Abstract
The BioCyc database collection is a set of 160 pathway/genome databases (PGDBs) for most eukaryotic and prokaryotic species whose genomes have been completely sequenced to date. Each PGDB in the BioCyc collection describes the genome and predicted metabolic network of a single organism, inferred from the MetaCyc database, which is a reference source on metabolic pathways from multiple organisms. In addition, each bacterial PGDB includes predicted operons for the corresponding species. The BioCyc collection provides a unique resource for computational systems biology, namely global and comparative analyses of genomes and metabolic networks, and a supplement to the BioCyc resource of curated PGDBs. The Omics viewer available through the BioCyc website allows scientists to visualize combinations of gene expression, proteomics and metabolomics data on the metabolic maps of these organisms. This paper discusses the computational methodology by which the BioCyc collection has been expanded, and presents an aggregate analysis of the collection that includes the range of number of pathways present in these organisms, and the most frequently observed pathways. We seek scientists to adopt and curate individual PGDBs within the BioCyc collection. Only by harnessing the expertise of many scientists we can hope to produce biological databases, which accurately reflect the depth and breadth of knowledge that the biomedical research community is producing.
INTRODUCTION
How should biological knowledge repositories be created and updated in the post-genome era to maximize the accuracy of our rapidly evolving knowledge about the genome and biochemical network of each organism? Without a clear roadmap for this vast information space, chaos will reign among the hundreds of competing and overlapping databases that are arising. We propose a strategy based on the following tenets.
For every organism whose genome has been sequenced and that has a significant experimental community, it is critical that an organism-specific database be created, and curated on an ongoing basis, to provide an up-to-date, authoritative, central resource on the evolving knowledge about the genome and the biochemical network of that organism. Often called model organism databases, or organism-specific databases, examples include EcoCyc for Escherichia coli (1), PseudoCyc for Pseudomonas aeruginosa (2), PlasmoCyc for Plasmodium falciparum (3), SGD for Saccharomyces cerevisiae (4), TAIR for Arabidopsis thaliana (5), MGD for Mus musculus (mouse) (6) and FlyBase for Drosophila melanogaster (7).
No one group can curate all the world's genomes. Therefore, it is imperative to involve many scientists in the updation of each database, ideally centred around scientific communities sharing an interest in a specific organism, or a related group of organisms (8). Pathway/genome databases (PGDBs) provide a mechanism to integrate genome information into higher-order biochemical or gene networks, such as metabolic pathways and transcription units (9,10). They also express a scientific theory within a formal ontology and make it available for computational analysis (11).
The BioCyc collection described herein is partly a mechanism for bootstrapping this process. We originally demonstrated the value of this approach by constructing a PGDB for Haemophilus influenzae (12), the first species whose genome was deciphered, 10 years ago (13). By creating draft versions of many organism-specific PGDBs that can then be adopted by outside groups for curation, we are lowering the barrier to initiating such curation efforts. Because most of the BioCyc PGDBs are openly available, outside groups can use, update and redistribute them, without intellectual property restrictions. We request (although we do not require) that adopters make the modified versions of their PGDBs openly available as well. Adopters are free to publish PGDBs on their own websites by using the same Pathway Tools software that powers the BioCyc website (14). We further propose that when adopters insert new experimentally determined metabolic pathways in an adopted PGDB, they also kindly submit those pathways for inclusion in the MetaCyc database (15). The set of reference metabolic pathways in MetaCyc will thus continue to expand by including community contributions, enabling more accurate pathway predictions in the future.
Pathway Tools provides a suite of graphical interactive editing tools (such as a pathway editor and an operon editor) that allow multiple geographically distributed authors to concurrently update a single PGDB through the Internet. This approach is our recommended mechanism for collaboration among multiple authors; concurrent updating of multiple copies of a single organism's PGDB will yield divergent databases that will be difficult to reconcile or merge.
As well as bootstrapping curation efforts by other groups, the BioCyc database collection is a useful resource in its own right. It provides a collection of metabolic pathway reconstructions and (where applicable) operon predictions for many organisms, available as a reference source through the BioCyc website. Each database can serve as a resource for the analysis of gene expression, proteomics and metabolomics data (alone or in combination) by using the Pathway Tools Omics Viewer, a web-based tool for painting overlaying datasets onto the Cellular Overview diagram—a wiring diagram for the metabolic network of the cell (see http://BioCyc.org/expr-examples/animation.html for an example).
The BioCyc collection is also an important tool for both scientific research and technology development on a genome-wide scale (16). For instance, EcoCyc has been used to profile properties of metabolic enzymes and pathways (17) and their relationships to protein families (18). EcoCyc has also been used to explore the conservation of a well-defined set of metabolic enzymes across all domains of life (19). Examples of method development include the recognition of enzymes from sequence (20) or their genome and pathway context (21). Other uses include the benchmarking of automatic annotation projects (22) and metabolic reconstructions (23). For instance, we are in the process of comparing the imported annotations for BioCyc against automatically derived annotations using a consistent sequence comparison strategy across all species, to investigate whether imported annotations reflect the most up-to-date information from public databases of functional annotation (Goldovsky,L., Ahrén,D., Tsoka,S., Darzentas,N., Ouzounis,C.A., unpublished data).
The 160 genome-specific databases within the BioCyc collection are organized into three different tiers according to the quality of their database content, which is a function of the degree of manual curation they have undergone.
Tier 1 BioCyc databases are of the highest quality. They have undergone multiple person-years of curation and are updated on an ongoing basis, with regular releases. The Tier 1 PGDBs are EcoCyc, MetaCyc and the BioCyc Open Chemical Database (BOCD) (see http://biocyc.org/open-compounds.shtml). Note that MetaCyc and BOCD are not organism-specific databases, thus yielding the 160 total databases in the BioCyc collection.
Tier 2 BioCyc databases were computationally generated by the PathoLogic program and have undergone less than one person-year of manual curation to review and polish their contents. BioCyc contains 17 PGDBs in Tier 2, including HumanCyc (24), AgroCyc and FrantCyc (25).
Tier 3 BioCyc databases are those emphasized in this paper. They were computationally generated by the PathoLogic program and have undergone neither manual curation nor review. For example, we have not manually reviewed the PathoLogic pathway predictions, nor have we refined the contents of Tier 3 PGDBs, such as by manually adding additional experimentally known metabolic pathways for that organism. BioCyc contains 142 PGDBs in Tier 3.
The remainder of this paper describes the methods used to create the Tier 3 BioCyc databases.
MATERIALS AND METHODS
Data sources
Creation of the Tier 3 BioCyc PGDBs began with annotated genomes for each organism. Unlike the approaches used by other groups, our approach does not reannotate each genome but instead builds new layers of knowledge on the basis of existing genome annotation. Annotations for most Tier 3 prokaryotic genomes were obtained from the Comprehensive Microbial Resource (CMR) (26) in the version available on November 13, 2004 (species/strain names in Supplement 1). For those genomes not included in CMR, annotations were imported from the UniProt database (which includes the curated Swiss-Prot database and the automatically generated TrEMBL supplement) (27). Most of the latter annotations were indeed imported from TrEMBL and directly reflect the original function assignments deposited in the corresponding GenBank files by the original genome sequencing projects (28).
Inclusion criteria and input format
Annotations for species not available in CMR were obtained from the UniProt database (27). Only those species with high coverage in UniProt have been considered (>90% of total number of proteins). To determine this level of coverage, all protein sequences from the COGENT database (29) were cross-referenced to the corresponding UniProt entries using the MagicMatch algorithm (30), an efficient algorithm that matches identical sequences across databases. The corresponding UniProt files were checked for species names to ensure that the same species is considered.
Although metabolic map reconstruction is robust and can be achieved even with partial genome information (31), we decided not to include any organisms that are not available in CMR and with ≤90% coverage of their genome by protein-coding genes in UniProt (Table 1), as mentioned above.
Table 1.
A list of the 16 species not available in CMR
| Genus and species name | Strain name | Coverage (%) | Status |
|---|---|---|---|
| Anabaena sp. | PCC 7120 | >90 | UniProt |
| Anopheles gambiae | PEST | 39 | Excluded |
| Ashbya gossypii | na | >90 | Deleteda |
| Caenorhabditis briggsae | na | 0 | Excluded |
| Caenorhabditis elegans | na | 73 | Excluded |
| Cyanidioschyzon merolae | 10D | 0 | Excluded |
| Drosophila melanogaster | na | >90 | UniProt |
| Encephalitozoon cuniculi | na | >90 | UniProt |
| Leptospira interrogans | L1-130 | >90 | UniProt |
| Listeria monocytogenes | F2365 | >90 | UniProt |
| L.monocytogenes | F6854 | 40 | Excluded |
| L.monocytogenes | H7858 | 48 | Excluded |
| Mus musculus | na | 59 | Excluded |
| Nanoarchaeum equitans | Kin4-M | 78 | Excluded |
| Neurospora crassa | na | >90 | UniProt |
| Schizosaccharomyces pombe | na | >90 | UniProt |
Columns: genus/species name, strain name, coverage in UniProt and status in BioCyc (Tier 3 or exclusion). na (for strain name): non-applicable; >90% coverage in UniProt, included (7 cases).
aA.gossypii poor in annotations, despite high coverage in UniProt, deleted (1 case); ≤90% coverage in UniProt, excluded (8 cases).
Of the 16 species not in CMR but available in the COGENT database (29), the breakdown is as follows. Only seven species have >90% coverage in UniProt and are not in any other tier, and thus included in Tier 3 (see Table 1 for details). One species (Ashbya gossypii) contains very few annotations in the form of EC numbers and was excluded from the current release, despite >90% coverage in UniProt. Finally, the remaining eight species whose coverage is ≤90% and are not supported by other resources were not included (see Table 1). It is conceivable that, depending on community requests, PGDBs for some of these species can be readily created, if our criteria are relaxed, because of the significance of these organisms, such as the mosquito Anopheles gambiae, the two Caenorhabditis species or mouse. Thus, of all genomes available at the time of this project (September 2004), only nine genomes were not ultimately incorporated into the BioCyc collection of PGDBs (Table 1).
The Pathway Tools engine for generating new PGDBs, called PathoLogic, accepts as its input a set of files describing the annotated genome of an organism. It accepts a file format called PathoLogic format, in which each file describes all genes within one replicon. For each gene, the file specifies its name, base pair position, product type (e.g. protein and rRNA), functional description of the gene product and EC number.
CMR data transformation
We transformed each CMR genome to PathoLogic format using a two-step process. In Step 1 we loaded the entire CMR into BioWarehouse, an Oracle-based bioinformatics database warehouse system (see http://bioinformatics.ai.sri.com/biowarehouse/). For genomes sequenced at TIGR, CMR provides two alternative annotations of the genome: one produced by the sequencing centre that sequenced the genome and one produced by TIGR using an automated annotation pipeline. We always loaded the former annotation into BioWarehouse. CMR also provides a reannotation of the genome using a combination of automated and manual analyses. In Step 2 we developed a Lisp-based program that issued SQL queries to BioWarehouse to retrieve the CMR data for each genome and wrote that data out in PathoLogicformat.
While validating the preceding programs, we discovered that CMR does not contain most RNA-coding genes for many genomes. Therefore, those genes are lacking from many of the CMR-derived genomes in BioCyc version 9.0. We expect these genes to be present in later versions of CMR and in later generated versions of BioCyc. For scientists wishing to adopt a CMR-derived genome, SRI will generate anew a PGDB from the original GenBank entry for the genome, thus providing a complete PGDB for curation.
In the future, because of the complexity of the CMR database and its omission of RNA-coding genes, other databases such as COGENT (29) and RefSeq (32) will have to be considered.
UniProt data transformation
UniProt annotations in the form of description line and EC number were imported into PathoLogic-format files for further processing. The following fields were extracted from UniProt: description line and EC number for function assignment as well as gene and species name for the derivation of the corresponding entities in PGDBs.
PathoLogic execution
The Tier 3 BioCyc PGDBs were created by using the PathoLogic program (14). PathoLogic computationally creates a new PGDB from the annotated genome of an organism. Its first step is to transform the input genome (in the form of a GenBank file or a PathoLogic-format file) into a set of objects within the Ocelot object database system (14). Each replicon, gene and protein within the input file is converted to an Ocelot object that represents those replicons, genes and proteins.
Its second step is to infer the metabolic pathway complement of the organism by reference to the MetaCyc database, copying appropriate metabolic pathway, reaction and small-molecule objects from MetaCyc to the new PGDB. PathoLogic matches enzymes in the annotated genome against metabolic pathways in the MetaCyc database. The matching is based on EC number and on the enzyme name assigned in the annotated genome. PathoLogic computes a score for each pathway, which indicates the likelihood that it is present in the organism, based on the number of enzymes present in each pathway and on their uniqueness to the pathway. Pathways with a score above a given threshold are predicted as present and copied into a PGDB. The score is selected to err on the side of more false-positive predictions to ensure that possible pathways are brought to the attention of the user. The PathoLogic operon predictor (33) was executed to populate the microbial PGDBs with objects describing predicted operons. All computationally predicted pathways and operons are marked with computational evidence codes, which are displayed on the BioCyc websites as computer icons in the upper right side of each display page.
PathoLogic execution time on a SunBlade-1500 (1.06 GHz/2 GB RAM) workstation is ∼15 min per microbial genome.
MetaCyc
The MetaCyc database (http://MetaCyc.org/) is a collection of metabolic pathways and enzymes from a wide variety of organisms, primarily microorganisms and plants (15). The goal of MetaCyc is to contain a representative sample of each experimentally elucidated pathway, and thereby to catalogue the universe of metabolism. MetaCyc also describes reactions, chemical compounds and genes. As of version 9.0, MetaCyc contains 547 pathways elucidated in more than 340 organisms, 5000 reactions, 2062 enzymes, 3900 compounds and 5400 literature citations. In addition, 55% of the pathways and 90% of the enzymes contain comments that can help the user to understand the physiological role of a predicted pathway. More than 120 species have two or more pathways represented in MetaCyc, with E.coli and A.thaliana represented by the greatest number of pathways, 169 and 59, respectively.
Database implementation of BioCyc
The entire collection of BioCyc databases is stored within the same database management system, the Ocelot object database system (14). Ocelot employs an object-oriented data model that includes a taxonomic hierarchy of classes that define the database schema, a set of instances of those classes that represent biological entities and a set of slots that define attributes of (and relationships among) those entities.
All BioCyc databases share the same database schema, namely the Pathway Tools schema (34). By sharing the same schema among all databases, we ensure that the same software environment can be used to manipulate the databases and that comparisons can be computed consistently across all databases. The schema consists of 1350 class definitions that define data types (such as biochemical reactions, small molecules, genes, promoters, operons and metabolic pathways) and taxonomic classification systems (34). For example, the Pathway Tools schema includes a classification system for pathways, for small molecules, for biochemical reactions (the Enzyme Commission system) and for genes (35).
RESULTS
Shared aspects of BioCyc databases
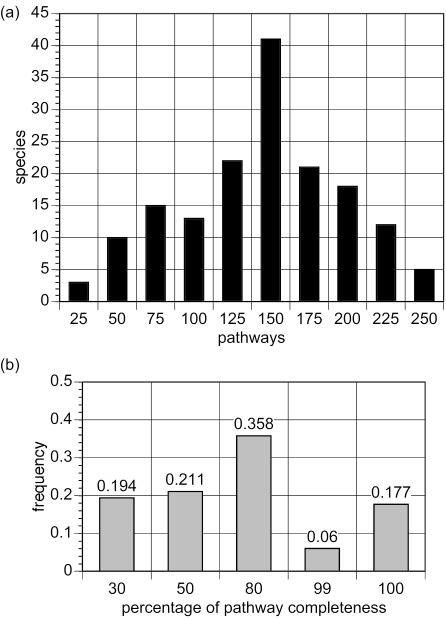
The entire collection of PGDBs encompasses 21 187 pathways distributed across 160 species, thus resulting in a mean value of 132.4 (SD 52) and a median value of 137.5 pathways per species (Figure 1a). Thus, most metabolic reconstructions generate a substantial amount of pathway information from the imported annotations. There are three species with 25 or fewer pathways—namely Mycobacterium avium paratuberculosis (5 pathways), Ralstonia solanacearum GMI1000 (14 pathways) and Pyrococcus horikoshii shinkaj OT3 (15 pathways)—and three species with more than 230 pathways—namely Bradyrhizobium japonicum USDA 110 (231 pathways), Streptomyces avermitilis MA-4680 (231 pathways) and Mesorhizobium loti MAFF303099 (234 pathways). The organisms with very small numbers of pathways seem to be artefacts caused by very sparse genome annotations or annotations in which the gene functions are provided in unusual formats.
Figure 1.
Distribution of BioCyc pathways across species. (a) Frequency analysis: the x-axis shows the number of detected pathways and the y-axis the number of species containing those pathways. (b) Completeness analysis: the x-axis shows the percentage of pathway completeness and the y-axis the frequency of pathways with the corresponding degree of completeness—more than 60% of pathways are more than 50% complete in the BioCyc collection of PGDBs.
Some of the most conserved pathways (19) are fully recovered in the BioCyc PGDB collection, such as glycolysis, nucleotide biosynthesis and amino acid (e.g. glycine, tryptophan, lysine) biosynthesis (Figure 1b and Table 2). This observation might be biased by the sample of organisms under consideration, yet it uncovers some of the most conserved (or, possibly, well-annotated) segments of metabolism. The high occurrence of one of these pathways, the Calvin cycle, is probably due to false-positive predictions because the Calvin cycle shares many reactions with the TCA cycle. Thus, the presence of TCA cycle reactions provides evidence for the Calvin cycle. Modification of our pathway prediction algorithms to reduce false-positive predictions is an ongoing work.
Table 2.
The 30 pathways that occur most frequently across the BioCyc databases, and their frequency (f) of occurrence
| PATHWAY-UNIQUE ID | Pathway description | f |
|---|---|---|
| PWY0-162 | De novo biosynthesis of pyrimidine ribonucleotides | 153 |
| GLYCOLYSIS | Glycolysis I | 152 |
| TRNA-CHARGING-PWY | tRNA charging pathway | 152 |
| DENOVOPURINE2-PWY | Purine nucleotides de novo biosynthesis I | 152 |
| PWY0-166 | De novo biosynthesis of pyrimidine deoxyribonucleotides | 151 |
| PHOSLIPSYN-PWY | Phospholipid biosynthesis I | 150 |
| GLYSYN-PWY | Glycine biosynthesis I | 149 |
| HEMESYN2-PWY | Biosynthesis of proto- and sirohaeme | 146 |
| P1-PWY | Salvage pathways of purine and pyrimidine nucleotides | 144 |
| FASYN-INITIAL-PWY | Fatty acid biosynthesis—initial steps | 144 |
| 1CMET2-PWY | formylTHF biosynthesis | 144 |
| PWY0-163 | Salvage pathways of pyrimidine ribonucleotides | 142 |
| P106-PWY | Serine-isocitrate lyase pathway | 142 |
| THIOREDOX-PWY | Thioredoxin pathway | 140 |
| ARO-PWY | Chorismate biosynthesis | 139 |
| P124-PWY | Glucose fermentation to lactate II | 139 |
| CALVIN-PWY | Calvin cycle | 136 |
| FOLSYN-PWY | Tetrahydrofolate biosynthesis | 136 |
| PWY0-662 | PRPP biosynthesis I | 136 |
| RIBOKIN-PWY | Ribose degradation | 133 |
| TRPSYN-PWY | Tryptophan biosynthesis | 133 |
| PEPTIDOGLYCANSYN-PWY | Peptidoglycan biosynthesis | 133 |
| PWY0-901 | Selenocysteine biosynthesis | 132 |
| FASYN-ELONG-PWY | Fatty acid elongation—saturated | 131 |
| PWY-841 | Purine nucleotides de novo biosynthesis II | 131 |
| RIBOSYN2-PWY | Riboflavin and FMN and FAD biosynthesis | 130 |
| P61-PWY | UDP-glucose conversion | 130 |
| DAPLYSINESYN-PWY | Lysine biosynthesis I | 130 |
| ILEUSYN-PWY | Isoleucine biosynthesis I | 130 |
| ACETATEUTIL-PWY | Acetate utilization | 129 |
It is worth noting that degradation pathways (with the exception of ribose degradation) appear to be much less conserved (Supplementary Data, complete table).
Distribution patterns of pathways across 160 genomes
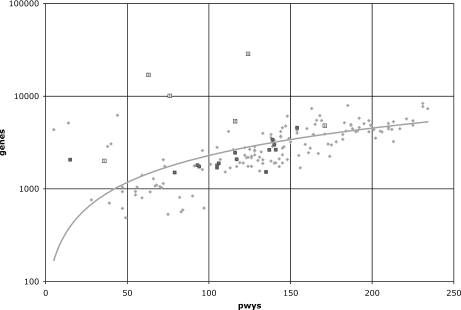
When correlated to genome size (as the number of protein-coding genes), pathways in the BioCyc collection appear to follow a trend (Figure 2). In particular, Bacteria follow a highly consistent monotonic relationship, with a few exceptions. Archaea also follow the pattern of Bacteria, with the exception of P.horikoshii shinkaj OT3. Finally, eukaryotes do not exhibit such a consistent pattern, with the exception of Schizosaccharomyces pombe (Figure 2).
Figure 2.
Relationship between number of pathways (x-axis) and number of genes (y-axis) for all species in the BioCyc collection. Bacterial species are shown in light grey, archaeal species in open-grey squares and eukaryotes in black. The fitted line—a linear regression curve—refers to Bacteria only; most Archaea exhibit a similar relationship. The two outlier bacterial species with fewer than 25 pathways can be seen on the left part of the graph: Mycobacterium avium paratuberculosis and Ralstonia solanacearum GMI1000. The three largest eukaryotic genomes with >10 000 genes show a significant underrepresentation of pathways for their genome size.
DISCUSSION
Scientists interested in adopting one or more of the BioCyc PGDBs should contact the authors. We will supply data files for the PGDBs as well as the Pathway Tools software. Pathway Tools supports updating, querying, analysis and Web publishing of PGDBs in the following manner.
SRI has established a central repository of publicly available PGDBs, similar to peer-to-peer file sharing in spirit. PGDB creators can register their PGDBs in the central repository for potential downloading by interested parties. The registry of shared PGDBs created by SRI and by third parties is available (see http://BioCyc.org/registry.html).
The Pathway/Genome Editors within Pathway Tools are graphical interactive tools for updating information within a PGDB. For example, given new findings in the literature, a scientist could add a new metabolic pathway to a PGDB or alter an existing pathway. They can alter the annotation of a gene, and add commentary and literature citations. They can annotate features on proteins, such as enzyme active sites or phosphorylation sites. They can also update the description of the genetic network of an organism by defining new operons, promoters and transcription-factor binding sites, and by defining regulatory interactions between transcription factors and their binding sites. These are the same tools used by the curators who update the EcoCyc and MetaCyc databases.
Pathway Tools is also the software used to power the BioCyc.org website. We encourage PGDB adopters to publish their PGDBs on their websites by using Pathway Tools and thus make them available to the scientific community.
We plan to regenerate the unadopted Tier 3 BioCyc PGDBs every 6 months to incorporate improvements in the data sources from which they were generated and improvements in MetaCyc and the Pathway Tools that will provide improved inferences.
RELATED WORK
The WIT database (36) has not been available on the web for more than 1 year, and appears to have been succeeded by the PUMA2 system (see http://compbio.mcs.anl.gov/puma2/cgi-bin/index.cgi). It contains metabolic reconstructions for more than 1000 organisms, but because of the absence of publications about the PUMA2 methodology we cannot perform a detailed comparison with BioCyc.
Version 35.0 of the KEGG database contains predicted metabolic maps for 333 organisms (37). Both KEGG and the BioCyc collection predict pathways by comparing the enzymes within a given genome against a known set of reference pathways. But many differences between the two methodologies exist.
One difference in pathway prediction methodology is that the two resources (KEGG reference maps and MetaCyc) use different reference pathway databases as the basis for prediction. MetaCyc contains extensive comments that describe individual pathways and enzymes; KEGG has very few such comments. MetaCyc cites the primary literature sources from which pathway and enzyme data were obtained. KEGG contains very few literature citations. KEGG maps are very much larger than MetaCyc pathways, and are mosaics that combine pathways and reactions from many organisms, whereas MetaCyc pathways describe single metabolic pathways that have been experimentally elucidated in single organisms.
In addition, the KEGG methodology has been shown to erroneously assign enzymes to pathway reactions based on matching on incomplete EC numbers such as ‘1.2.3.-’, resulting in many incorrect enzyme–reaction associations within KEGG (38).
The KEGG pathway prediction process also appears to be different from that used by PathoLogic. It appears to begin by computing new functions for all gene products within a genome, thus replacing a genome annotation that is often derived from significant manual work by scientists with one that is purely computationally derived, and thus potentially of lower quality (because the KEGG publications are unclear, we note that there is uncertainty in how this genome reannotation is performed). Our work builds from the originally submitted genome annotation. In addition, the KEGG algorithm does not actually predict pathways—it simply colours a set of static KEGG map diagrams to indicate which enzymes within a map are present within a given genome. This approach avoids actually predicting whether a pathway is present or absent—that decision is left to the user—whereas our PathoLogic algorithm does call each pathway in MetaCyc as present or absent in the organism it is analysing.
Neither KEGG nor PUMA2 make their pathway databases openly available for adoption and curation by experts (the PUMA2 website does not have a downloads section). Furthermore, the software environment underlying KEGG does not support the rich level of curation and annotation as does the graphical editing environment and schema within the Pathway Tools software underlying BioCyc. For example, the KEGG software does not allow the editing of pathway information, to remove erroneous reactions that are not present in a given organism, or to add organism-specific reactions to a pathway. The Pathway Tools schema supports 220 different database fields to capture a broad array of information from genomes to pathways; we are unaware of a description of the KEGG schema that provides comparable information.
BioCyc AVAILABILITY
All BioCyc PGDBs are accessible online through the Web at http://BioCyc.org/ for interactive querying. The website is freely available to all users. The Pathway Tools software is freely available to academics. More information on downloading the software and databases is available at http://biocyc.org/download.shtml.
We provide several mechanisms by which computational or experimental biologists can compute with the data within the BioCyc PGDBs (39). BioCyc PGDBs in Tiers 1 and 2 are queryable via application program interfaces (APIs) and are available by data file download. We plan to make Tier 3 PGDBs available for download, upon publication. In any event, we will make any Tier 3 PGDBs available for download for adoption based on requests to the authors.
Pathway Tools APIs allow users to query BioCyc databases in the Java, Perl and Lisp languages (described at http://bioinformatics.ai.sri.com/ptools/ptools-resources.html). All three APIs provide extremely comprehensive and easy-to-use query facilities. The APIs access the data through a binary executable program that bundles the Pathway Tools software with all the BioCyc databases and runs on Linux, Windows and Sun workstations. This executable runs as a desktop application and also supports local installation of the BioCyc website on an intranet.
Flatfile versions of BioCyc PGDBs can be downloaded in four formats including SBML (see sbml.org) and BioPAX (see biopax.org).
Most BioCyc PGDBs are freely and openly available and may be redistributed. A fee applies to commercial installations of some BioCyc PGDBs and to the Pathway Tools software.
Acknowledgments
This work was supported by grant GM70065 from the National Institutes of Health, by SRI International, and by EMBL-EBI. N.L.B. is supported by a long-term Fellowship from the Human Frontiers Science Program and S.T. by a Career Development Award from the UK Medical Research Council. This financial support does not constitute an endorsement of the views expressed herein. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM70065.
Conflict of interest statement. None declared.
REFERENCES
- 1.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero P., Karp P. PseudoCyc, a pathway-genome database for Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 2003;5:230–239. doi: 10.1159/000071075. [DOI] [PubMed] [Google Scholar]
- 3.Yeh I., Hanekamp T., Tsoka S., Karp P.D., Altman R.B. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14:917–924. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie K.R., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., et al. The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–D475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drysdale R.A., Crosby M.A., Gelbart W., Campbell K., Emmert D., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., et al. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouzounis C.A., Karp P.D. The past, present and future of genome-wide re-annotation. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-comment2001. COMMENT2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karp P.D. Metabolic databases. Trends Biochem. Sci. 1998;23:114–116. doi: 10.1016/s0968-0004(98)01184-0. [DOI] [PubMed] [Google Scholar]
- 10.Karp P.D., Krummenacker M., Paley S., Wagg J. Integrated pathway-genome databases and their role in drug discovery. Trends Biotechnol. 1999;17:275–281. doi: 10.1016/s0167-7799(99)01316-5. [DOI] [PubMed] [Google Scholar]
- 11.Karp P.D. Pathway databases: a case study in computational symbolic theories. Science. 2001;293:2040–2044. doi: 10.1126/science.1064621. [DOI] [PubMed] [Google Scholar]
- 12.Karp P.D., Ouzounis C., Paley S. HinCyc: a knowledge base of the complete genome and metabolic pathways of H. influenzae. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1996;4:116–124. [PubMed] [Google Scholar]
- 13.Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.F., Dougherty B.A., Merrick J.M., et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 14.Karp P.D., Paley S., Romero P., editors. The Pathway Tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 15.Krieger C.J., Zhang P., Mueller L.A., Wang A., Paley S., Arnaud M., Pick J., Rhee S.Y., Karp P.D. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32:D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsoka S., Ouzounis C.A. Metabolic database systems for the analysis of genome-wide function. Biotechnol. Bioeng. 2003;84:750–755. doi: 10.1002/bit.10881. [DOI] [PubMed] [Google Scholar]
- 17.Ouzounis C.A., Karp P.D. Global properties of the metabolic map of Escherichia coli. Genome Res. 2000;10:568–576. doi: 10.1101/gr.10.4.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsoka S., Ouzounis C.A. Functional versatility and molecular diversity of the metabolic map of Escherichia coli. Genome Res. 2001;11:1503–1510. doi: 10.1101/gr.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peregrin-Alvarez J.M., Tsoka S., Ouzounis C.A. The phylogenetic extent of metabolic enzymes and pathways. Genome Res. 2003;13:422–427. doi: 10.1101/gr.246903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.des Jardins M., Karp P.D., Krummenacker M., Lee T.J., Ouzounis C.A. Prediction of enzyme classification from protein sequence without the use of sequence similarity. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:92–99. [PubMed] [Google Scholar]
- 21.Green M.L., Karp P.D. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karp P.D., Paley S., Krieger C.J., Zhang P. An evidence ontology for use in pathway/genome databases. Pac. Symp. Biocomput. 2004;9:190–201. doi: 10.1142/9789812704856_0019. [DOI] [PubMed] [Google Scholar]
- 23.Paley S.M., Karp P.D. Evaluation of computational metabolic-pathway predictions for Helicobacter pylori. Bioinformatics. 2002;18:715–724. doi: 10.1093/bioinformatics/18.5.715. [DOI] [PubMed] [Google Scholar]
- 24.Romero P., Wagg J., Green M.L., Kaiser D., Krummenacker M., Karp P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson P., Oyston P.C., Chain P., Chu M.C., Duffield M., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nature Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 26.Peterson J.D., Umayam L.A., Dickinson T., Hickey E.K., White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen P., Enright A.J., Audit B., Cases I., Goldovsky L., Harte N., Kunin V., Ouzounis C.A. COmplete GENome Tracking (COGENT): a flexible data environment for computational genomics. Bioinformatics. 2003;19:1451–1452. doi: 10.1093/bioinformatics/btg161. [DOI] [PubMed] [Google Scholar]
- 30.Smith M., Kunin V., Goldovsky L., Enright A.J., Ouzounis C.A. MagicMatch—cross-referencing sequence identifiers across databases. Bioinformatics. 2005;21:3429–3430. doi: 10.1093/bioinformatics/bti548. [DOI] [PubMed] [Google Scholar]
- 31.Ahren D.G., Ouzounis C.A. Robustness of metabolic map reconstruction. J. Bioinform. Comput. Biol. 2004;2:589–593. doi: 10.1142/s021972000400079x. [DOI] [PubMed] [Google Scholar]
- 32.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romero P.R., Karp P.D. Using functional and organizational information to improve genome-wide computational prediction of transcription units on pathway-genome databases. Bioinformatics. 2004;20:709–717. doi: 10.1093/bioinformatics/btg471. [DOI] [PubMed] [Google Scholar]
- 34.Karp P.D. An ontology for biological function based on molecular interactions. Bioinformatics. 2000;16:269–285. doi: 10.1093/bioinformatics/16.3.269. [DOI] [PubMed] [Google Scholar]
- 35.Serres M.H., Riley M. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics. 2000;5:205–222. doi: 10.1089/omi.1.2000.5.205. [DOI] [PubMed] [Google Scholar]
- 36.Overbeek R., Larsen N., Pusch G.D., D'Souza M., Selkov E., Jr, Kyrpides N., Fonstein M., Maltsev N., Selkov E. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 2000;28:123–125. doi: 10.1093/nar/28.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green M.L., Karp P.D. Genome annotation errors in pathway databases due to semantic ambiguity in partial EC numbers. Nucleic Acids Res. 2005;33:4035–4039. doi: 10.1093/nar/gki711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krummenacker M., Paley S., Mueller L., Yan T., Karp P.D. Querying and computing with BioCyc databases. Bioinformatics. 2005;21:3454–3455. doi: 10.1093/bioinformatics/bti546. [DOI] [PMC free article] [PubMed] [Google Scholar]