylated tRNA strongly prefers binding to the P/P instead of the P/E-site of the mutant ribosomes. Thus, tRNA binding to the P/E-site correlates with uniformly efficient and accurate translocation, as was found by the stepwise translation of MFK-encoding mRNA, monitored by toe-printing. The E-site participate in translocation both prior to GTP hydrolysis, when formation of the P/E hybrid state triggers the activity of EF-G, and later, when the E-site accepts the deacylated tRNA. Binding of deacylated tRNA to the E-site should make the translocation more energetically favourable, thus increasing its efficiency. At the same time, formation of the hybrid P/E-site should lower the activation barrier for translocation, increasing its speed. The cells, carrying the C2394G mutation are cold sensitive, which speaks in favour of the higher activation barrier for translocation, introduced by damage to the E-site. An alternative explanation for the cold sensitivity might rest on higher elongation rate at the higher temperature. It might be that the A-site occupation happens before the deacylated tRNA leaves the E-site, even if the affinity to the E-site is reduced by the C2394G mutation.
A further intriguing question is how the allosteric signal from the E- or P/E-site can reach the binding site of the elongation factor, which is located 70–100 Å away. We have demonstrated here that a mutation in the large subunit of the ribosome can affect the function of the EF-G, although the latter binds at more than half-a-ribosome distance away, thus underscoring the importance of this allosteric link.
Acknowledgments
The authors are very grateful to Drs T. Ueda, C. Squires and A.E. Dahlberg for providing us with strains and plasmids, and to Drs A. Mankin, K. Nierhaus and M. Rodnina for their comments on the manuscript. This work was supported by grants from the Howard Hughes Medical Institute #55000303, the Russian Foundation for Basic Research 04-06-49505, 05-04-49712, Leading Scientific Schools 1707.2003.4 and the Deutsche Forschungsgemeinschaft. Funding to pay the Open Access publication charges for this article was provided by Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Warner J.R., Rich A. The number of soluble RNA molecules on reticulocyte polyribosomes. Proc. Natl Acad. Sci. USA. 1964;51:1134–1141. doi: 10.1073/pnas.51.6.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson J.D. The synthesis of proteins upon ribosomes. Bull. Soc. Chim. Biol. 1964;46:1399–1425. [PubMed] [Google Scholar]
- 3.Noll H. Chain initiation and control of protein synthesis. Science. 1966;151:1241–1245. doi: 10.1126/science.151.3715.1241. [DOI] [PubMed] [Google Scholar]
- 4.Rheinberger H.J., Sternbach H., Nierhaus K.H. Three tRNA binding sites on Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1981;78:5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saruyama H., Nierhaus K.H. Evidence that the three-site model for the ribosomal elongation cycle is also valid in the archeobacterium Halobacterium halobium. Mol. Gen. Genet. 1986;204:221–228. [Google Scholar]
- 6.Triana-Alonso F.J., Chakraburtty K., Nierhaus K.H. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 1995;270:20743–20748. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal R.K., Penczek P., Grassucci R.A., Burkhardt N., Nierhaus K.H., Frank J. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J. Biol. Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- 8.Schmeing T.M., Moore P.B., Steitz T.A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 10.Moazed D., Noller H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 11.Bocchetta M., Xiong L., Shah S., Mankin A.S. Interactions between 23S rRNA and tRNA in the ribosomal E site. RNA. 2001;7:54–63. doi: 10.1017/s1355838201001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grajevskaja R.A., Ivanov Y.V., Saminsky E.M. 70-S ribosomes of Escherichia coli have an additional site for deacylated tRNA binding. Eur. J. Biochem. 1982;128:47–52. doi: 10.1111/j.1432-1033.1982.tb06929.x. [DOI] [PubMed] [Google Scholar]
- 13.Lill R., Lepier A., Schwagele F., Sprinzl M., Vogt H., Wintermeyer W. Specific recognition of the 3′-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J. Mol. Biol. 1988;203:699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg J.S., Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc. Natl Acad. Sci. USA. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gnirke A., Geigenmuller U., Rheinberger H.J., Nierhaus K.H. The allosteric three-site model for the ribosomal elongation cycle. Analysis with a heteropolymeric mRNA. J. Biol. Chem. 1989;264:7291–7301. [PubMed] [Google Scholar]
- 16.Rheinberger H.J., Sternbach H., Nierhaus K.H. Codon-anticodon interaction at the ribosomal E site. J. Biol. Chem. 1986;261:9140–9143. [PubMed] [Google Scholar]
- 17.Semenkov Y.P., Rodnina M.V., Wintermeyer W. The ‘allosteric three-site model’ of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc. Natl Acad. Sci. USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nierhaus K.H., Junemann R., Spahn C.M. Are the current three-site models valid descriptions of the ribosomal elongation cycle? Proc. Natl Acad. Sci. USA. 1997;94:10499–10500. doi: 10.1073/pnas.94.20.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nierhaus K.H. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 20.Rheinberger H.J., Nierhaus K.H. Allosteric interactions between the ribosomal transfer RNA-binding sites A and E. J. Biol. Chem. 1986;261:9133–9139. [PubMed] [Google Scholar]
- 21.Nierhaus K.H. Solution of the ribosome riddle: how the ribosome selects the correct aminoacyl-tRNA out of 41 similar contestants. Mol. Microbiol. 1993;9:661–669. doi: 10.1111/j.1365-2958.1993.tb01726.x. [DOI] [PubMed] [Google Scholar]
- 22.Marquez V., Wilson D.N., Tate W.P., Triana-Alonso F., Nierhaus K.H. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Spahn C.M., Nierhaus K.H. Models of the elongation cycle: an evaluation. Biol. Chem. 1998;379:753–772. doi: 10.1515/bchm.1998.379.7.753. [DOI] [PubMed] [Google Scholar]
- 24.Lill R., Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J. Mol. Biol. 1987;196:137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- 25.Robertson J.M., Wintermeyer W. Mechanism of ribosomal translocation. tRNA binds transiently to an exit site before leaving the ribosome during translocation. J. Mol. Biol. 1987;196:525–540. doi: 10.1016/0022-2836(87)90030-1. [DOI] [PubMed] [Google Scholar]
- 26.Chinali G., Parmeggiani A. Differential modulation of the elongation-factor-G GTPase activity by tRNA bound to the ribosomal A-site or P-site. Eur. J. Biochem. 1982;125:415–421. doi: 10.1111/j.1432-1033.1982.tb06699.x. [DOI] [PubMed] [Google Scholar]
- 27.Lill R., Robertson J.M., Wintermeyer W. Binding of the 3′-terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voigt J., Nagel K. Regulation of elongation factor G GTPase activity by the ribosomal state. The effects of initiation factors and differentially bound tRNA, aminoacyl-tRNA, and peptidyl-tRNA. J. Biol. Chem. 1993;268:100–106. [PubMed] [Google Scholar]
- 29.Nagel K., Voigt J. Regulation of the uncoupled GTPase activity of elongation factor G (EF-G) by the conformations of the ribosomal subunits. Biochim. Biophys. Acta. 1993;1174:153–161. doi: 10.1016/0167-4781(93)90109-q. [DOI] [PubMed] [Google Scholar]
- 30.Zavialov A.V., Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 31.Joseph S., Noller H.F. EF-G-catalyzed translocation of anticodon stem–loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 33.Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 34.Borowski C., Rodnina M.V., Wintermeyer W. Truncated elongation factor G lacking the G domain promotes translocation of the 3′ end but not of the anticodon domain of peptidyl-tRNA. Proc. Natl Acad. Sci. USA. 1996;93:4202–4206. doi: 10.1073/pnas.93.9.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dabrowski M., Spahn C.M., Schafer M.A., Patzke S., Nierhaus K.H. Protection patterns of tRNAs do not change during ribosomal translocation. J. Biol. Chem. 1998;273:32793–32800. doi: 10.1074/jbc.273.49.32793. [DOI] [PubMed] [Google Scholar]
- 36.Powers T., Noller H.F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc. Natl Acad. Sci. USA. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sergiev P.V., Bogdanov A.A., Dahlberg A.E., Dontsova O. Mutations at position A960 of E.coli 23S ribosomal RNA influence the structure of 5S ribosomal RNA and the peptidyltransferase region of 23S ribosomal RNA. J. Mol. Biol. 2000;299:379–389. doi: 10.1006/jmbi.2000.3739. [DOI] [PubMed] [Google Scholar]
- 38.Asai T., Zaporojets D., Squires C., Squires C.L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl Acad. Sci. USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connor M., Dahlberg A.E. Mutations at U2555, a tRNA-protected base in 23S rRNA, affect translational fidelity. Proc. Natl Acad. Sci. USA. 1993;90:9214–9218. doi: 10.1073/pnas.90.19.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cupples C.G., Miller J.H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blaha G., Stelzl U., Spahn C.M., Agrawal R.K., Frank J., Nierhaus K.H. Preparation of functional ribosomal complexes and effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Methods Enzymol. 2000;317:292–309. doi: 10.1016/s0076-6879(00)17021-1. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 43.Leonov A.A., Sergiev P.V., Bogdanov A.A., Brimacombe R., Dontsova O.A. Affinity purification of ribosomes with a lethal G2655C mutation in 23S rRNA that affects the translocation. J. Biol. Chem. 2003;278:25664–25670. doi: 10.1074/jbc.M302873200. [DOI] [PubMed] [Google Scholar]
- 44.Kao C., Zheng M., Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartz D., McPheeters D.S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 46.Ehrenberg M., Bilgin N., Kurland C.G. In: The Ribosomes and Protein Synthesis: A Practical Approach. Spedding G., editor. Oxford, UK: Oxford University Press; 1990. pp. 101–129. [Google Scholar]
- 47.Moine H., Dahlberg A.E. Mutations in helix 34 of Escherichia coli 16S ribosomal RNA have multiple effects on ribosome function and synthesis. J. Mol. Biol. 1994;243:402–412. doi: 10.1006/jmbi.1994.1668. [DOI] [PubMed] [Google Scholar]