Abstract
The oligonucleotide specificity for microarray hybridization can be predicted by its sequence identity to non-targets, continuous stretch to non-targets, and/or binding free energy to non-targets. Most currently available programs only use one or two of these criteria, which may choose ‘false’ specific oligonucleotides or miss ‘true’ optimal probes in a considerable proportion. We have developed a software tool, called CommOligo using new algorithms and all three criteria for selection of optimal oligonucleotide probes. A series of filters, including sequence identity, free energy, continuous stretch, GC content, self-annealing, distance to the 3′-untranslated region (3′-UTR) and melting temperature (Tm), are used to check each possible oligonucleotide. A sequence identity is calculated based on gapped global alignments. A traversal algorithm is used to generate alignments for free energy calculation. The optimal Tm interval is determined based on probe candidates that have passed all other filters. Final probes are picked using a combination of user-configurable piece-wise linear functions and an iterative process. The thresholds for identity, stretch and free energy filters are automatically determined from experimental data by an accessory software tool, CommOligo_PE (CommOligo Parameter Estimator). The program was used to design probes for both whole-genome and highly homologous sequence data. CommOligo and CommOligo_PE are freely available to academic users upon request.
INTRODUCTION
Microarrays are a powerful and versatile tool for genome-wide expression analysis (1–5) and environmental studies (6,7). Two types of microarrays, cDNA arrays and oligonucleotide arrays, are widely used. Oligonucleotide arrays have become more and more popular because they offer a number of advantages, including better specificity and easy construction (8,9). In addition, oligonucleotide arrays provide practical solutions to more complicated problems. For example, it is very difficult to construct a comprehensive functional gene array representing diverse meta-genomic sequences from environmental samples (such as groundwater, soil or subsurface sediments) using the PCR amplification approach because obtaining all the diverse environmental clones and bacterial strains from various sources as templates for amplification can be a big challenge (7,9,10).
One major challenge for designing oligonucleotide arrays is how to identify optimum probes for each gene in a group of sequences or in a whole genome. Oligonucleotide probe design programs differ in criteria to define optimal probes and algorithms for probe selection. The probes designed based on the same set of sequences could be quite different.
Previously, we experimentally demonstrated that the combination of different probe design criteria is needed to obtain optimally specific probes (11). A combination of multiple criteria will allow more liberal cutoffs for each criterion and thus be able to find more specific probes and fewer non-specific probes. Although the existing programs have used different criteria for probe design, no programs have considered the three criteria, sequence identity, free energy and continuous stretch together. And also for computation of oligonucleotide specificity, most programs primarily rely on BLAST for local alignment or suffix array for exact string search, which do not always reflect the overall identity between an oligonucleotide and its non-targets. Therefore, those criteria and algorithms used may lead to selection of ‘false’ specific oligonucleotides and/or exclusion of ‘true’ optimal probes. Moreover, most available programs have been developed to design probes for whole-genome sequences in which the proportion of homologous sequences are generally low. However, in environmental studies, a large number of sequences are highly homologous (7). In this case, it is very difficult for those programs to select specific probes for such highly homologous sequences. On the other hand, it is well known that data from microarray experiments have inherent high variations. It is very unlikely that a fixed set of parameter values can be used for all experimental conditions. Therefore, tools to estimate parameter thresholds from experimental data are desirable.
In this study, a software tool called CommOligo has been developed. The program uses global alignments for more accurate calculations of identity and free energy, and a search for the optimal interval of melting temperature. Then all probe candidates are ranked using a combination of user-configurable piece-wise linear functions and an iterative process. The cutoff values for identity, stretch and free energy are automatically estimated from experimental data by another software tool CommOligo parameter estimator (CommOligo_PE). The program has been used to design probes for whole-genome sequences and highly homologous sequences.
ALGORITHMS
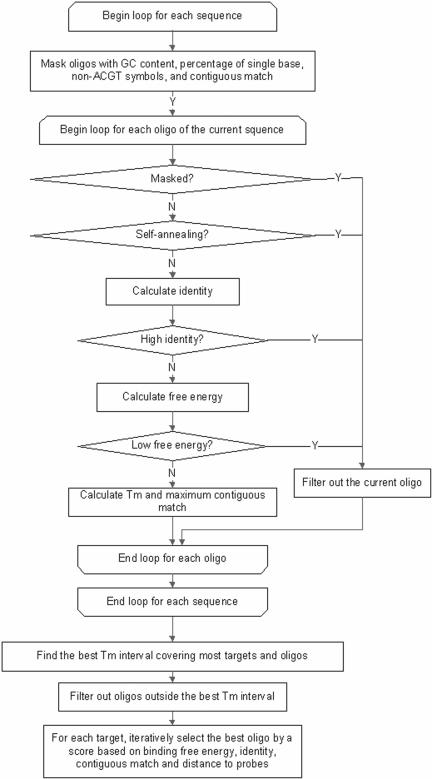
A flowchart of the program is shown in Figure 1. A series of filters are used to check each oligonucleotide from the beginning to the end of each sequence. First, for each sequence, oligonucleotides are masked using distance to the 3′-UTR, GC content, the maximum percentage of each single base (A, T, C or G), non-ACGT symbols and maximum length of continuous matches to non-targets. Then self-annealing, identity to non-targets, and binding free energy to non-targets are calculated for unmasked oligonucleotides. The optimal interval of Tm is obtained from the oligonucleotides that have passed the above filters. Probe optimization is an iterative process based on a quality score. CommOligo is implemented in C++ and runs under Microsoft Windows. The limit of distance to the 3′-UTR can be defined in number of nucleotides or percentage of the target length. All thresholds and parameters are user-adjustable through graphic user interfaces.
Figure 1.
Flowchart for CommOligo.
Maximum length of continuous matches to non-targets and self-annealing
In CommOligo, the maximum length of continuous matches is used not only as a filter but also for picking the best probes after filtering. The process to check continuous matches (stretches) of a given oligonucleotide to its non-target sequences is similar to the method used in OligoPicker (12). A table of words of length 10 is constructed by scanning all sequences, and the table has a size of 410. To search continuous matches >10 bases, multiple words from the oligonucleotide, where the last word may overlap with its preceding word, are checked against the word table for occurrences in other sequences. When a stretch longer than the user-specified cutoff is detected, oligonucleotides containing the stretch are masked. For unfiltered oligonucleotides, the maximum number of continuous matches to non-target sequences is calculated.
For self-annealing measurement, if an oligonucleotide has continuous matches longer than a user-specified threshold within itself, it will be filtered out.
Sequence identity
Most preexisting software tools calculate identities using local alignment algorithm such as BLAST, but actual hybridization is performed on a global identity scenario (13). In CommOligo, identities between an oligonucleotide and its non-targets are calculated as the percentage of matches in their optimal gapped alignment generated from Myers' bit-vector algorithm (14), which is considered the fastest for generic global alignment. The identity of an oligonucleotide is the maximum identity with its non-targets. Oligonucleotides with identities higher than a user-specified threshold are filtered out.
Binding free energy
Calculation of the exact minimum binding free energy between a probe and a sequence would require a complete comparison of all possible alignments, which is too slow to be applied in probe design. In CommOligo, we only calculate free energy values for global alignments with high identities. An oligonucleotide is filtered out if the binding free energy to its non-targets is less than a user-specified threshold. The binding free energy of an oligonucleotide is the minimal free energy to its non-targets.
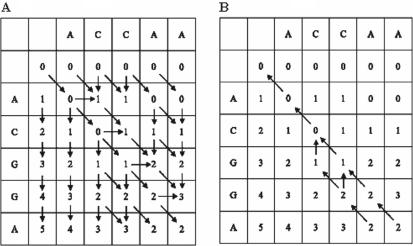
Global alignment algorithms produce a dynamic programming matrix or equivalent with mismatch/gap scores. Given the number of mismatches/gaps, the end positions of alignments can be located in the matrix. However, a further step is needed to generate the alignments between individual nucleotides. To generate all alignments with high identities, we traverse the dynamic programming matrix from elements with small number of mismatches/gaps in the bottom row to the top row (Figure 2). An element in the matrix is calculated from its adjacent top-left, top and/or left element, and the matrix can be considered a directed graph. The edges of the graph connect an element from its preceding elements where its value is derived. The traversal algorithm is essentially a breadth first search on a directed graph. It utilizes the bit vectors generated in Myers algorithm during identity calculation. Let the oligonucleotide be O = o1o2 … on, a non-target sequence be S = s1s2 … sm, horizontal positive bit vector be Ph, vertical bit vector be Pv, diagonal equal bit vector be Eq and mismatch vector of the bottom row be D, the algorithm outputs VCAlignment, a vector of alignments with high identities. An alignment corresponds to a path from the bottom row with high identities to the top row and is denoted as a vector of points (x, y), where x ∈ {o1, o2, …, on, −} and y ∈ {s1, s2, …, sn, −} with ‘−’ as a gap. As the dynamic programming matrix is traversed, the current column and row position of each path is recorded as (c, r).
Figure 2.
Directed graph of the dynamic programming matrix for alignment of sequence ACCAA and ACGGA (A), and the traversal of the dynamic programming matrix for alignment of sequence ACCAA and ACGGA with two mismatches (B).
Traversal algorithm is as follows:
For i = 1 to m
If D[i] - lowest mismatch/gaps the in D < T, where T > 0 is a threshold
append an empty path to the path vector VCAlignment, set its current position to (i, m)
TraversePath (the last path)
TraversePath (current path)
If the current row r of the current path = 1, return
- Calculate vcDelta: a vector of (deltaX, deltaY), where deltaX and deltaY is the difference in column and row between the current position and the next position.
- if the Eq bit of the current position (c,r) is set, append (1,1)
- if the Ph bit of (c,r) is set, append (1,0)
- if the Pv bit of (c,r) is set, append (0,1)
- From the second to the last element in vcDelta
- append a copy of the current path to vcPath
- for the last path (newly appended)
- If deltaX = 1 and deltaY = 1, append (or, sc)
- If deltaX = 1 and deltaY = 0, append (−, sc)
- If deltaX = 0 and deltaY = 1, append (or, −)
- c−=deltaX, r−=deltaY
- TraversePath (the appended path)
- For the first element in vcDelta
- If deltaX = 1 and deltaY = 1, append (or, sc) to current path
- If deltaX = 1 and deltaY = 0, append (−, sc) to current path
- If deltaX = 0 and deltaY = 1, append (or, −) to current path
- c−=deltaX, r−=deltaY
- TraversePath (current path)
Given an alignment between an oligonucleotide and a sequence, the binding free energy is calculated using the method very similar to the MFOLD program (15,16). Free energy for a loop/bulge is calculated using parameters from MFOLD (15). Free energy of the matches and single mismatches is calculated using the nearest-neighbor model with established parameters (17–23). It should be noted that the free energy value is calculated at 37°C rather than the actual hybridization temperature.
Calculation and optimization of melting temperature (Tm)
Tm is calculated for each unfiltered oligonucleotide using the nearest-neighbor model with parameters from SantaLucia (24) and a fixed DNA concentration of 10 µM. We try to design probes for maximum number of sequences within a user-specified range, range. In other words, the best Tm interval [t1, t1 + range] is the one with maximum number of sequences that have probes. If multiple Tm intervals have the maximum number of sequences with probes inside, the interval with maximum number of probes is selected. The algorithm is as follows: first, unfiltered oligonucleotides are sorted according to their Tm values. Second, a Tm range window is moved from the first oligonucleotide to the last and the number of sequences ns with oligonucleotides and the number of oligonucleotides no inside the window are calculated. Finally, the best Tm interval is chosen according to ns and no. Oligonucleotides with Tm outside the best Tm interval are filtered out.
Probe quality score and probe optimization
After all filters are applied, a sequence may have more probes than it requires. In this case, an optimization step will ensure that the best probes are selected. CommOligo picks probes according to two criteria: (i) probe candidates with lowest cross-hybridization and located in different regions in the target are picked first; (ii) for the same target, identities between any two probes must be less than a user-specified threshold. To measure how good a probe is, a quality score valued between 0 and 1 is assigned. Cross-hybridization is more complicated than that a single measurement can predict, and the definition of good probe can be subjective and application dependent. The score combines four individual scores between 0 and 1, each measuring continuous matches, sequence identity, and binding free energy between the oligonucleotide and its non-targets, and its distance to other probes for the same target, respectively.
A score for identity with non-targets of an oligonucleotide Si is calculated using a piece-wise linear function. Oligonucleotides with identities more than the filter threshold tf (e.g. 85%) have been filtered out. An oligonucleotide with identity tf is assigned a score sf (e.g. 0.5). On the other hand, it can be assumed there will be no cross-hybridization if the identity is sufficiently low (e.g. <40%) or less than a saturation threshold. Thus an oligonucleotide with an identity less than the saturation threshold ts is assigned score 1.0. A user-specified ‘middle’ point is used to partition the interval between the filter threshold and saturation threshold for flexibility. The score function is as follows:
where x is the identity of the oligonucleotide to its non-targets.
The score Sc for continuous match are calculated in the same way as Si. Score Sd measures its distance to other probes of the same target and score Se for free energy is calculated in a similar way, except their tf < ts and their equations are non-decreasing.
The quality score S for an oligonucleotide can be a weighted average
where we, wd, wi and wc are user-configurable weights, or S = min(Se, Sd, Si, Sc).
The filter point, saturation point and middle point are user-configurable. The score function for identity should reflect the relationship between hybridization intensity and the identity. Assuming there are sufficient experimental data (relative intensity) with a known sequence identity, a scatter plot can be drawn to show the relationship between the hybridization intensity and sequence identity. Ideally, the score for identity should follow the trend line of the scatter plot. The score for other parameters could be determined in the same way. Filter thresholds are hard limits in previous sections. The saturation point defines a minimal requirement for ‘best’ probe or no cross-hybridization. The ‘middle’ points are provided to achieve non-linear effect between the filter threshold and saturation threshold. If the relationship between hybridization and the measurement is linear between filter and saturation point, ((ts + tf/2), (1 + sf/2)) can be used as the middle point.
The minimum distance and the maximum identity to other probes of the same target actually depend on probes that have been already picked. Initially, distance scores of all oligonucleotides of the target are assigned to 1.0 and the oligonucleotide with the highest combination score is picked. Then the identities of other oligonucleotides to the selected probe are calculated and oligonucleotides with identities higher than user-specified threshold are removed from the probe candidates. Distances of other oligonucleotides to the selected probe are determined, their new scores are calculated and the oligonucleotide with the best combined score is selected. This process is repeated until all oligonucleotides are checked or the probes picked for the sequence meets the user-specified number.
Automatic estimation of cutoff values for cross-hybridization
CommOligo predicts cross-hybridization using three criteria. In order to systematically determine the thresholds for those criteria based on experimental data, an accessory tool, CommOligo_PE has been developed. CommOligo_PE uses a user-specified threshold T to identify the low signal spots. A probe is considered no cross-hybridization if its signal is less than or equal to T. Our goal is to find the cutoff values to cover the low signal spots using two indexes. One is the negative predictive value (NPV):
The other is probe coverage (C):
NPV measures the correctness of a prediction and C measures the completeness of prediction. The ideal criteria should have an NPV of 1.0 and a C of 1.0. Thus our goal is to maximize both NPV and C. However, the maximum NPV and the maximum C often contradict and there is no universal solution to this dilemma. In CommOligo_PE, users can specify an optimization goal to either maximize C or NPV. Since maximizing NPV freely may lead to very low C and maximizing C freely may lead to very low NPV, users can specify a minimal NPV value and/or a minimal C value as constraints. With an experimental data file, an optimization goal and optimization constraints, CommOligo_PE tries to find the best cutoff values. This is done by an exhaust search in full parameter space. For maximizing speed, the search range for identity, stretch and free energy are from 50 to 96%, from 10 to 40, and from −70 to 0, respectively. For all possible cutoff values, CommOligo_PE calculates NPV and C values. The best cutoff values are those that maximize the optimization goal and satisfy constraints.
In a typical case, a user can specify T, a minimal NPV (e.g. 95%), and an optimization goal (e.g. to maximize C), and CommOligo_PE will output the best cutoffs for identity, stretch and free energy.
For some applications, maximizing NPV around the maximal C is more desirable than maximizing C. Since the maximal C depends on data and T, CommOligo_PE provides an automatic optimization goal, which is a two-step process. First, the maximum C is sought and then the best NPV with C >90% of the best C generated in the first step is used for optimization, both under user-specified constraints. Users can run the estimation process in two separate steps as well.
CommOligo_PE can optimize cutoff values for five sets of criteria: (i) identity, (ii) stretch, (iii) free energy, (iv) identity and stretch, and (v) identity, stretch and free energy. It should be noted CommOligo_PE maximizes C or NPV under given constraints for training data. To help users investigate results for testing data and compare different sets of criteria, CommOligo_PE provides an option for cross-validation. In cross-validation, the dataset is partitioned into 10 equally sized subsets randomly and the calibration process is run 10 times, each time one subset is used for testing and the rest for training. The average values of all cutoff values, NPV and C for both training and testing data are outputted.
RESULTS
Automatic estimation of thresholds for identity, stretch and free energy
Experimental data from Rhee et al. (10) and He et al. (11) for 50mer oligonucleotides were used to estimate thresholds for sequence identity, stretch and free energy. Signal was expressed as the ratio to its perfect match. The data file is provided in Supplementary Data.
Table 1 shows the estimated cutoffs for five sets of criteria: (i) identity only, (ii) stretch only, (iii) free energy only, (iv) identity and stretch combined, and (v) identity, stretch and free energy combined. It can be seen that the criteria combining identity, stretch and free energy had a higher C than others when the signal threshold was 8 and 10%. Actually, under the experimental conditions examined, spots with signal <10% generally had an SNR <3.0 (11), and thus a signal threshold of 10% is recommended. For example, for 50mer oligonucleotides, with a maximal cross-hybridization of 10%, a minimal NPV of 95%, and a C of 70%, the thresholds were determined to be 87% identity, 17-base stretch and −29 kcal/mol of free energy. This suggests that combination of the three can yield a better coverage.
Table 1.
Cutoff values for five sets of criteria estimated by CommOligo_PE
| Signal threshold (%) | Criteria | NPV (%) | Coverage (%) |
|---|---|---|---|
| 5 | Identity | ||
| Stretch | |||
| Energy ≥ −12.00 | 96.2 | 30.1 | |
| Identity and stretch | |||
| Identity ≤ 0.85, stretch ≤ 13 and energy ≥ −12.00 | 96.2 | 30.1 | |
| 8 | Identity ≤ 0.77 | 96.6 | 28.3 |
| Stretch | |||
| Energy ≥ −19.00 | 95.8 | 46.5 | |
| Identity ≤ 0.81 and stretch ≤ 12 | 97.4 | 38.4 | |
| Identity ≤ 0.87, stretch ≤ 17 and energy ≥ −24.00 | 95.0 | 57.6 | |
| 10 | Identity ≤ 0.77 | 96.6 | 27.5 |
| Stretch ≤ 11 | 95.7 | 43.1 | |
| Energy ≥ −19.00 | 95.8 | 45.1 | |
| Identity ≤ 0.87 and stretch ≤ 11 | 95.7 | 43.1 | |
| Identity ≤ 0.87, stretch ≤ 17 and energy ≥ −29.00 | 96.0 | 69.6 |
Optimization goal was set to an automatic mode. The minimal NPV was set to 95%. Blank cells indicate no cutoff values were found in search range under the constraints. Data from Rhee et al. (10) and He et al. (11) in the Supplementary Data were used for training. When the optimization goal was changed to maximizing coverage, ‘identity ≤ 0.81 and stretch ≤ 12’ was changed to ‘identity ≤ 0.81 and stretch ≤ 18’, and ‘identity ≤ 0.87, stretch ≤ 17 and energy ≥ −29.00’ was changed to ‘identity ≤ 0.87, stretch ≤ 17 and energy ≥ −32.00’, while the others remained the same.
To further compare the five sets of criteria, CommOligo_PE was run for cross-validation. The same dataset was randomly partitioned into 10 subsets. CommOligo_PE was run 10 times, each time using one subset for testing and the rest for training. The average cutoff values, average NPV and average C values for both testing and training data for a run are shown in Table 2. Since not all criteria had cutoff values in the search range under the constraints, the average values were calculated using the actual number of values, and the number of times that cutoff value was generated is listed in Table 2 as well. The average cutoff values and the NPV and C values for training data were close to the values derived with all data for training. The combination of identity, stretch and free energy had NPV ranked in the middle among the five sets of criteria for all signal thresholds and had the highest C for 8 and 10% signal threshold. The NPVs are close to each other among different sets of criteria because the constraint limits it to 95% or higher for training data. In general, the results in Table 2 suggest that the criteria combining the three had a better C with a similar NPV. This is consistent with results trained from all data shown in Table 1.
Table 2.
Estimated criteria with cross-validation
| Signal threshold (%) | Criteria | Training NPV (%) | Training C (%) | Testing NPV (%) | Testing C (%) | N of runs with cutoffs generated |
|---|---|---|---|---|---|---|
| 5 | Identity | 0 | ||||
| Stretch | 0 | |||||
| Energy ≥ −12.40 | 95.9 | 31.2 | 93.3 | 30.8 | 10 | |
| Identity ≤ 0.81 and stretch ≤ 11 | 96.2 | 34.1 | 70.8 | 39.1 | 2 | |
| Identity ≤ 0.85, stretch ≤ 13 and energy ≥ −13.00 | 95.9 | 32.0 | 91.7 | 30.9 | 10 | |
| 8 | Identity ≤ 0.77 | 96.3 | 31.3 | 82.5 | 31.3 | 10 |
| Stretch ≤ 11 | 95.2 | 43.9 | 66.7 | 33.3 | 2 | |
| Energy ≥ −20.00 | 95.8 | 48.8 | 91.7 | 55.0 | 10 | |
| Identity ≤ 0.81 and stretch ≤ 14 | 96.5 | 42.5 | 85.0 | 38.7 | 10 | |
| Identity ≤ 0.83, stretch ≤ 17 and energy ≥ −29.20 | 96.8 | 58.1 | 89.1 | 58.6 | 10 | |
| 10 | Identity ≤ 0.77 | 96.0 | 28.8 | 98.0 | 30.3 | 10 |
| Stretch ≤ 11.00 | 95.9 | 43.2 | 96.3 | 44.5 | 10 | |
| Energy ≥ −20.10 | 95.9 | 47.9 | 91.7 | 48.6 | 10 | |
| Identity ≤ 0.84 and stretch ≤ 13 | 96.0 | 44.2 | 91.3 | 36.0 | 10 | |
| Identity ≤ 0.87, stretch ≤ 17 and energy ≥ −30.70 | 95.7 | 72.2 | 93.3 | 72.6 | 10 |
Data were partitioned into 10 subsets. Values shown are averages. Settings were the same as in Table 1.
Probe design datasets
Both whole-genome sequences and groups of highly homologous sequences were used to examine the performance of CommOligo. Here, we show the results for two datasets. The first was the whole-genome CDS sequences of Methanococcus maripaludis with 1766 ORFs; the second was groups of dissimilatory nitrite reductase genes, nirS and nirK, from our earlier collections with a total of 842 sequences, which were highly homologous (9). The homology of the two sets of sequences was measured by CD-HIT (25). At the threshold of 85% identity, CD-HIT produced 1754 clusters for M.maripaludis and 399 clusters for nirS and nirK. In other words, CD-HIT found that 0.7% of M.maripaludis and 52.6% of nirS and nirK sequences were redundant at 85% identity. At the cutoff of 50% identity, CD-HIT found that 4.0% of M.maripaludis and 83.1% of nirS and nirK sequences are redundant. This indicates the group sequences were much more homologous than the whole-genome sequences.
Probe design by CommOligo
50mer oligonucleotide probes were designed using the fitted thresholds, 87% of sequence identity, 17-base stretch and −29 kcal/mol of binding free energy with non-targets for the above two datasets. The maximal identity and the maximal length of continuous stretch were calculated for the first probe of each sequence. Identities of an oligonucleotide to its non-targets were calculated using ungapped alignment in a Perl script. Continuous stretches of a probe matching non-targets were first identified using BLAST (26), and then stretch lengths were determined using a Perl script. Free energy was calculated by CommOligo.
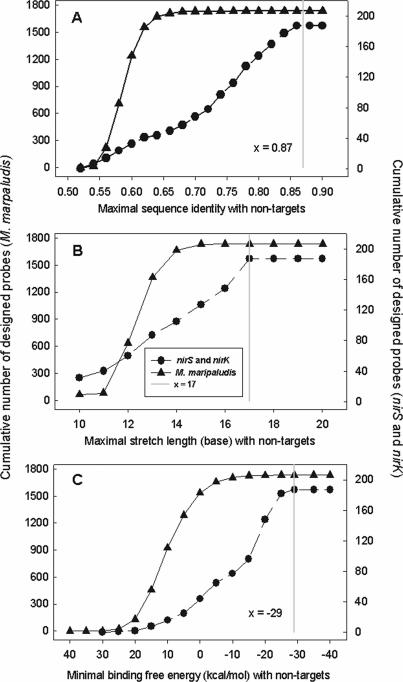
The relationships between sequence identity, stretch length or binding free energy and the number of designed probes were shown in Figure 3, which was a cumulative graph, with y value denoting the number of probes at no more than x identity. The maximum number of probes for a sequence was set to one when the probes were designed. Thus the number of probes equals the number of sequences with a probe designed. For the M.maripaludis genome sequences, CommOligo selected 1734 unique probes, and most probes (99.8%) with sequence identities <75%, 4 (0.2%) probes at sequence identities 75–87%, and 0 at sequence identities >87%. For the group of nirS and nirK sequences, the program selected 187 probes, and 97 probes with sequence identities <75%, 90 between sequence identities 75 and 87%, and 0 with sequence identities >87% (Figure 3A). For the stretch length of a designed probe to its non-targets, CommOligo chose 65, 15, 557, 727, 300, 67, 3 and 0 unique probes at the maximal stretch lengths of ≤10, 11, 12, 13, 14, 15, 16 or 17 bases, respectively for the whole-genome sequences. For the group of nirS and nirK sequences, the program selected 31, 9, 20, 27, 18, 22, 21 and 39 unique probes at stretch lengths of ≤10, 11, 12, 13, 14, 15, 16 or 17 bases, respectively (Figure 3B). For the binding free energy of a probe to its non-targets, CommOligo chose 1553, 172, 23, 6 and 0 probes at binding free energy values of ≥0, 0 to −10, −10 to −20, −20 to −29, and <−29 kcal/mol, respectively for the M.maripaludis genome sequences. For nirS and nirK group sequences, the program selected 44, 33, 71, 32 and 0 unique probes at binding free energy values of ≥0, 0 to −10, −10 to −20, −20 to −29 and <−29 kcal/mol, respectively (Figure 3C). Compared with previous design criteria determined by experiments, 85% identity, 15-base stretch and −30 kcal/mol of free energy (11), the number of designed probes and distributions of their identities and stretches did not change (only three more probes selected at relatively high similarities and stretches) for the whole-genome sequences. However, for nirS and nirK sequences, 44 more probes were selected, and most of them were distributed at relatively high similarity and/or long stretch levels (data not shown).
Figure 3.
The relationships between sequence identity (A), stretch length (B) or binding free energy (C) and the number of designed probes. Vertical lines x = 0.87, x = 17 and x = −29 indicate the fitted thresholds for probe design criteria.
Probe design by other software
Since different programs use different criteria, it is challenging to compare the probe design results. Here, maximal sequence identities and maximal lengths of continuous stretches to non-targets were calculated for designed probes. Those two criteria were experimentally established by Kane et al. (13), and then modified by Tiquia et al. (9), Rhee et al. (10) and He et al. (11). Four other programs, OligoArray (27) and OligoArray 2.0 (28), OligoPicker 2.3.8 (12) and ArrayOligoSelector (29) (http://sourceforge.net/projects/arrayoligosel/), were used to design probes for the same two datasets, M.maripaludis and nirS and nirK. Those programs were set to use the same cutoff values when the options were available. Otherwise, the closest values or default values were used. For the whole-genome sequences, the results from the four programs were generally good. ArrayOligoSelector, OligoArray, OligoArray 2.0 and OligoPicker selected probes for 1759, 1698, 1698 and 1724 sequences, respectively, and 1463, 1670, 1506 and 1721 with identities ≤87% and stretches ≤17 bases. However, those software tools also selected some probes with sequence identity >87% and/or continuous stretch length >17 bases. For example, ArrayOligoSelector had 296 with sequence identity >87% or/and maximal stretch length >17 bases, which may cross-hybridize with non-targets (Table 3). The average and standard deviation of identity to non-targets were 60.8 and 4.7% for OligoArray, 62.0 and 5.2% for OligoArray 2.0, 63.0 and 3.7% for OligoPicker, and 64.1 and 4.7% for ArrayOligoSelector, respectively, compared with 59.9 and 2.7% for CommOligo.
Table 3.
Number and quality of designed probes by different programs
| Programs used | Whole-genome sequences of M.maripaludis (1766 ORFs) | Group sequences of nirS and nirK (842 gene sequences) | ||||||
|---|---|---|---|---|---|---|---|---|
| ORFs rejected | Probes designed | Sim. ≤87% and Str. ≤17 | Sim. >87% or Str. >17 | ORFs rejected | Probes designed | Sim. ≤87% and Str. ≤17 | Sim. >87% or Str. >17 | |
| ArrayOligoSelector | 7 | 1759 | 1463 | 296 | 0 | 842 | 146 | 696 |
| OligoArray | 68 | 1698 | 1670 | 28 | 35 | 807 | 112 | 695 |
| OligoArray 2.0 | 68 | 1698 | 1506 | 292 | 51 | 791 | 55 | 736 |
| OligoPicker | 42 | 1724 | 1721 | 3 | 673 | 169 | 162 | 7 |
| CommOligo | 32 | 1734 | 1734 | 0 | 655 | 187 | 187 | 0 |
For the nirS and nirK sequences, OligoPicker, ArrayOligoSelector, OligoArray and OligoArray 2.0 selected 162, 146, 112 and 55 probes with identity ≤87% and continuous match ≤17 bases, respectively compared with 187 for CommOligo, but they also generated a different number (7–736) of probes with sequence identity >87% and/or maximal stretch length >17 bases (Table 3). The results indicated that all other software might not be suitable for selecting probes for groups of highly homologous genes.
DISCUSSION
The best oligonucleotide probe should have maximum hybridization with its target and minimum cross-hybridization with its non-targets. Although the nearest-neighbor model (24) has been widely used to study the binding free energy in solution, the hybridization kinetics on microarray slides seems to be more complicated. Kane et al. (13) suggested that an oligonucleotide probe showing >75–85% identity with non-targets may cause cross-hybridization. Their studies also showed that a probe, which had 15- and 20-base stretches with non-targets over 50 bases had detectable cross-hybridization. Previous studies showed that no significant cross-hybridization was observed when an identity was 85–88% for 50mer (9,10), and 80% for 60mer (30) oligonucleotide probes. Another study showed that significant cross-hybridization could happen when free energy <−35 kcal/mol for 70mer oligonucleotides (31). In our recent study, the relationships between hybridization signal and sequence identity, continuous stretch, binding free energy or mismatch position were examined, and a set of criteria were experimentally suggested for 50 and 70mer oligonucleotide probe design (11). The experimental data demonstrated that it was difficult to exclude all experimentally verified non-specific oligonucleotides using a single criterion, and that an appropriate combination of multiple criteria could exclude all non-specific probes (11). By combining multiple criteria, more liberal cutoffs can be used for each criterion. Indeed, the estimated parameter thresholds, identity of ≤87%, continuous stretch of ≤17 bases, and free energy of ≥−29 kcal/mol showing the best choice for designing 50mer oligonucleotides at a maximal cross-hybridization of 10%, are also very consistent with our experimentally established criteria: ≤85%, ≤15 bases and ≥−30 kcal/mol, respectively. Users can select their thresholds by choosing their tolerance of cross-hybridization, the minimal NPV and the minimal coverage using CommOligo_PE. Besides the above three criteria, other factors (GC content, the percentage of continuous single base and Tm) are also used to reject oligonucleotide candidates in CommOligo. To ensure probe sensitivity, an oligonucleotide probe must be accessible by its target. One of the most important characteristics is that an oligonucleotide probe should have no strong secondary structures. In CommOligo, an oligonucleotide probe is checked by self-annealing. In addition, oligonucleotide specificity and sensitivity may be affected by experimental conditions, such as hybridization temperatures, and the percentages of formamide used.
It is not surprising that all four other programs produced some probes with sequence identity >87% or continuous stretch >17 bases because they do not implement both criteria. For example, OligoArray, OligoArray 2.0 and ArrayOligoSelector do not use the continuous stretch as a criterion. On the other hand, CommOligo combines three criteria and more probes may be excluded than it uses two criteria. It should be also noted that CommOligo uses gapped alignment while the testing script uses ungapped alignment. OligoPicker selected a close number of probes to CommOligo for M.maripaludis (1721–1734), or nirS and nirK (162–187) sequences with identity ≤87% and continuous stretch ≤17 bases because both programs implement identity and stretch criteria. However, OligoPicker uses a BLAST-based approach, while CommOligo uses a global alignment algorithm. In addition, the probes from OligoPicker may not have free energy less than −29 kcal/mol, because CommOligo adopts the free energy as a cross-hybridization criterion while OligoPicker does not. These results suggest that most of preexisting software have been developed for designing oligonucleotides for whole-genome sequences, not for highly homologous sequences, and that CommOligo can be used for both types of sequences.
Most of probe design programs use BLAST to calculate a local sequence identity (12,27,32,33). BLAST reports the most similar regions between two sequences, while the length of those similar regions cannot be controlled. In most cases, a local alignment does not really reflect the overall identity between an oligonucleotide and its non-targets. Therefore, a filter based on local identity may filter out true specific probes and keep false-specific probes. ProbeSelect (34) selects oligonucleotides with words occurring least frequently in non-target sequences using a suffix array and a sequence landscape. However, the word frequency has no clear association with the sequence identity. PROBEmer (35) uses a suffix array to check exact matches and n-off matches. We believe that the global identity between a probe and non-target sequences is more relevant to probe specificity. Thus the identity is calculated using global alignment in CommOligo. An accurate measurement of sequence identity can be obtained for both filtering and probe optimization.
Because the calculation of the exact minimum binding free energy between a probe and a sequence is too slow, probe design programs only calculate the free energy for highly homologous regions. ArrayOligoSelector (29) and OligoArray 2.0 (28) use BLAST to find those regions. ProbeSelect (34) uses Myers' global alignment to locate the end positions of an alignment with the given number of mismatches and uses a heuristic algorithm to test alternatives. In CommOligo, we propose a traversal algorithm to generate all alignments for more accurate free energy calculation. This ensures that all alignments with high global identities are evaluated.
It is ideal to keep the melting temperatures of all probes in a narrow range. Some software tools, such as OligoArray (27) and PROBEmer (35) use a user-defined interval. OligoPicker (12) automatically filters probes around the median Tm with a user-defined interval. OligoWiz (32) tries to find the best length of probes with the minimum deviation from the center of melting temperature of all positions. These methods may filter out some specific probes because the Tm calculation is based on all possible oligonucleotides, especially when sequences have high identity and only few probes can be picked. By searching only the probe candidates that passed all other filters, CommOligo is able to design probes for maximum number of sequences against Tm interval.
Optimization of probe candidates will ensure that the best oligonucleotides are selected for a gene when the number of designed oligonucleotides is more than that required by a user. OligoDesign (33) uses a sigmoid function to calculate a score for each property (maximum number of matching nucleotides by BLAST, longest continuous stretch, Tm, self-annealing and secondary structures) of an oligonucleotide and then calculates the weighted average of scores of all properties. CommOligo uses a piece-wise linear function to calculate a score for individual measurements of an oligonucleotide and the overall score is the weighted average or the minimum of all individual scores. Piece-wise linear functions are chosen because they are consistent with hard-limit filters and all parameters are easy to set by the users according to their applications and experiences. By utilizing an iterative process, the specificity and distances between probes of the same target are taken into account.
Commonly used oligonucleotide probes are between 15 and 70mers. Our effort has been focused on long oligonucleotide probes from 40 to 80mers. CommOligo comes with a set of default parameters for 50mer probes. All parameters are user adjustable and one can turn off a filter by setting its parameters to a considerably large or small value. However, setting the criteria for the selection of oligonucleotides with a particular length is still a challenge. This requires experimental data to support, such as those described by Kane et al. (13), Tiquia et al. (9), Rhee et al. (10), Bozdech et al. (31) and He et al. (11), or sufficient knowledge about their hybridization. Our software can compute probes from 10 to 128mers. However, long oligonucleotide probes (>80mer) are rarely used because they may cause less probe availability for a given dataset, synthesis difficulty and higher possibility of secondary structures. For a probe <15mer, the signal intensity is too weak so that it may not be practically useful for detection. For short oligonucleotide probes, some further measurements for sensitivity may be needed. In addition, while a mismatch match (MM) probe (one mismatch at the middle position) is widely used along with a perfect match (PM) probe for probes <30mers, our software does not automatically select MM probes. One may have to select a PM probe first, and then replace the middle nucleotide of the PM probe to produce its MM probe.
Systematical estimation of parameter thresholds for oligonucleotide design is a new feature. Up to now, the selection of suitable cutoff values for probe design criteria remains a great challenge. Most thresholds are set arbitrarily, or based on individual experiments. Here, CommOligo_PE has been developed to determine thresholds for probe design criteria using available experimental data. However, it requires high quality and relatively large datasets to generate more accurate cutoff values. Unfortunately, publicly available spike-in large datasets are mainly for short oligonucleotide arrays. For long oligonucleotides, only a few datasets are publicly available. With an increase in experimental microarray data, it is predicted that this estimator will be able to produce more reliable thresholds for probe design. Since different hybridization protocols may result in significant differences in results, large datasets under the same condition are desirable. We used datasets from Rhee et al. (10) and He et al. (11), which were performed under very similar conditions.
CONCLUSIONS AND FUTURE WORK
CommOligo differs from other probe design software in several important ways. First, CommOligo is able to estimate parameter cutoffs from hybridization data. Second, to handle high homologous sequences, cross-hybridization is predicted with three measurements, maximum sequence identity, minimum binding free energy and maximum continuous stretch between a probe and its non-targets. Preexisting software only use one or two of them. Our testing shows a combination of the three measurements can generate better gene coverage at a similar NPV. Third, special attention is paid to the calculation of identity and binding free energy in a more accurate way. Our sequence identity is based on the best global gapped alignment between an oligonucleotide and its non-target sequences. This approach is different from BLAST-based local identity, or suffix tree for exact string detection, which have been used by most available software. This approach also ensures that only oligonucleotides with true high identities are filtered. A traversal algorithm is proposed to generate global alignments, allowing the most stable alignments to be compared for free energy calculation. Fourth, the optimal interval of Tm is based on probe candidates that have passed all other filters, rather than all possible oligonucleotides, which ensures that a maximal number of sequences have probes. Finally, three measurements and the distance between probes for the same target are combined for probe optimization using piece-wise linear functions in an iterative process. Our evaluation results on both whole-genome and highly homologous group sequences demonstrate that CommOligo performed well and promises to be a general oligonucleotide probe design tool for various types of sequences.
It should be noted that this program runs relatively slow. It took ∼18 h to design probes for the M.Maripaludis genome on a PC with 2.50 GHz CPU and 512 MB memory. For a group of highly homologous sequences, only a small fraction of genes have probes. Our future studies will focus on development of faster algorithms and selection of probes specific to a group of sequences for highly homologous genes.
Acknowledgments
This research was supported by the United States Department of Energy under Genomics:GTL program through the Virtual Institute of Microbial Stress and Survival (VIMSS; http://vimss.lbl.gov), the Natural and Accelerated Bioremediation Research Program, and Microbial Genome Program of the Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725. Funding to pay the Open Access publication charges for this article was provided by the US Department of Energy Genomics: GTL and NABIR programs.
Conflict of interest statement. None declared.
REFERENCES
- 1.DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 2.Wodicka L., Dong H., Mittmann M., Ho M.H., Lockhart D.J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 3.Richmond C.S., Glasner J.D., Mau R., Jin H., Blattner F.R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes T.R., Marton M.J., Jones A.R., Roberts C.J., Stoughton R., Armour C.D., Bennett H.A., Coffey E., Dai H., He Y.D., et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Zhou J., Omelchenko M., Beliaev A., Venkateswaran A., Stair J., Wu L., Thompson D.K., Xu D., Rogozin I.B., et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl Acad. Sci. USA. 2003;100:4191–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J., Thompson D.K. Challenge in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 2002;13:204–207. doi: 10.1016/s0958-1669(02)00319-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhou J. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 2003;6:288–294. doi: 10.1016/s1369-5274(03)00052-3. [DOI] [PubMed] [Google Scholar]
- 8.Relógio A., Schwager C., Richter A., Ansorge W., Valcarcel A. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acid Res. 2002;30:e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiquia S.M., Wu L., Chong S.C., Passovets S., Xu D., Xu Y., Zhou J. Evaluation of 50mer oligonucleotide arrays for detecting microbial populations in environmental samples. Biotechniques. 2004;36:664–675. doi: 10.2144/04364RR02. [DOI] [PubMed] [Google Scholar]
- 10.Rhee S.K., Liu X., Wu L., Chong S.C., Wan X., Zhou J. Detection of biodegradation and biotransformation genes in microbial communities using 50mer oligonucleotide microarrays. Appl. Environ. Microbiol. 2004;70:4303–4317. doi: 10.1128/AEM.70.7.4303-4317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Z., Wu L., Li X., Fields M.W., Zhou J. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 2005;71:3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 13.Kane M.D., Jatkoe T.A., Stumpf C.R., Lu J., Thomas J.D., Madore J.M. Assessment of the specificity and sensitivity of oligonucleotide (50mer) microarrays. Nucleic Acid Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers E.W. A fast bit-vector algorithm for approximate string matching based on dynamic programming. J. ACM. 1999;46:539–553. [Google Scholar]
- 15.Zuker M., Matthews D.H., Turner D.H. NATO ASI Series. Dordrecht, NL: Kluwer; 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. [Google Scholar]
- 16.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 17.Allawi H.T., SantaLucia J., Jr Nearest neighbor thermodynamic parameters for internal G–A mismatches in DNA. Biochemistry. 1998a;37:2170–2179. doi: 10.1021/bi9724873. [DOI] [PubMed] [Google Scholar]
- 18.Allawi H.T., SantaLucia J., Jr Nearest-neighbor thermodynamics of internal A–C–ismatches in DNA: sequence dependence and pH effects. Biochemistry. 1998b;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- 19.Allawi H.T., SantaLucia J., Jr Thermodynamics of internal C·T mismatches in DNA. Nucleic Acids Res. 1998c;26:2694–2701. doi: 10.1093/nar/26.11.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger J., Turner D.H., Zuker M. Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peritz A.E., Kierzek R., Sugimoto N., Turner D.H. Thermodynamic study of internal loops in oligonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry. 1991;30:6428–6436. doi: 10.1021/bi00240a013. [DOI] [PubMed] [Google Scholar]
- 22.Peyret N., Seneviratne P.A., Allawi H.T., SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A-A, C-C, G-G, and T-T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 23.Lyngso R.B., Zuker M., Pedersen C.N. Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics. 1999;15:440–445. doi: 10.1093/bioinformatics/15.6.440. [DOI] [PubMed] [Google Scholar]
- 24.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., Jaroszewski L., Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Rouillard J.-M., Herbert C., Zuker M. OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics. 2002;18:486–487. doi: 10.1093/bioinformatics/18.3.486. [DOI] [PubMed] [Google Scholar]
- 28.Rouillard J.-M., Zuker M., Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J., Bozdech Z., DeRisi J. 2003. ArrayOligoSelector program. Available at http://sourceforge.net/projects/arrayoligosel/ [Google Scholar]
- 30.Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 31.Bozdech Z., Zhu J., Joachimiak M.P., Cohen F.E., Pulliam B., DeRisi J.L. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparium with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen H.B., Wernersson R., Knudsen S. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 2003;31:3491–3496. doi: 10.1093/nar/gkg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolstrup N., Nielsen P., Kauppinen S. OligoDesign: design of LNA oligonucleotides for gene expression arrays. Nucleic Acids Res. 2003;31:3758–3762. doi: 10.1093/nar/gkg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F., Stormo G. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics. 2001;17:1067–1076. doi: 10.1093/bioinformatics/17.11.1067. [DOI] [PubMed] [Google Scholar]
- 35.Emrich S.J., Lowe M., Delcher A. PROBEmer: a web-based software tool for selecting optimal DNA oligos. Nucleic Acids Res. 2003;31:3746–3750. doi: 10.1093/nar/gkg569. [DOI] [PMC free article] [PubMed] [Google Scholar]