Abstract
Decoding is the key step during protein synthesis that enables information transfer from RNA to protein, a process critical for the survival of all organisms. We have used large-scale (2.64 × 106 atoms) all-atom simulations of the entire ribosome to understand a critical step of decoding. Although the decoding problem has been studied for more than four decades, the rate-limiting step of cognate tRNA selection has only recently been identified. This step, known as accommodation, involves the movement inside the ribosome of the aminoacyl-tRNA from the partially bound “A/T” state to the fully bound “A/A” state. Here, we show that a corridor of 20 universally conserved ribosomal RNA bases interacts with the tRNA during the accommodation movement. Surprisingly, the tRNA is impeded by the A-loop (23S helix 92), instead of enjoying a smooth transition to the A/A state. In particular, universally conserved 23S ribosomal RNA bases U2492, C2556, and C2573 act as a 3D gate, causing the acceptor stem to pause before allowing entrance into the peptidyl transferase center. Our simulations demonstrate that the flexibility of the acceptor stem of the tRNA, in addition to flexibility of the anticodon arm, is essential for tRNA selection. This study serves as a template for simulating conformational changes in large (>106 atoms) biological and artificial molecular machines.
Keywords: proofreading, protein synthesis, molecular dynamics simulations, RNA, high-performance computing
Accurate and rapid selection of tRNAs by the ribosome is critical for cell viability. Selection of cognate tRNAs occurs in two stages (initial selection and proofreading), which are separated by hydrolysis of GTP in the aminoacyl-tRNA·GTP·elongation factor Tu (EF-Tu) ternary complex (1-4). During initial selection, the ternary complex binds to the aminoacyl site (A site) of the small (30S) ribosomal subunit and the GTPase activation center (GAC) of the large (50S) ribosomal subunit producing the “A/T” state complex (5-7). It has also been suggested that codon-anticodon interactions at the exit site can affect selection (8).
During the proofreading step, the EF-Tu·GDP dissociates, and the aminoacyl-tRNA moves from the A/T to the fully bound A/A state, where the tRNA occupies the 30S and 50S A sites (accommodation). The ribosome must adjust its conformation to accommodate this change in the aminoacyl-tRNA (6). This accommodation step is rate-limiting for cognate tRNA (9). The mechanism of this important step is currently unknown. Because the ribosome performs the complex operation of transforming a 4-letter alphabet sequence into a 20-letter alphabet sequence, the decoding mechanism has implications for artificial information-processing molecular machines.
Structural biologists have made a significant contribution, determining the lower-resolution structures of the ribosome in the A/A (10, 11) and A/T states (12, 13), as well as the interactions of cognate tRNA analogs with the 30S and 50S subunits in atomic detail (14-17). More recently, higher-resolution cryo-EM data have shown that the aminoacyl-tRNA in the A/T state is kinked with respect to the A/A state (hereby referred to as the A/T kink), suggesting that the accommodation change involves a relaxation of the kinked tRNA that preserves the decoding-center interactions (6, 7, 18). The only significant change in the A/T state ribosome is the movement of the GAC (6), which occurs in two steps, corresponding to before and after GTP hydrolysis (7, 19). The structures of the A/A (10) and A/T (6) states of the 70S ribosome have established the initial and final states of the accommodation step; however, important questions remain concerning the accommodation pathway. In particular, (i) which proteins and 23S rRNA nucleotides does the tRNA interact with during accommodation? and (ii) how is it possible for the acceptor stem of the tRNA to move into the peptidyl transferase center, given the tight packing (20) of 23S rRNA in this region of the large subunit?
Molecular dynamics simulations and modeling studies have explored the decoding center (21-24), isolated tRNA (25), and the inner core of the 30S ribosomal subunit (26, 27). Adaptive-mesh refinement Poisson-Boltzmann studies based on phosphate/Cα structures have examined the electrostatic potential of the subunits (28). Coarse-grained studies have investigated the motions of the ribosome within the approximations of the Gaussian network and bead models (28-31). Real-space refinement has been combined with cryo-EM structures to study the beginning and end states of a conformational change that occurs during translocation (32). Explicit solvent molecular dynamics simulations have previously helped elucidate functional mechanisms of biomolecular systems (33-38) and been validated with experiments (39).
The Advanced Simulation and Computing (ASCI) Q machine at Los Alamos National Laboratory, together with the namd program (40), has enabled us to simulate the accommodation of tRNA into the 70S ribosome in explicit solvent (2.64 × 106 atoms), yielding significantly more accurate dynamics than implicit solvent methods (41) and the coarse-grained calculations described above. Computationally, the simulations represent a step forward in that the largest previous biomolecular dynamics simulations have been on the order of ≈4.2 × 105 atoms (42, 43). Because the accommodation rate of cognate tRNAs is ≈7/s (9), even enhanced sampling molecular dynamics simulation techniques yield insufficient sampling to observe spontaneous accommodation. Therefore, to examine accommodation, we have implemented the targeted molecular dynamics algorithm (35, 44, 45), which gradually reduces the root mean square deviation between the simulated structure and the final target structure (A/A state) while allowing thermal fluctuations at a given root mean square deviation. Targeted molecular dynamics simulates a single barrier-crossing event. Although the accommodation barrier-crossing time cannot be measured experimentally, it is thought to be several orders of magnitude smaller than the accommodation rate. Kinetic experiments have suggested that accommodation follows EF-Tu dissociation (1). Here, we assume that EF-Tu has already dissociated from the ribosome.
Rather than providing an exact energy landscape of the transition, our simulations produce stereochemically feasible pathways that can be tested experimentally. In particular, the goal of this work is to predict 23S rRNA bases and ribosomal protein residues that are important for accommodation. This approach is complementary to recent x-ray crystallography (10, 11, 14, 16, 17), cryo-EM (7, 12, 18), kinetic (1), and single-molecule (19) studies and helps establish a logical picture of the mechanism of tRNA selection. We tested the sensitivity of our results to initial conditions by performing seven 2-ns simulations with different initial velocities and the sensitivity to the simulated time scale by doubling and halving the simulation time, giving a total sampling of 20 ns for production simulations.
Methods
The A/T-state model structure is similar to the previous A/A-state model (46), with the exception of the tRNA and the GAC (23S rRNA helices 43-44 and L11), whose structures are based on the corresponding cryo-EM map (6). The solvated systems were minimized and equilibrated (namd2.5) for 1.6 ns. Performance studies demonstrated near-linear scaling to 768 processors (see Supporting Text, which is published as supporting information on the PNAS web site, for details of the methods used). The figures were created with vmd (47).
Results
The accommodation of the aminoacyl-tRNA in all simulations displays a bulk motion of the tRNA body into the large subunit A site (Movie 1 A, which is published as supporting information on the PNAS web site, and Fig. 1). The motion involves the relaxation of the A/T kink about tRNA positions 44-45 and 26, combined with a ≈45° rotation around the anticodon stem loop axis. Each simulation also shows the subsequent accommodation of the universally conserved 3′-CCA end into the peptidyl transferase center and convergence to the target structure. The entrance of the 3′-CCA end into the peptidyl transferase center is impeded with respect to the motion of the tRNA body because of the interaction with the 23S A-loop (positions 2552-2561). The codon-anticodon interactions, and the interactions of 16S rRNA G530, A1492, and A1493 with the tRNA and mRNA, remained intact throughout the accommodation simulations (Fig. 5, which is published as supporting information on the PNAS web site).
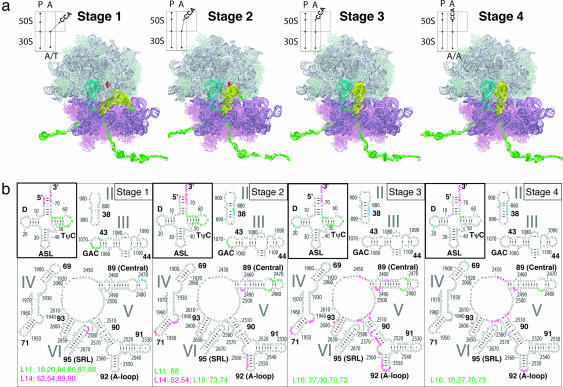
Fig. 1.
Accommodation simulation overview and aminoacyl-tRNA-50S interactions. (a) Time evolution of cognate tRNA accommodation into the ribosome for one of seven 2-ns simulations. Explicit waters, ions, and the top portion of the 70S ribosome are not shown so that the tRNAs are visible. The 23S rRNA (white), 50S proteins (light green), 16S rRNA (purple), 30S proteins (pink), mRNA (dark green), aminoacyl-tRNA (yellow) with Phe amino acid (dark green), peptidyl-tRNA (cyan), and 23S rRNA 2553 (red) are shown. The schematics are similar to that of Moazed and Noller (5) and depict the process of accommodation. A, aminoacyl site; P, peptidyl site. Stage 1 is represented by the initial production structure, after 1.6 ns of equilibration (t = 0 ns). Stage 2 shows the relaxation of the aminoacyl-tRNA body (i.e., all but the 3′-CCA portion) (t = 0.292 ns). In stage 3, the aminoacyl-tRNA body is accommodated (t = 0.65 ns). During stage 4, the aminoacyl-tRNA 3′-CCA end is accommodated into the peptidyl transferase center (2.2 ns). (b) Secondary structure diagrams color-coded by the region of tRNA that participates in the aminoacyl-tRNA-50S interaction. Green, interactions with TΨC loop; blue, interactions with D loop; magenta, interactions with 3′-CCA end. ASL, anticodon stem loop.
The interactions between the tRNA and the ribosome (Movie 1B and Fig. 2) are divided into four stages, describing A/T interactions (stage 1), relaxation of the tRNA body (stage 2), relaxation of the tRNA acceptor stem (stage 3), and A/A interactions (stage 4). The tRNA movement is characterized by the parameters θ1, φ1, and φ2 (defined in the Fig. 2 legend).
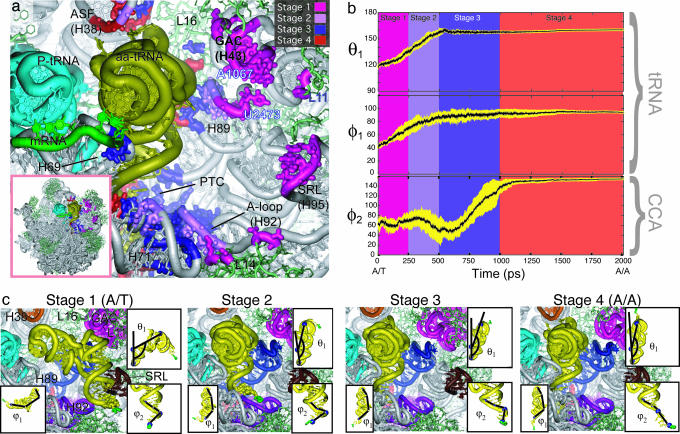
Fig. 2.
Stages of accommodation. (a) Aminoacyl-tRNA interaction regions (within 3.5 Å) colored according to the accommodation stage. (Inset) The context of the accommodation wall on the 50S ribosomal subunit. (b) Time evolution of parameters that describe the deformation of aminoacyl-tRNA. Average values (black) of θ1, φ1, and φ2 (averaged over seven trajectories) and corresponding variances (yellow) are shown. θ1 is the angle between O3′ atoms at positions 33, 40, and 55 of the tRNA. φ1 is the dihedral angle between O3′ atoms at positions 33, 40, 55 and 1. φ2 is the dihedral angle between O3′ atoms at positions 69, 71, and 73 and the O of the Phe. (c) Snapshots from stage 1 (t = 0 ns), stage 2 (t = 0.488 ns), stage 3 (t = 0.650 ns), and stage 4 (t = 2.2 ns).
Stage 1. During the initial stage of accommodation, the 3′-CCA end of the tRNA remains relatively unencumbered by the ribosome, apart from a slight deflection (Δφ2 ≈ 5°) because of interaction with ribosomal protein L14 (residues 52, 54, 89, and 90) (Fig. 2 and Fig. 6, which is published as supporting information on the PNAS web site). There is also a transient interaction between the acceptor arm and ribosomal protein S12. Initially, the phosphate of tRNA base U69 interacts with His-80 of S12. As the acceptor arm moves away from the A/T-state position, the phosphate of tRNA U68 briefly interacts with Gln-78.
The elbow region (tRNA bases 52-58) interacts with the GAC at base A1067, whose N6 position interacts consecutively with elbow bases (tRNA bases 55, 54, 53, 52, and 51) as the tRNA moves toward the A site (Figs. 2 and 3 and Fig. 7, which is published as supporting information on the PNAS web site). The elbow region also interacts with 23S rRNA helix 89 U2473, which makes consecutive interactions with tRNA bases 62, 63, and 64 as they pass by helix 89 (Fig. 3). The tRNA also interacts with 23S rRNA A2660-G2661 in the sarcin-ricin loop (SRL) at early times as it moves toward the A site (Fig. 3). Large subunit rRNA bases U2473, G2661, and A1067 appear to “monitor” the tRNA by interacting continuously with the backbone of the tRNA as the tRNA moves toward the A/A state.
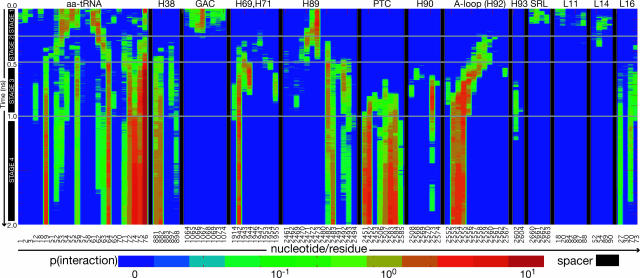
Fig. 3.
Time evolution of probability densities, p, of interactions (blue, no interaction; red, strong interaction). p is the number of interactions normalized by the number of simulations (defined in Fig. 7). Nucleotide/residue positions for the aminoacyl-tRNA, 23S rRNA, and 50S proteins participating in aminoacyl-tRNA interactions are shown along the x axis. Diagonal contours for the A-loop correspond to the aminoacyl-tRNA 3′-CCA end interacting with consecutive A-loop nucleotides as a function of time. Distinct regions are separated by vertical black bars.
Stage 2. In stage 2 (Figs. 1b and 2), helix 89 continues to interact with the tRNA. We see that at the end of stage 2 the tRNA body is nearly accommodated, with θ1 and φ1 within 10% of their final values (Fig. 2). The 3′-CCA end of the tRNA begins its interaction with the A-loop in this stage (Figs. 1b and 3); however, it is not accommodated by the end of stage 2 (Movie 1B and Fig. 2). Instead, it is impeded by the 23S A-loop, whose interaction causes a significant deflection (Δφ2 ≈ -25°) in the 3′-CCA end (Fig. 2).
Stage 3. In stage 3, after first interacting with the elbow of the tRNA, the relative orientation of H89 with respect to the tRNA causes the interaction point to move on the tRNA progressively toward the 3′-CCA end (Figs. 2 and 3). In the sense that H89 sterically restricts the motion of the tRNA, helix 89 appears to “guide” the aminoacyl-tRNA toward the 50S A site. When the tRNA reaches the A-loop, the 3′-CCA end moves along the A-loop and H90 toward the peptidyl transferase center, where the 3′-CCA end interacts consecutively with respect to time with 2560, 2559, 2558, 2557, 2556, 2555, 2554, and 2553 (Fig. 3).
During stage 3, both the 3′-CCA end and the A-loop flex slightly to allow accommodation of the 3′-CCA end through the A-loop toward the peptidyl transferase center (Movie 1D, Fig. 4 a-c, and Fig. 8 A-C, which is published as supporting information on the PNAS web site). The mutual flexibility between the tRNA 3′-CCA end and the 23S rRNA A-loop resolves the apparent steric clash present during accommodation.
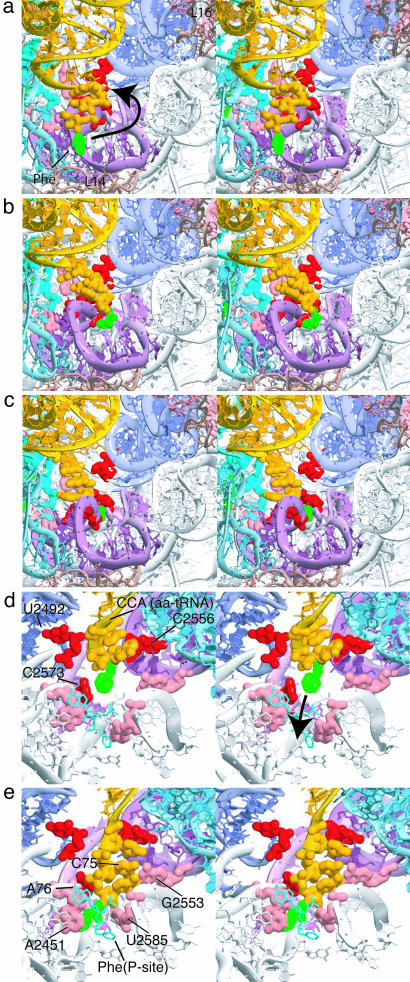
Fig. 4.
Close-up stereoview of the accommodation of the aminoacyl-tRNA 3′-CCA end by the peptidyl transferase center of the large ribosomal subunit. The aminoacyl-tRNA (yellow) and Phe amino acid (green), peptidyl-tRNA (cyan), 50S proteins (pink), 23S H69 and H71 (light blue), H92 A-loop (purple), H90 (light purple), H89 (dark blue), gate nucleotides (U2492, C2556, and C2573, red), and peptidyl transferase center nucleotides (A2451, C2452, G2553, and U2585, pink/peach) are shown. (a) The 3′-CCA end begins its interaction with the A-loop (t = 0.345 ns). (b) 3′-CCA end continues to move through the A-loop (t = 0.592 ns). (c) The 3′-CCA end enters the peptidyl transferase center (t = 0.670 ns). (d) View from the peptidyl transferase center (t = 0.676 ns). Strong interactions between the 3′-CCA end and universally conserved 23S rRNA U2492, C2556, and C2573 occur. (e) The 3′-CCA end reaches the target A/A state configuration where the CCA end forms the C75:G2553 base pair and close interaction with A2451 and P-tRNA Phe (t = 2.2 ns).
Strong interactions (probability density, p > 1, defined in the Fig. 7 legend) occur between the 3′-CCA end and universally conserved bases U2492, C2556, and C2573 (Movie 1 C and D and Fig. 3). Interaction of the 3′-CCA end with these three nucleotides coincides with the pausing of the 3′-CCA-Phe portion of the tRNA (i.e., the center of mass velocity of this portion decreases dramatically) just before entrance into the peptidyl transferase center (Movie 1C and Fig. 9, which is published as supporting information on the PNAS web site). In particular, the 3′-CCA end pauses upon interaction with U2492 and C2556 until the U2492/C2556 gate opens (i.e., the U2492 O2′-C2556 O2′ distance increases significantly) at which point the CCA end proceeds through (Fig. 9). The CCA end then immediately interacts with C2573, causing it to pause a second time (Fig. 9). These characteristics are consistent with a 3-nt, 3D gate that impedes the 3′-CCA-Phe portion before entrance into the peptidyl transferase center.
Finally, L16, H38, and H69 are positioned to prevent the aminoacyl-tRNA from overshooting its target position and interacting with the peptidyl-tRNA. Thus, during stage 3, the tRNA is sterically guided, paused, and stopped by 23S rRNA.
Stage 4. By stage 4, the tRNA body has been accommodated by the ribosome, and the 3′-CCA end enters the peptidyl transferase center (Figs. 4 D and E and 8 D and E). During this stage, the 3′-CCA end moves into its final configuration [based on the higher resolution x-ray data (16)], forming the C75:G2553 base pair and proper interactions with peptidyl transferase bases A2451, C2452 and U2585 (Movie 1E and Figs. 4 and 8). The phosphate root mean square deviation values between the final simulated structure and the A/A-state x-ray structures are 1.21, 1.27, and 2.41 Å for the 30S A and peptidyl (P) sites (14), the 50S A and P sites (16), and the 70S complex (10), respectively (A and P sites are defined by all rRNA nucleotides and protein residues within 4.5 Å of the tRNA analogs in the higher-resolution x-ray structures). The larger root mean square deviation for the 70S is likely caused by the lower resolution of the 70S x-ray structure upon which our 70S A/A state is based.
Discussion
Of the 68 nucleotides in the 23S rRNA that were observed to interact with the aminoacyl-tRNA during our simulations, 28 nucleotides (41%) are ≥95% conserved. Eighteen nucleotides are “universally conserved” in the sense that they are as highly conserved (≈99%) or more highly conserved than 16S rRNA base A1493 (48). The universal 23S rRNA nucleotides that interact with the aminoacyl-tRNA are 1943, 1953, and 1955 (H71 region); 2492 (H89); 2506, 2451, 2452, 2583, 2584, and 2585 (peptidyl transferase center); 2552, 2553, and 2556 (A-loop H92); 2508 and 2573 (H90); 2602 (H93); and 2662-3 (SRL). The remaining 10 bases that are ≥95% conserved are 1074 (GAC); 2469, 2470, 2472, 2482, and 2493 (H89); 2554, 2555, and 2559 (A-loop H92); and 2660 (SRL).
The focus of this investigation is the transition from the A/T to the A/A state, rather than the A/T and A/A states themselves. Of the tRNA interactions that occur between these two states, the A-loop and H89 display the strongest interactions (Fig. 3) and also have the largest number of highly conserved nucleotides. In H89, purine mutations of U2492 will change the U2492:U2460 pair [present in the x-ray structure (16) and in our simulations] to the more stable G:U or A:U pairs; however, the larger purine nucleotide will cause the backbone of the 2492 region to protrude into the pathway of the tRNA, narrowing the accommodation corridor and, thus, increasing the steric barrier. The mutation of C2573 to a purine may also result in increased steric inhibition and corresponding hyperaccurate phenotypes.
Previous studies showed that mutations of 23S U2492 suppress frameshifting (49), even though U2492 does not interact with the tRNA in the A/T or A/A states. This fact is consistent with U2492 interacting with the tRNA during accommodation, but not in A/T or A/A states, as observed in our simulations. Mutations of U2555 exhibit a ribosomal ambiguity phenotype (49, 50), whereas mutations in G2583 result in increased translational accuracy (51). Interestingly, both of these nucleotides display strong interactions with the tRNA in our simulations. Finally, changes in the conformation of the D-loop are thought to play a key role in decoding as shown by rapid kinetics and D-loop mutations (52). Performing simulations of available mutants in addition to WT systems may provide greater insight into experimental data.
We note that if the structure of the rRNA-tRNA interaction regions differs significantly in other species, the accommodation pathway may differ. However, we expect the accommodation pathway to be highly conserved because the backbone conformations of the interaction regions (e.g., H92 and H89) appear to be highly conserved in known x-ray structures (10, 11, 15, 17) and the specific nucleotides are highly conserved across all species. Although the specific orientation of the 3′ end of the tRNA differs in Haloarcula marismortui and Deinococcus radiodurans, we expect it only affects the final minor structural adjustments made during stage 4 of the accommodation simulation. We expect our conclusions, which were drawn mainly from stages 2 and 3, to remain the same.
The successful movement of the system from the A/T to the A/A states (Fig. 5) for seven sets of initial velocities demonstrates the stereochemical feasibility of maintaining similar codon-anticodon-16S geometries throughout accommodation, as proposed by Ramakrishnan and colleagues (14) and Frank and colleagues (6). The maintenance of the codon-anticodon-16S geometry is made possible by the flexibility of the tRNA and manifested in part by a change of the variable loop conformation and reorganization in the T-loop/D-loop stacking interactions. The decoding-center interactions, which are constrained in the simulations, have been shown by x-ray crystallography to be critical for cognate tRNA recognition (14). Molecular dynamics studies suggest that the G530-A1492 interaction cannot occur for purine-purine near-cognate codon-anticodon interactions (21).
Within the context of the decoding process, the favorable interactions of the ternary complex with the ribosome likely overcome the energetic cost of inducing the A/T kink in the aminoacyl-tRNA. Once EF-Tu dissociates, the tRNA is allowed to relax to the A/A state (which more closely resembles the native tRNA state) while its anticodon stem loop remains tightly bound to the 30S A site. The 3′-CCA end, however, is deterred mainly by the A-loop. The flexibility of the tRNA 3′-CCA end facilitates its movement into the peptidyl transferase center. The A-loop constitutes a corridor through which the amino acid may pass if the tRNA and the A-loop are sufficiently flexible and the tRNA is properly aligned. The large available volume in the corridor (i.e., below the Phe amino acid in Figs. 4B and 8B) makes this pathway a candidate for the accommodation of all other amino acids. The U2492/C2556/C2573 gate presents an obvious mechanism to prevent the 3′-CCA end from escaping the peptidyl transferase center, increasing the probability of C75:G2553 base-pairing and proper orientation of the amino acid.
Because proper accommodation requires a properly aligned anticodon stem loop, a near-cognate tRNA with near-cognate codon-anticodon interaction (21, 53) may significantly affect the accommodation trajectory, potentially decreasing the chances of entry of the 3′-CCA end into the peptidyl transferase center. If the 3′-CCA end does not enter the peptidyl transferase center, the tRNA dissociation rate will likely increase.
Although x-ray and cryo-EM studies (6, 14) have established the structural interactions that occur during cognate tRNA recognition, the recent kinetic results (9) have demonstrated the importance of kinetic activation barriers in decoding and established accommodation as the rate-limiting step for cognate tRNA selection. We have taken the next step by obtaining candidate transition structures for the accommodation of the cognate tRNA. In the future, examination of near-cognate tRNAs will be critical to uncover the mechanism by which ribosomes are able to reject incorrect tRNAs, which is an essential part of the translation of genetic messages by the ribosome.
Supplementary Material
Acknowledgments
We thank Steve Harvey, Angel Garcia, Chris Jarzynski, Jason Feinberg, and Steve Phelps for useful discussions and critical reading of the manuscript; and Andy White, Manuel Vigil, and Hal Marshal for their support and input. This work was performed under the auspices of the U.S. Department of Energy under Contract W-7405-ENG-36. K.Y.S. was supported by National Institutes of Health Grant R01-GM072686, and S.J. was supported by National Science Foundation Grant MCB-0078322. The Ribosome Project is generously supported by the Los Alamos National Laboratory Institutional Computing Program.
Author contributions: K.Y.S. designed research; K.Y.S. performed research; C.-S.T. contributed new reagents analytic tools; K.Y.S. and S.J. analyzed data; and K.Y.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: A site, aminoacyl site; GAC, GTPase activation center; EF-Tu, elongation factor Tu; SRL, sarcin-ricin loop.
References
- 1.Rodnina, M. V. & Wintermeyer, W. (2001) Annu. Rev. Biochem. 70, 415-435. [DOI] [PubMed] [Google Scholar]
- 2.Ninio, J. (1974) J. Mol. Biol. 84, 297-313. [DOI] [PubMed] [Google Scholar]
- 3.Hopfield, J. J. (1974) Proc. Natl. Acad. Sci. USA 71, 4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson, R. C. & Stone, P. J. (1977) Proc. Natl. Acad. Sci. USA 74, 198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moazed, D. & Noller, H. F. (1989) Nature 342, 142-148. [DOI] [PubMed] [Google Scholar]
- 6.Valle, M., Zavialov, A., Li, W., Stagg, S. M., Sengupta, J., Nielsen, R. C., Nissen, P., Harvey, S. C., Ehrenberg, M. & Frank, J. (2003) Nat. Struct. Biol. 10, 899-906. [DOI] [PubMed] [Google Scholar]
- 7.Frank, J., Sengupta, J., Gao, H., Li, W., Valle, M., Zavialov, A. & Ehrenberg, M. (2005) FEBS Lett. 579, 959-962. [DOI] [PubMed] [Google Scholar]
- 8.Nierhaus, K. H. (1990) Biochemistry 29, 4997-5008. [DOI] [PubMed] [Google Scholar]
- 9.Gromadski, K. B. & Rodnina, M. V. (2004) Mol. Cell 13, 191-200. [DOI] [PubMed] [Google Scholar]
- 10.Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H. D. & Noller, H. F. (2001) Science 292, 883-896. [DOI] [PubMed] [Google Scholar]
- 11.Vila-Sanjurjo, A., Ridgeway, W. K., Seymaner, V., Zhang, W., Santoso, S., Yu, K. & Cate-Doudna, J. H. (2003) Proc. Natl. Acad. Sci. USA 100, 8682-8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark, H., Rodnina, M. V., RinkeAppel, J., Brimacombe, R., Wintermeyer, W. & vanHeel, M. (1997) Nature 389, 403-406. [DOI] [PubMed] [Google Scholar]
- 13.Frank, J., Heagle, A. B. & Agrawal, R. K. (1999) J. Struct. Biol. 128, 15-18. [DOI] [PubMed] [Google Scholar]
- 14.Ogle, J. M., Brodersen, D. E., Clemons, W. M., Tarry, M. J., Carter, A. P. & Ramakrishnan, V. (2001) Science 292, 897-902. [DOI] [PubMed] [Google Scholar]
- 15.Schmeing, T. M., Seila, A. C., Hansen, J. L., Freeborn, B., Soukup, J. K., Scaringe, S. A., Strobel, S. A., Moore, P. B. & Steitz, T. A. (2002) Nat. Struct. Biol. 9, 225-230. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, J. L., Schmeing, T. M., Moore, P. B. & Steitz, T. A. (2002) Proc. Natl. Acad. Sci. USA 99, 11670-11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashan, A., Agmon, I., Zarivach, R., Schluenzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H. A., et al. (2003) Mol. Cell 11, 91-102. [DOI] [PubMed] [Google Scholar]
- 18.Valle, M., Sengupta, J., Swami, N. K., Grassucci, R. A., Burkhardt, N., Nierhaus, K. H., Agrawal, R. & Frank, J. (2002) EMBO J. 21, 3557-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchard, S. C., Gonzalez, R. L., Kim, H. D., Chu, S. & Puglisi, J. D. (2004) Nat. Struct. Mol. Biol. 11, 1008-1014. [DOI] [PubMed] [Google Scholar]
- 20.Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905-920. [DOI] [PubMed] [Google Scholar]
- 21.Sanbonmatsu, K. Y. & Joseph, S. (2003) J. Mol. Biol. 328, 33-47. [DOI] [PubMed] [Google Scholar]
- 22.Lim, V. I. & Curran, J. F. (2001) RNA 7, 942-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VanLoock, M. S., Agrawal, R. K., Gabashvili, I. S., Qi, L., Frank, J. & Harvey, S. C. (2000) J. Mol. Biol. 304, 507-515. [DOI] [PubMed] [Google Scholar]
- 24.VanLoock, M. S., Easterwood, T. R. & Harvey, S. C. (1999) J. Mol. Biol. 285, 2069-2078. [DOI] [PubMed] [Google Scholar]
- 25.Auffinger, P., LouiseMay, S. & Westhof, E. (1999) Biophys. J. 76, 50-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W., Ma, B. & Shapiro, B. (2003) Nucleic Acids Res. 31, 629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mears, J. A., Cannone, J. J., Stagg, S. M., Gutell, R. R., Agrawal, R. K. & Harvey, S. C. (2002) J. Mol. Biol. 321, 215-234. [DOI] [PubMed] [Google Scholar]
- 28.Trylska, J., Konecny, R., Tama, F., Brooks, C. L., 3rd & McCammon, J. A. (2004) Biopolymers 74, 423-431. [DOI] [PubMed] [Google Scholar]
- 29.Chacon, P., Tama, F. & Wriggers, W. (2003) J. Mol. Biol. 326, 485-492. [DOI] [PubMed] [Google Scholar]
- 30.Tama, F., Valle, M., Frank, J. & Brooks, C. L., 3rd (2003) Proc. Natl. Acad. Sci. USA 100, 9319-9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trylska, J., Tozzini, V. & McCammon, J. A. (2005) Biophys. J. 89, 1455-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao, H., Sengupta, J., Valle, M., Korostelev, A., Eswar, N., Stagg, S. M., Van Roey, P., Agrawal, R. K., Harvey, S. C., Sali, A., et al. (2003) Cell 113, 789-801. [DOI] [PubMed] [Google Scholar]
- 33.Karplus, M. & McCammon, J. A. (2002) Nat. Struct. Biol. 9, 646-652. [DOI] [PubMed] [Google Scholar]
- 34.Berneche, S. & Roux, B. (2001) Nature 414, 73-77. [DOI] [PubMed] [Google Scholar]
- 35.Young, M. A., Gonfloni, S., Superti-Furga, G., Roux, B. & Kuriyan, J. (2001) Cell 105, 115-126. [DOI] [PubMed] [Google Scholar]
- 36.Bockmann, R. A. & Grubmuller, H. (2002) Nat. Struct. Biol. 9, 198-202. [DOI] [PubMed] [Google Scholar]
- 37.McCammon, J. A., Gelin, B. R. & Karplus, M. (1977) Nature 267, 585-590. [DOI] [PubMed] [Google Scholar]
- 38.Tajkhorshid, E., Nollert, P., Jensen, M. O., Miercke, L. J., O'Connell, J., Stroud, R. M. & Schulten, K. (2002) Science 296, 525-530. [DOI] [PubMed] [Google Scholar]
- 39.Sporlein, S., Carstens, H., Satzger, H., Renner, C., Behrendt, R., Moroder, L., Tavan, P., Zinth, W. & Wachtveitl, J. (2002) Proc. Natl. Acad. Sci. USA 99, 7998-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kale, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K. & Schulten, K. (1999) J. Comput. Phys. 151, 283-312. [Google Scholar]
- 41.Nymeyer, H. & Garcia, A. (2003) Proc. Natl. Acad. Sci. USA 100, 13934-13939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tieleman, D. P. (2004) BMC Biochem. 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sotomayor, M., Corey, D. P. & Schulten, K. (2005) Structure (London) 13, 669-682. [DOI] [PubMed] [Google Scholar]
- 44.Ma, J., Sigler, P. B., Xu, Z. & Karplus, M. (2000) J. Mol. Biol. 302, 303-313. [DOI] [PubMed] [Google Scholar]
- 45.Schlitter, J., Engels, M. & Kruger, P. (1994) J. Mol. Graphics 12, 84-89. [DOI] [PubMed] [Google Scholar]
- 46.Tung, C. S. & Sanbonmatsu, K. Y. (2004) Biophys. J. 87, 2714-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Humphrey, W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14, 33-38. [DOI] [PubMed] [Google Scholar]
- 48.Cannone, J. J., Subramanian, S., Schnare, M. N., Collett, J. R., D'Souza, L. M., Du, Y., Feng, B., Lin, N., Madabusi, L. V., Muller, K. M., et al. (2002) BMC Bioinformatics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Connor, M. & Dahlberg, A. E. (1995) J. Mol. Biol. 254, 838-847. [DOI] [PubMed] [Google Scholar]
- 50.O'Connor, M. & Dahlberg, A. E. (1993) Proc. Natl. Acad. Sci. USA 90, 9214-9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saarma, U. & Remme, J. (1992) Nucleic Acids Res. 20, 3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yarus, M., Valle, M. & Frank, J. (2003) RNA 9, 384-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogle, J. M., Murphy, F. V., Tarry, M. J. & Ramakrishnan, V. (2002) Cell 111, 721-732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.