Abstract
Spx, a global transcription regulator in Bacillus subtilis, interacts with the C-terminal domain of the α subunit (αCTD) of RNA polymerase to control gene expression under conditions of disulfide stress, which is sensed by disulfide bond formation between Spx residues C10 and C13. Here, we describe the crystal structure of the B. subtilis αCTD bound to oxidized Spx. Analysis of the complex reveals interactions between three regions of “anti-alpha” Spx and helix α1 and the “261” determinant of αCTD. The former contact could disrupt the interaction between αCTD and activator proteins or alter the DNA-bound conformation of αCTD, thereby repressing activator-stimulated transcription. Binding to the 261 determinant would prevent interaction between αCTD and region 4 of σA. Intriguingly, the Spx disulfide bond is far from the αCTD–Spx interface, suggesting that Spx regulates transcription allosterically or through the redox-dependent creation or destruction of binding sites for additional components of the transcription machinery.
Keywords: ArsC family, global transcription regulation, oxidative/disulfide stress
Bacillus subtilis Spx is a global transcription factor that regulates the transcription of multiple genes in response to disulfide stress. The spx gene was first identified as a second site suppressor locus of clpP and clpX mutations (1). ClpX is an ATPase that acts in concert with the ClpP protease to unfold and degrade targeted proteins. Spx has been shown to be a substrate for this protease, and in wild-type cells under normal growth conditions Spx is nearly undetectable because of its degradation (2). The importance of the removal of Spx by ClpXP from cells during normal growth conditions was suggested by the findings that mutations in the clpP and clpX genes caused growth defects and blocked genetic competence and sporulation. Spx also blocks transcription of the srf operon, which encodes an essential competence regulatory gene (3, 4) and is positively regulated by the ComP–ComA two-component regulatory system (1, 5).
Additional suppressor mutations of the clpX phenotype were mapped to codon substitutions in the rpoA gene, which encodes the α subunit of RNA polymerase (6). These point mutants, termed cxs for ClpX suppressor, corresponded to residues V260 and Y263, which are located in helix α1 of the C-terminal domain of the α subunit of RNA polymerase (αCTD). Helix α1 has been shown to be involved in activator stimulated transcription and RNA polymerase binding to UP elements as well as sequence-nonspecific DNA interactions (7–10). Furthermore, in vitro transcription assays with native and cxs mutant containing RNA polymerase revealed that a direct interaction between Spx and αCTD is required for the repression of the srf operon, and hence, Spx has been termed an “anti-alpha” factor (2).
Microarray analysis revealed that, along with repression of transcription of the srf operon and multiple other genes, transcription of the thioredoxin (trxA) and thioredoxin reductase (trxB) genes is induced when Spx is expressed in wild-type cells but not in cells containing the rpoAcxs-1 mutation, suggesting that Spx acts as a global regulator under conditions of disulfide stress (11). More recently, Zuber and coworkers (12) demonstrated that both trxA and trxB are induced in wild-type cells but not in spx null or rpoAcxs-1 cells upon treatment with diamide, a reagent that induces oxidation of intracellular cysteines. In vitro transcription experiments revealed that Spx-stimulated transcription of trxA or trxB is abolished in the presence of reducing agent, which is consistent with the hypothesis that Spx acts to regulate genes involved in thiol homeostasis in response to disulfide stress conditions.
Spx is a monomeric, 15-kDa protein and belongs to the arsenate reductase (ArsC) family of proteins (13, 14). The crystal structure of Escherichia coli ArsC bound to arsenate has been determined and revealed that the protein contains a four-stranded mixed β-sheet that is surrounded by α-helices (15). Members of the ArsC family contain a conserved cysteine residue that is located in a loop between the first β-strand and the first α-helix and forms a covalent adduct with arsenate. In Gram-positive bacteria, Spx and its homologues contain a conserved CXXC motif, the first cysteine of which aligns with the conserved arsenate-ligating cysteine of ArsC (16). Under normal reducing conditions, these cysteine residues in Spx are in their sulfhydryl form because electrospray–MS experiments with iodoacetamide-treated Spx under reducing and oxidizing conditions confirmed the alkylation of Spx under reducing but not oxidizing conditions (12). Thus, the oxidation state of the Spx cysteines acts as a sensor of disulfide stress.
On the basis of accumulated genetic and biochemical evidence, a major role of Spx is to function as a global transcription regulator to restore the thiol balance of the cell under conditions of disulfide stress. Spx is unusual among bacterial transcriptional regulators in that it does not bind DNA as a part of its transcription regulatory role (12). Rather, at the least, a direct interaction between Spx and αCTD is required for Spx to exert its regulatory effects at certain promoters. To understand the structural mechanism of transcription regulation by Spx via its interaction with the α subunit of RNA polymerase, we determined the x-ray crystal structure of oxidized Spx in complex with αCTD to 1.5 Å resolution. The structure reveals direct interaction between Spx and αCTD residues, which are involved in the DNA, transcription activator, and σ subunit binding functions of this RNA polymerase subunit.
Materials and Methods
Protein Purification and Crystallization. Spx was overexpressed in E. coli as described in ref. 2 and purified by Ni2+-NTA affinity column chromatography (Qiagen). After elution from the Ni2+-NTA resin, Spx was incubated with tobacco-etch virus protease (TEV) to cleave the N-terminal His6-tag. Uncleaved Spx, along with the TEV protease, was removed by repassing the sample over a Ni2+-NTA column. Purified, cleaved Spx was dialyzed into 25 mM Pipes, pH 6.5/100 mM KCl for use in subsequent crystallization trials.
αCTD containing residues 245–314 from RpoA was cloned into the IMPACT self-cleavable intein vector (New England Biolabs) as described in ref. 2. The protein was purified by using the IMPACT self-cleavable affinity purification system (New England Biolabs). Purified αCTD was dialyzed into 25 mM Pipes, pH 6.5/100 mM KCl. The Spx–αCTD complex was formed by mixing equimolar amounts of purified Spx protein and αCTD. The complex was concentrated to 12 mg/ml.
Selenomethionine-substituted Spx and αCTD were expressed by inhibiting the methionine biosynthetic pathway and supplementing with selenomethionine (17). Both proteins were purified as described above with the addition of 10 mM 2-mercaptoethanol to all purification buffers.
Crystals of both native and selenomethionine-substituted oxidized Spx–αCTD were grown by the hanging-drop vapor diffusion method. Two microliters of 0.5 mM Spx–αCTD was mixed with 2 μl of reservoir solution containing 1.6–2.0 M Li2SO4, 0.1 M sodium citrate (pH 5.6), and 0.1 M ammonium sulfate. Crystals appeared within 2–3 days and reached maximum dimensions of 0.2 × 0.05 × 0.05 mm3 in ≈1 week. The crystallization solution was suitable as a cryoprotectant and crystals were flash-frozen in a nitrogen stream at -180°C. X-ray intensity data for native crystals were collected on beamline 8.2.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA), and data were processed by using mosflm (18) as implemented in the ccp4 suite of programs (19) (Table 1). For crystals of the selenomethionine-substituted protein, x-ray intensity data were collected at beamline 4.2.2 at the Advanced Light Source, and data were processed and scaled by using d*trek (20) (Table 1). Native crystals took space group R3 with cell dimensions a = b = 96.4 Å, c = 57.1 Å, α = β = 90°, and γ = 120°. The selenomethionine-substituted crystals also took space group R3 with cell dimensions a = b = 97.9 Å, c = 57.4 Å, α = β = 90°, and γ = 120°.
Table 1. Selected crystallographic data and statistics.
| Native | SeMet (λ1) | SeMet (λ2) | SeMet (λ3) | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength, Å | 1.0003 | 0.9794 | 0.9796 | 0.9952 |
| Resolution, Å | 33.0–1.5 | 15.0–2.3 | 15.0–2.3 | 15.0–2.3 |
| No. of observed reflections | 275,058 | 41,223 | 41,163 | 40,465 |
| No. of unique reflections | 31,461 | 17,789 | 17,777 | 17,567 |
| Completeness, % (last shell) | 99.2 (98.5) | 97.6 (83.0) | 97.5 (82.4) | 96.2 (76.3) |
| I/σI (last shell) | 12.5 (2.5) | 9.6 (2.8) | 10.8 (3.5) | 15.3 (4.3) |
| Rsym,* % (last shell) | 4.2 (31.1) | 6.2 (28.5) | 5.5 (24.4) | 3.8 (18.2) |
| MAD phasing | ||||
| Resolution, Å | 15.0–2.5 | |||
| Selenium sites | 6 | |||
| Figure of merit† | 0.56 | |||
| Refinement | ||||
| Resolution, Å | 20.0–1.5 | |||
| Reflection, working set/test set | 28,327/3,108 | |||
| Protein atoms | 1,512 | |||
| Solvent molecules | 143 | |||
| Rcryst/Rfree,‡ % | 19.7/22.1 | |||
| rmsd bond lengths, Å | 0.006 | |||
| rmsd bond angles, ° | 1.28 | |||
| Average B-factor, Å2 | 17.3 |
rmsd, rms deviation.
Rsym = ΣΣ|Ihkl–Ihkl(j)|/ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity
Figure of merit = 〈|ΣP(α)eiα/ΣP(α)|〉, where α is the phase and P(α) is the phase probability distribution
Rwork = Σ|Fobs|–|Fcalc|/Σ|Fobs| and Rfree = Σ|Fobs|–|Fcalc|/Σ|Fobs|, where all reflections belong to a test set of 10% randomly selected data
Structure Determination, Model Building, and Refinement. The structure of the selenomethionine-substituted Spx–αCTD complex was solved by multiple-wavelength anomalous diffraction methods (MAD), using data collected at three wavelengths (Table 1). Using data from 15.0–2.5 Å resolution, all six selenium sites were located and initial phases were calculated with solve (21, 22). Phases were improved by density modification, and a partial structure was built automatically by using resolve (22, 23). Manual fitting of the remaining residues resulted in the placement of amino acid residues 1–118 of Spx and residues 245–311 of αCTD into the electron density map by using o (24), with the positions of the selenomethionine residues as reference points. The initial protein model was subjected to rigid body refinement, followed by simulated annealing and positional and B-factor refinement in cns (25). After the addition of several water molecules, the Rwork and Rfree were 25.4% and 29.6%, respectively, and the model was further refined against native intensity data to 1.50 Å resolution. Further rounds of model building and solvent addition followed by positional and B-factor refinement resulted in a final model that contained residues 1–118 of Spx and residues 245–311 of αCTD, 143 water molecules, and 4 sulfate molecules. The final Rwork was 19.7%, and the final Rfree was 22.1%. The stereochemistry of the final model was assessed with procheck (26), which revealed 95.2% of all φ/Ψ angles in the most favored regions of the Ramachandran plot and none in the generously allowed or disallowed regions.
Results and Discussion
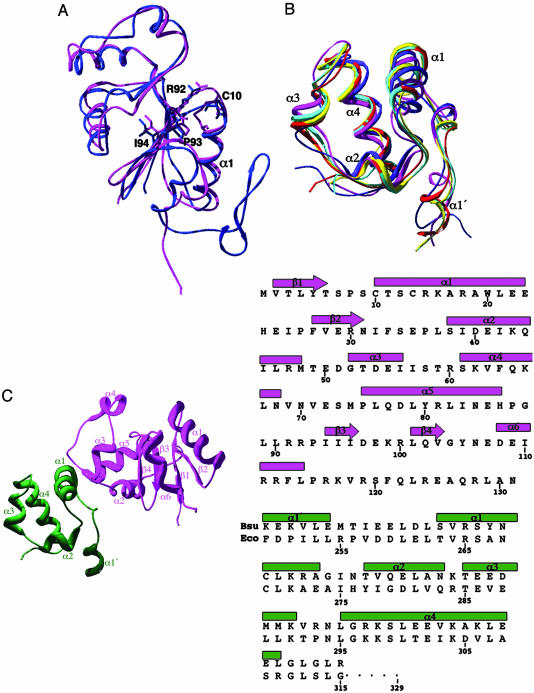
Structure of Spx and αCTD. The global structure of Spx consists of a mixed α/β topology with a central four-stranded β-sheet, which is flanked by two 310-helices and four α-helices. The last 13 residues (119–131) are missing from the structure because of structural disorder. The C-terminal tail is necessary for recognition by the ClpXP protease, and its unstructured nature is not surprising (11). The topology of the Spx structure is β1-α1-β2-α2-α3(310)-α4-α5-β3-β4-α6(310) (Fig. 1). As predicted, the overall fold of Spx is structurally homologous to that of the E. coli ArsC with a root mean square deviation (rmsd) of 2.37 Å over residues 2–114 (Fig. 1 A). ArsC is 9 residues longer with the last 20 residues forming an additional two-stranded β-sheet.
Fig. 1.
The structures of the anti-alpha protein Spx and αCTD, and their complex. (A) An overlay of Spx and structural homologue ArsC [Protein Data Bank (PDB) entry 1JZW]. Spx is shown in pink, and ArsC is blue. The conserved CXXC motif is shown, as is the structurally conserved cysteine of ArsC. The residues of the RPI motif (R92, P93, and I94) are shown as sticks for both Spx and ArsC. (B) Overlay of currently publishedαCTD structures. B. subtilisαCTD is colored red, E. coliαCTD bound to CAP–DNA is colored cyan, and E. coli αCTD bound to UP element DNA is colored yellow (PDB entry 1LB2). The NMR structures of E. coli (PDB entry 1COO) and T. thermophilus αCTD (PDB entry 1DOQ) are colored pink and blue, respectively. (C) The structure of the Spx–αCTD complex. Spx is shown in pink, andαCTD is shown in green. The helices of Spx labeled α3 and α6 are 310-helices. (Right) Alignment of the sequences and secondary structure elements of Spx (pink) and αCTD (green).
The CTD of the B. subtilis (Bsu) RNA polymerase α subunit is composed of residues 246–311 and forms an independently folded domain of five loosely packed α-helices (Fig. 1 B and C). The equivalent residues of E. coli αCTD are numbered from 250 to 315, and for clarity the E. coli numbering will be provided in parentheses throughout the text (Fig. 1C Right). This is the highest resolution structure of an αCTD determined to date, as well as the first structure of an αCTD from a Gram-positive bacterium. As expected, the Bsu αCTD is structurally homologous to the previously determined x-ray (27) and NMR structures (28, 29) of other αCTDs with an rmsd over all Cα atoms of 0.90–1.98 Å (Fig. 1B). It is of interest to note here that solvent-accessible residue Y263 (267) of the Bsu αCTD, which is necessary for the direct interaction of Spx and αCTD, is strictly conserved in all low G+C Gram-positive species but not in Gram-negative organisms where the corresponding residue is often an alanine (14).
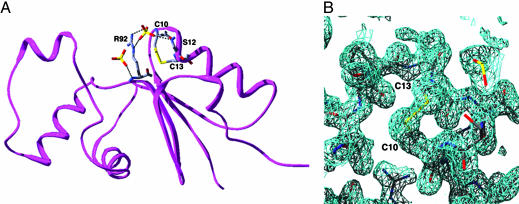
As shown in previous genetic and biochemical studies (12), the conserved cysteines at positions 10 and 13 of oxidized Spx form a disulfide bond, which is located within the first turn of α1 (Fig. 2 A and B). Interestingly, three Spx residues, R92, P93, and I94 (the RPI motif, Fig. 1 A), are conserved in all Spx homologues and in ArsC (14). The RPI motifs of Spx and ArsC are located at the N terminus of β3 and are proximal to the reactive cysteines of each protein (Figs. 1 A and 2 A). In both the Spx and ArsC structures, residue P93 takes the less favorable cis configuration. The guanidinium side chain of Spx residue R92 is 3.6 Å from the sulfur atom of residue C10 and 5.0 Å from the sulfur atom of C13. The corresponding arginine in ArsC is hypothesized to lower the pKa of the active site cysteine, which is equivalent to Spx residue C10, that ligates the arsenate ion (15). A similar role for R92 of Spx is envisioned in that by lowering the pKa of residue C10 or C13 under normal aerobic conditions the nucleophilic thiolate would form more readily at physiological pH and hence be an effective oxidative stress sensor. Intriguingly, in both the ArsC and Spx crystal structures, there is a sulfate molecule originating from the crystallization solution that interacts with this arginine (Fig. 2 A). The possibility that this sulfate-binding site has biological relevance has been bolstered by recent findings of Zuber and coworkers (30) that reveal that when cells are grown with sulfate as the sole sulfur source, Spx regulates transcription of sulfur assimilation genes. Perhaps this conserved RPI motif is involved in both modulating the reactivity of C10 or C13 and binding sulfate in vivo.
Fig. 2.
Disulfide bridge and sulfate binding sites of oxidized Spx. (A) The disulfide bridge is shown as yellow sticks. The sulfate, which is found in both Spx and ArsC, and may be involved in transcription regulation by Spx, is bound by the guanidinium moiety of residue R92 and the main chain carbonyl oxygen of S12. A second sulfate ion, which is found only in the Spx–αCTD structure, is shown to the left of R92. Interactions between the sulfates and protein are depicted by dashed line. (B) A representative 2Fo - Fc simulated annealing composite omit map contoured at 1σ shows the electron density for the disulfide bridge of Spx.
Spx–αCTD Complex. In B. subtilis the regulation of transcription by Spx under conditions of disulfide stress requires a direct interaction between Spx and αCTD (11). Surprisingly, αCTD interacts with Spx on a face that is distal from the disulfide bridge (the closest distance is 26 Å), whereby α1 of αCTD and its preceding loop pack against the N terminus of α2 and α5 and a loop region between α2 and α3 of Spx (Fig. 1C). The interface between the two proteins is relatively small and polar and buries 1,311 Å2 of surface area per heterodimer out of a total accessible surface area of 10,564 Å2. Residues N264 (268), K267 (271), and R268 (272) from α1 of αCTD hydrogen bond with residues T53(NH and γ-OH), R47(CO), and D54(COO-) of Spx, respectively, which are located on α2 and α3 (Fig. 3A). Interestingly, these Bsu αCTD residues overlap with a region of the E. coli αCTD identified as the interaction surface of several positively regulated transcription factors (31–34). Residues Q77 and Y80, which are located within the N terminus of α5 of Spx, are engaged in hydrogen bonds to αCTD residues E254(E258) (CO) and L256(L260) (CO), which are located in the loop region N-terminal to α1 (Fig. 3B). In addition, the polypeptide backbone carbonyl oxygen and amide nitrogen of residues R261(R265) (NH) and L258(L262) (CO) of αCTD make hydrogen bonds to the polypeptide backbone amide nitrogen and carbonyl oxygens of residues E72(CO) and L76(NH) of Spx, respectively.
Fig. 3.
The Spx–αCTD heterodimer interface. (A and B) Hydrogen bonding interactions at the dimer interface. The backbones of Spx and αCTD are colored pink and green, respectively. Interacting residues are depicted as sticks and are colored according to atom type. Hydrogen bonds are depicted as dashes. (C) Interactions of cxs mutants of αCTD (V260 and Y263) and Spx (G52). These residues cluster and are shown as blue sticks.
In previous work, Zuber and coworkers (1, 2, 6) showed that substitution of αCTD residues V260(V264) with alanine or Y263(A267) with cysteine or residue G52 of Spx with arginine resulted in loss of transcription activation or repression by Spx and that this loss of function was due to disruption of the direct interaction between Spx and αCTD. The structure of the Spx–αCTD complex provides the atomic basis for these observations. Residues V260 and Y263 are found within the N-terminal portion of α1, which forms part of the interface with Spx (Fig. 3C). The side chain of V260(V264) resides within a strictly hydrophobic environment and makes multiple van der Waals contacts with Spx residues T53, E72, M74, and L79. The phenolic moiety of Y263(A267) sits just above V260(V264) in the pocket and is in van der Waals contact with residues L46 and G52 of Spx. The side chains of Y263(A267) and N264(N268) lie within 3.8 Å and 3.6 Å of G52, respectively, thereby restricting the identity of position 52 to glycine to maintain the tightly packed dimer interface because substitution of G52 by an arginine would interfere sterically with the formation of the complex.
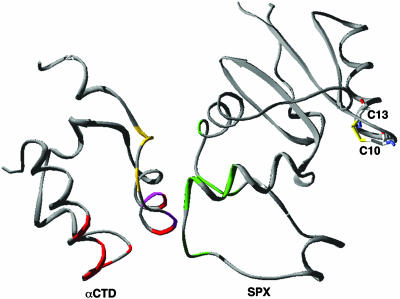
Possible Mechanisms for Transcription Regulation by Spx. Spx has been termed an “anti-alpha” factor because this protein is involved in disrupting activator-stimulated transcription at some promoters (14). Activator-stimulated transcription at both class I and class II promoters involves a direct interaction between the activator protein and αCTD, which acts to recruit RNA polymerase to the promoter (35, 36). Several determinants on αCTD have been implicated in both activator and UP element-dependent transcription activation in E. coli (7, 9). In addition, the crystal structure of the αCTD–CAP–DNA complex has elucidated further the molecular details of the interaction of αCTD with DNA at class I and UP element promoter sites (27). α–DNA interactions in B. subtilis, as in E. coli, have been shown to enhance transcription both in the presence and absence of activators (37, 38). αCTD interacts with the DNA minor groove at A/T-rich sequences, and several residues from α1, α3, and the turn following α3 make DNA backbone contacts. Bsu residues R261 and N264, the equivalent E. coli residues of which (R265 and N268) make direct contacts to DNA in the αCTD–DNA and αCTD–CAP–DNA structures, are involved in interactions with Spx in the Spx–αCTD complex (Fig. 3 A and B). However, residues R268, G292, K294, and S295, the equivalent E. coli residues of which (R272, G296, K298, and S299) are all involved in binding DNA in the αCTD–CAP–DNA structure, are still available to interact with DNA in the αCTD–Spx complex. Moreover, residue N264, although engaged in hydrogen bonds to Spx, could still make a hydrogen bond with the DNA backbone. Thus, Spx binding to αCTD does not preclude DNA binding by this RNA polymerase subunit. Indeed, electrophoretic mobility shift assays with an idealized UP-element promoter sequence show that Spx does not disrupt the αCTD–DNA interaction but rather appears to increase the affinity of αCTD for this DNA sequence (see Fig. 5, which is published as supporting information on the PNAS web site). As described, Spx also interacts with the “261” determinant of αCTD, which encompasses residues E254 (258), E255 (259), and D257 (261). In E. coli the 261 determinant is important for interaction between αCTD and region 4 of σ70 (9) (Fig. 4). Recent mutational studies have shown that disruption of αCTD–σ4 interactions abrogates both UP element- and class I activator-stimulated transcription (39, 40). Thus, Spx might repress activator-stimulated transcription at some promoters by using one or more mechanisms including disruption of αCTD interactions with an activator, region 4 of the RNA polymerase σA subunit, or both.
Fig. 4.
Ribbon diagram of αCTD and Spx, which depicts the determinants of αCTD that are important for transcription regulation and DNA binding. αCTD residues involved in DNA binding are colored red [V264, N294, G296, K298, S299, and E302 (E. coli numbering)], and residues involved in interaction with σ4 are shown in yellow (E258, E259, and D261). The residues of Spx that interact with αCTD are shown in green (R47, G52, T53, D54, E72, L76, Q77, and Y80), and the cxs mutants of αCTD are shown in pink (V264, Y267). Residues C10 and C13 and the disulfide bridge found between them are shown as sticks.
The mechanism(s) by which Spx acts to repress activator-stimulated transcription is readily envisioned, but the present crystal structure does not present a clear understanding of how the interaction between oxidized Spx, i.e., the disulfide bond form, and αCTD could act to enhance transcription from other promoters (12). Zuber and colleagues (14) have shown both biochemically and genetically that under oxidizing conditions, a direct interaction of Spx with αCTD is necessary for activation of several genes, including trxA and trxB, and repression of transcription of others. Footprinting experiments of the trxA and trxB promoters revealed that RNA polymerase alone was able to protect a potential UP element positioned near nucleotide -60 of these promoters but was not sufficient for activation of transcription. Upon addition of Spx the footprint extended downstream to the -30 and -10 promoter regions (12). These data suggest that in some promoter-specific instances, Spx acts in concert with the RNA polymerase holoenzyme to mediate formation of an active promoter complex. Perhaps Spx forms an additional interaction with other subunits particularly the sigma subunit or the αNTD to position RNA polymerase optimally for transcription from these promoters. Indeed, Zuber and coworkers (personal communication) have recently shown that mutations in region 4.2 of σA affect transcription regulation by Spx.
The structure of the oxidized Spx–αCTD complex provides the first view of an “anti-alpha” factor and insight into several aspects of the multiple transcription regulation functions of this protein. A key question that remains is how does oxidation of residues C10 and C13 and their subsequent formation of a disulfide bond act to modulate the activity of Spx as a transcriptional regulator, especially given the distal location of the disulfide bond from the αCTD–Spx heterodimerization interface (Fig. 4). One possibility is that formation of the disulfide bridge causes a conformational change that diverts Spx from the ClpXP proteolytic machinery, hence invoking a default functional state. This scenario is unlikely because Western blot analysis of cells expressing a C10A/C13A double mutant of Spx, which is incapable of sensing disulfide stress, has shown no differences in protein levels as compared with those of wild-type cells (P.Z., unpublished results). Alternatively, a conformational change, which is associated with reduction, might be transmitted through the Spx structure to disrupt or alter the Spx–αCTD interaction. However, initial isothermal titration calorimetry experiments suggest no differences in the affinity of Spx for αCTD in the presence or absence of reducing agent (K.J.N., unpublished results). Perhaps an interaction between Spx and the σ4A region is needed for its transcriptional regulation under oxidizing conditions; or alternatively, reduced Spx may be recognized and sequestered by another cellular factor such as MecA, which has been shown to interact with Spx (41). Additional biochemical, genetic, and structural studies are needed to explore this and other possibilities to understand fully the mechanisms of transcription regulation by Spx.
Supplementary Material
Acknowledgments
We thank Dr. Cori Ralston for her help with data collection at beamline 8.2.1 and Drs. Ed Westbrook and Jay Nix for their help with multiple-wavelength anomalous diffraction data collection at beamline 4.2.2. Portions of this research were carried out at the Macromolecular Crystallography Facility at the Advanced Light Source. The Advanced Light Source is supported by the Director, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under Contract DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. This work was supported by National Institutes of Health Grant GM045898 (to P.Z.). R.G.B. is the Richard T. Jones Professor of Structural Biology. K.J.N. is supported by a postdoctoral fellowship from the American Heart Association, Pacific Mountain Affiliate.
Author contributions: K.J.N. and R.G.B. designed research; K.J.N. performed research; S.N. and P.Z. contributed new reagents/analytic tools; K.J.N. and R.G.B. analyzed data; and K.J.N., P.Z., and R.G.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The structure coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1Z3E).
References
- 1.Nakano, M. M., Hajarizadeh, F., Zhu, Y. & Zuber, P. (2001) Mol. Microbiol. 42, 383-394. [DOI] [PubMed] [Google Scholar]
- 2.Nakano, S., Nakano, M. M., Zhang, Y., Leelakriangsak, M. & Zuber, P. (2003) Proc. Natl. Acad. Sci. USA 100, 4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Souza, C., Nakano, M. M. & Zuber, P. (1994) Proc. Natl. Acad. Sci. USA 91, 9397-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamoen, L. W., Eshuis, H., Jongbloed, J., Venema, G. & van Sinderen, D. (1995) Mol. Microbiol. 15, 55-63. [DOI] [PubMed] [Google Scholar]
- 5.Nakano, S., Zheng, G., Nakano, M. M. & Zuber, P. (2002) J. Bacteriol. 184, 3664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano, M. M., Zhu, Y., Liu, J., Reyes, D. Y., Yoshikawa, H. & Zuber, P. (2000) Mol. Microbiol. 37, 869-884. [DOI] [PubMed] [Google Scholar]
- 7.Gourse, R. L., Ross, W. & Gaal, T. (2000) Mol. Microbiol. 37, 687-695. [DOI] [PubMed] [Google Scholar]
- 8.Busby, S. & Ebright, R. H. (1999) J. Mol. Biol. 293, 199-213. [DOI] [PubMed] [Google Scholar]
- 9.Savery, N. J., Lloyd, G. S., Busby, S. J., Thomas, M. S., Ebright, R. H. & Gourse, R. L. (2002) J. Bacteriol. 184, 2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross, W. & Gourse, R. L. (2005) Proc. Natl. Acad. Sci. USA 102, 291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano, S., Küster-Schock, E., Grossman, A. D. & Zuber, P. (2003) Proc. Natl. Acad. Sci. USA 100, 13603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakano, S., Erwin, K. N., Ralle, M. & Zuber, P. (2005) Mol. Microbiol. 55, 498-510. [DOI] [PubMed] [Google Scholar]
- 13.Gladysheva, T. B., Oden, K. L. & Rosen, B. P. (1994) Biochemistry 33, 7288-7293. [DOI] [PubMed] [Google Scholar]
- 14.Zuber, P. (2004) J. Bacteriol. 186, 1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, P., DeMel, S., Shi, J., Gladysheva, T., Gatti, D. L., Rosen, B. P. & Edwards, B. F. (2001) Structure (Cambridge, MA) 9, 1071-1081. [DOI] [PubMed] [Google Scholar]
- 16.Zuber, P. (2002) Front. Biosci. 7, d1857-d1866. [DOI] [PubMed] [Google Scholar]
- 17.Doublié, S. (1997) Methods Enzymol. 276, 523-530. [PubMed] [Google Scholar]
- 18.Leslie, A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography.
- 19.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 20.Pflugrath, J. W. (1999) Acta Crystallogr. D 55, 1718-1725. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terwilliger, T. (2004) J. Synchrotron Radiat. 11, 49-52. [DOI] [PubMed] [Google Scholar]
- 23.Terwilliger, T. C. (1999) Acta Crystallogr. D 55, 1863-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 25.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta. Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski, R. A., Moss, D. S. & Thornton, J. M. (1993) J. Mol. Biol. 231, 1049-1067. [DOI] [PubMed] [Google Scholar]
- 27.Benoff, B., Yang, H., Lawson, C. L., Parkinson, G., Liu, J., Blatter, E., Ebright, Y. W., Berman, H. M. & Ebright, R. H. (2002) Science 297, 1562-1566. [DOI] [PubMed] [Google Scholar]
- 28.Wada, T., Yamazaki, T. & Kyogoku, Y. (2000) J. Biol. Chem. 275, 16057-16063. [DOI] [PubMed] [Google Scholar]
- 29.Ebright, R. H. & Busby, S. (1995) Curr. Opin. Genet. Dev. 5, 197-203. [DOI] [PubMed] [Google Scholar]
- 30.Erwin, K. N., Nakano, S. & Zuber, P. (2005) J. Bacteriol. 187, 4042-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giffard, P. M. & Booth, I. R. (1988) Mol. Gen. Genet. 214, 148-152. [DOI] [PubMed] [Google Scholar]
- 32.Grainger, D. C., Belyaeva, T. A., Lee, D. J., Hyde, E. I. & Busby, S. J. (2004) Mol. Microbiol. 51, 1311-1320. [DOI] [PubMed] [Google Scholar]
- 33.Aiyar, S. E., McLeod, S. M., Ross, W., Hirvonen, C. A., Thomas, M. S., Johnson, R. C. & Gourse, R. L. (2002) J. Mol. Biol. 316, 501-516. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, M. S. & Glass, R. E. (1991) Mol. Microbiol. 5, 2719-2725. [DOI] [PubMed] [Google Scholar]
- 35.Busby, S. & Ebright, R. H. (1994) Cell 79, 743-746. [DOI] [PubMed] [Google Scholar]
- 36.Browning, D. F. & Busby, S. J. (2004) Nat. Rev. Microbiol. 2, 57-65. [DOI] [PubMed] [Google Scholar]
- 37.Meijer, W. J. & Salas, M. (2004) Nucleic Acids Res. 32, 1166-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fredrick, K., Caramori, T., Chen, Y. F., Galizzi, A. & Helmann, J. D. (1995) Proc. Natl. Acad. Sci. USA 92, 2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, H., Tang, H. & Ebright, R. H. (2003) Mol. Cell 11, 1621-1633. [DOI] [PubMed] [Google Scholar]
- 40.Ross, W., Schneider, D. A., Paul, B. J., Mertens, A. & Gourse, R. L. (2003) Genes Dev. 17, 1293-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano, M. M., Nakano, S. & Zuber, P. (2002) Mol. Microbiol. 44, 1341-1349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.