Abstract
The adenoviral E1A oncogene sensitizes mammalian cells to tumor necrosis factor-α (TNF-α), in part by repressing the nuclear factor-κ B (NF-κB)-dependent defense against this cytokine. Other E1A activities involve binding to either p300/cyclic AMP response element-binding protein (CBP) or retinoblastoma (Rb)-family proteins, but the roles of E1A interactions with these transcriptional regulators in sensitizing cells to TNF-α are unclear. E1A expression did not block upstream events in TNF-α-induced activation of NF-κB in NIH 3T3 cells, including degradation of IκB-α, nuclear translocation of NF-κB subunits, and their dimeric binding to κB sequences in the nucleus. However, E1A markedly repressed NF-κB-dependent transcription and sensitized cells to TNF-αinduced apoptosis. These E1A effects were selective for κB-dependent transcription and for the function of the NF-κB p65/RelA subunit. A four amino acid E1A deletion that eliminates binding to Rb-family proteins blocked both repression of TNF-α-induced transcription and sensitization to apoptosis. In contrast, mutations that eliminate E1A binding to p300/CBP (coactivators of p65/RelA) did not affect either E1A activity. These data suggest that E1A-Rb-binding blocks the NF-κB-dependent activation response to TNF-α by altering the function of p65/RelA at a stage after formation of the transcription factor-enhancer complex. These observations also open questions about the general role of Rb-family proteins in modulation of NF-κB-dependent transcription.
One E1A oncogene effect on mammalian cells is to sensitize them to immunological injuries by cytolytic lymphocytes and activated macrophages—key components of the innate cellular immune response (1–11). E1A-induced cytolytic-susceptibility correlates well with tumor-cell rejection by immunocompetent animals (12–17). The mechanisms by which E1A converts cells to a cytolytic-susceptible, nontumorigenic phenotype are independent of expression of the p53 tumor-suppressor gene (18) but are otherwise unknown. Most E1A-induced cellular phenotypes are mediated by transcription modulation through binding between first exon-encoded regions of E1A and two families of transcriptional regulatory proteins, p300/cyclic AMP response element-binding protein (CBP) and retinoblastoma (Rb). E1A-induced susceptibility to lysis by natural killer cells requires the combined effect of p300-binding domains in the E1A first exon plus an undefined second exon activity, but does not require Rb binding (19). In contrast, the mechanisms through which E1A sensitizes cells to killing by macrophage-produced tumor necrosis factor-α (TNF-α) are not well understood. There have been conflicting reports about whether E1A domains required for binding to p300/CBP, Rb, or both are required for this activity (5, 20–22).
One cellular defense that protects against TNF-α-induced apoptotic death is the nuclear factor-κB (NF-κB) activation response that is mediated primarily by the p65/RelA NF-κB subunit (23–25). TNF-α triggers activation of NF-κB inhibitor (IκB) kinase (IKK) and consequent turnover of cytoplasmic IκB, freeing NF-κB subunits to travel to the nucleus, where they dimerize and bind to κB enhancers and stimulate transcription (26–28). NF-κB coactivation by p300/CBP and binding to core promoter components are important for optimal transcription responses (29–35). Despite many activities ascribed to Rb-family proteins, no direct role has been reported for Rb in enhancement of NF-κB activation. Conversely, Rb can directly stimulate binding of a transcriptionally inactive form of NF-κB p50 homodimer (36).
There are few reports on E1A repression of NF-κB responses. Janaswami et al. (37) reported that E1A blocks TNF-α-induced, NF-κB activation by altering the quality of NF-κB dimers in the nucleus. Shao et al. (22) reported that E1A blocks a proximal step in the NF-κB activation pathway by repressing IKK activation and IκB turnover, thereby preventing NF-κB trafficking to the nucleus. Neither study defined a specific E1A interaction with p300/CBP or Rb in the mechanism of transcription repression.
We report that the Rb-binding domain of E1A represses TNF-α-induced NF-κB activation at a stage after formation of transcription factor–enhancer complexes in the nucleus. The results suggest the existence of cellular control mechanisms through which E1A-Rb interactions repress the function of NF-κB p65/RelA in response to cytokine stimulation and block the NF-κB-dependent cellular defense against apoptosis.
Materials and Methods
Cell Lines and Plasmids.
NIH 3T3 mouse and U20S human cells were obtained from American Type Culture Collection. Cells were maintained in DMEM with antibiotics and 5–10% calf serum. κB-luciferase (κB-LUC) reporters (κB from IκB, HIV LTR, and H-2Kb), cytomegalovirus (CMV) expression plasmids for p65/RelA, p50/KBF1, c-Rel, β-galactosidase, and an empty CMV construct were provided by Robert Scheinman (Univ. of Colorado Health Sciences Center, Denver) (38). The dual luciferase reporter system (Promega) was used to normalize transfection efficiency when different cell types were compared. E2F-luciferase reporter was purchased from CLONTECH. The following CMV-promoted E1A constructs were obtained: E1A 12Swt (nonmutant E1A), 12S.RG2 (Arg-to-Gly change in the second E1A amino acid) (provided by Betty Moran, Temple Univ. School of Medicine, Philadelphia; ref. 39); E1A 12S/dl1101 (amino acids 4–25 deleted), 12S/dl1104 (amino acids 48–60 deleted), 12S/dl1106 (amino acids 90–105 deleted), and 12S/dl1108 (amino acids 124–127 deleted) (provided by Stanley Bayley, McMaster Univ., Hamilton, Ontario, Canada; ref. 40). E1A protein expression in transfected cells was compared by Western analysis (41). Our preliminary studies and the data presented here showed that CMV-promoted E1A gene expression in NIH 3T3 and U2OS cells occurred in a gene dose-dependent manner, despite high level (>95%) repression of cellular NF-κB activity and, therefore, is relatively NF-κB-independent in these cells.
NF-κB Pathway Activation.
Recombinant mouse TNF-α (R&D Systems) was used at 20 ng/ml. IκBα turnover was assessed by Western blotting by using IκB antibody (Santa Cruz Biotechnology). DNA probes used in electrophoretic mobility-shift assays (EMSA) were 5′-ATCGCTGGGGATTCCCCA-3′ (H2TF1 κB; 38); 5′-GCCATTGGGGATTTCCTCT-3′ (ELAM κB; 42); and 5′-ATCGGGGGATTTCCT (mutant ELAM κB). Probes were labeled with [γ32P]ATP by using T4 polynucleotide kinase (Invitrogen). Cells treated with TNF-α for 30 min at 37°C were lysed (20 mM Hepes, pH 7.9/300 mM sucrose/10 mM KCl/1.5 mM MgCl2), nuclear pellets were washed repeatedly and lysed [20 mM Hepes, pH 7.9/300 mM sucrose/420 mM NaCl2/1.5 mM MgCl2/0.5% Nonidet P-40 containing 3 μl PMSF (0.333 M), 6 μl aprotinin (1.9 mg/ml), 2 μl leupeptin (5 mg/ml), 1 μl DTT (1.0 M), and 50 μl Nonidet P-40 (10%) per ml] and nuclear lysates were clarified and stored at −70°C. For EMSAs, 10 μg of nuclear protein was incubated with 1 μg of poly(dI-dC) (Sigma) and 10,000 cpm of labeled probe for 30 min at room temperature. Anti-p65/RelA or -p50/KBF1 antibody (Santa Cruz Biotechnology) supershifts and competition with unlabeled probes (500-fold excess) were tested by additions for 30 min at room temperature before adding labeled probe. Free and protein-bound oligonucleotides were resolved on 4% nondenaturing polyacrylamide gels and detected by fluorography.
NF-κB-Dependent Transcription.
Cells were cotransfected with 50 ng of κB-LUC reporter and E1A or NF-κB expression plasmids in triplicate under endotoxin-free conditions by using Lipofectamine Plus and Opti-Mem medium (Invitrogen). After 24 h, cells were split into duplicate samples. After 48 h, one sample was treated with TNF-α (4 h at 37°C), lysed (reporter buffer, Promega), clarified and assayed for luciferase activity (Monolight 3010; PharMingen), and normalized to protein concentration by using bicinchoninic acid (BCA; Pierce) or the dual-luciferase system (Promega) for comparisons of different cell lines. Data for test samples are reported as the percentage of activity in sham-transfected cells (reporter only). CMV promoter concentration was kept constant for all comparisons by using empty CMV vector to control for squelching. NF-κB signal specificity was confirmed by using a mutant κB-LUC reporter that showed no TNF-α-induced activity.
Apoptosis Assays.
Duplicate 60-mm dishes of cells were transfected with an empty vector (sham), E1A or p65/RelA plasmids plus pCMV-β-gal. A 3:1 E1A:β-gal ratio was used to favor E1A expression in all β-gal-marked cells. After 48 h at 37°C, cells were treated with TNF-α for 18 h, fixed [16% (wt/vol) paraformaldehyde], and stained overnight with X-gal [1 mg/ml in 4 mM K4Fe(CN)6 and 4 mM K3Fe(CN)6]. Surviving cells were counted and averaged for duplicate cultures by using a microscopic grid in five different 20× fields. Apoptotic death of TNF-treated, E1A-positive cells was >95% as reported (9, 18), which was indicated by characteristic nuclear morphology of dying cells (43) and confirmed by quantitating nuclear release of low-molecular weight DNA (9).
The significance of differences in transcription and apoptosis assays was estimated by using Student's t test.
Results
E1A Repression of the TNF-Induced, NF-κB Activation Response.
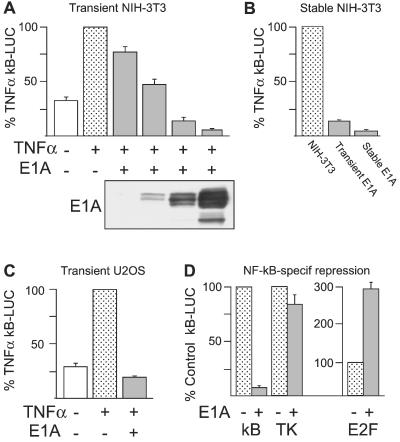
E1A repressed the TNF-α-induced NF-κB activation response in transiently transfected NIH 3T3 mouse fibroblasts in an oncoprotein expression-dependent manner (Fig. 1A; P < 0.05 for all levels of E1A expression). Comparable E1A-induced repression of NF-κB activation was observed with NIH 3T3 cells stably transfected with E1A contrasted with transiently transfected cells (Fig. 1B) and with transiently transfected human, U2OS osteosarcoma cells (Fig. 1C). In contrast, high-level E1A expression in NIH 3T3 did not repress a thymidine kinase reporter and enhanced expression of an E2F reporter (Fig. 1D). Therefore, E1A repression of NF-κB activation was detected in both mouse and human cells and did not result from a general repression of transcription.
Figure 1.
E1A repression of TNF-α-induced κB-LUC activity in mouse and human cells. (A) NIH 3T3 cells transfected with empty vector alone (sham transfection; white and stippled bars) or plus increasing doses of E1A 12S plasmid (gray bars; n = 4). (B) Comparison of NIH 3T3 cells transiently transfected with the second highest E1A 12S plasmid dose shown in A or stably expressing E1A 12S protein vs. sham-transfected cells (n = 3). (C) Effect of transient transfection of E1A 12S plasmid on κB-LUC response of human U2OS cells (n = 4). (D) Comparison of E1A effect on TNF-α-induced κB-LUC activity vs. E1A effect on TK-LUC and E2F-LUC activity in transiently transfected NIH 3T3 cells (n = 3). κB-LUC activity in all assays is presented as the percentage (mean ± SEM of n experiments) of the TNF-treated, sham-transfected control (stippled bars). TK-LUC and E2F-LUC activity in E1A-transfected cells in D is presented as the percentage activity of the E1A-negative control. κB-LUC activity was significantly repressed in all E1A-positive samples (P < 0.05). In D, the E1A effect on TK-LUC activity is not significant, and E1A significantly enhanced E2F-LUC activity (P < 0.05). Comparative E1A protein expressions by cells from sister cultures from a representative E1A transfection in A were tested by Western analysis. E1A expression was seen at the lowest transfection level upon prolonged exposure.
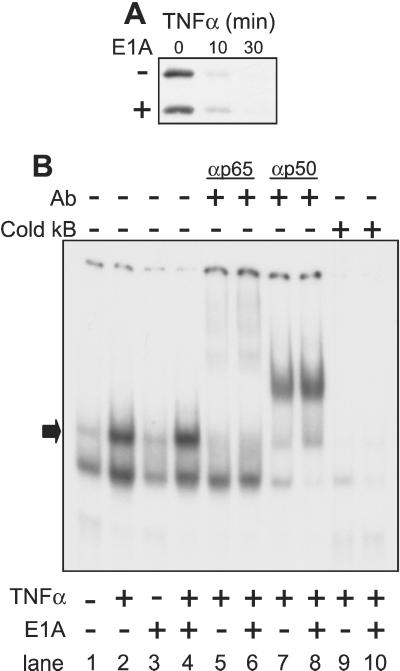
The effect of E1A on the stages of the NF-κB activation response was tested in NIH 3T3 cells. The initial key segment of the NF-κB activation pathway involves TNF-α-induced IKK activation, IKK phosphorylation of IκBα, and consequent IκBα degradation (reviewed in ref. 44). In time-response experiments, IκBα degradation was evident by 10 min after TNF-α treatment of both E1A-positive and E1A-negative cells and was complete by 30 min (Fig. 2A). The second key segment of the NF-κB activation pathway is nuclear translocation of NF-κB subunits and their dimerization and binding to the κB enhancer. EMSAs were used to test this NF-κB activation stage. There were no E1A-related differences in basal expression of either NF-κB p65/RelA or NF-κB p50/KBF-1 subunits (not shown). Fig. 2B is representative of four different EMSAs testing three different κB probes. Similar TNF-α-induced retardation of κB probe migration was observed for nuclear lysates from E1A-negative (lanes 1 and 2) and E1A-positive (lanes 3 and 4) cells. NF-κB specificity of EMSA activity was tested three ways. First, Ab specific for p65/RelA (lanes 5 and 6) or p50/KBF1 (lanes 7 and 8) supershifted TNF-α shifted bands from both E1A-negative (lanes 5 and 7) and E1A-positive (lanes 6 and 8) cells, implicating κB probe binding by the p50/p65 NF-κB heterodimer (arrow). Second, competition for nuclear protein binding to the labeled κB probe using excess unlabeled κB probe indicated that shifted bands were κB-specific (lanes 9 and 10). Third, κB-specificity was confirmed by the lack of binding competition by an excess of unlabeled mutant κB probe (not shown). Shifted p50/p65-binding activities from E1A-negative and E1A-positive nuclei were quantitated by phosphorimager and were similar when using two different probes (shifted activity of E1A-negative vs. E1A-positive as a percentage of total activity: H2TF1 κB probe, 30 vs. 34%, respectively; ELAM κB probe, 21 vs. 24%, respectively). Therefore, high level E1A expression in NIH 3T3 did not block TNF-α-induced turnover of IκB, NF-κB subunit expression or trafficking to the nucleus, or NF-κB p50/p65 heterodimerization and binding to κB enhancer sequences.
Figure 2.
TNF-α-induced activation of the upstream NF-κB activation pathway in E1A-negative vs. stably transfected, E1A-positive cells. (A) Expression of IκB α-protein detected by Western analysis with time after treatment with TNF-α. (B) EMSA of TNF-α-induced nuclear protein binding to labeled κB-oligomer probe. As indicated, some samples were pretreated with antibodies against NF-κB p65/RelA or p50/KBF1 or with excess (500×) unlabeled (cold) κB probe.
NF-κB p65/RelA Blockade of both E1A Repression of the NF-κB Activation Response and E1A-Induced Cellular Sensitivity to TNF-α.
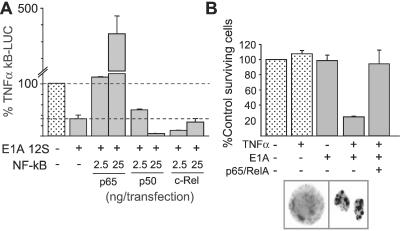
Three NF-κB subunits—p65/RelA, p50/KBF-1 and c-Rel—were tested in overexpression assays for relief of E1A-induced repression of the NF-κB activation response. Cotransfection of p65/RelA eliminated E1A repression in a dose-dependent manner, whereas p50/KBF1 and c-Rel did not (Fig. 3A). p65/RelA blockade of E1A-induced transcription repression was eliminated by increasing E1A expression in cotransfected cells (not shown). Therefore, E1A repressed both ectopic and endogenous (Fig. 1A) p65/RelA activity. We tested the prediction from these data that p65/RelA overexpression would block E1A-induced cellular sensitivity to TNF-α-induced apoptosis (Fig. 3B). Initial E1A titrations were done to ensure that the oncoprotein expression level tested had no independent, adverse affect on cell survival (Fig. 3B, bar 3 vs. bar 1). TNF treatment of sham-transfected (empty vector) cells did not reduce cell survival (Fig. 3B, bar 2 vs. bar 1). In contrast, 75–80% of E1A-expressing cells died by apoptosis when treated with TNF-α (Fig. 3B, bar 4 vs. bars 1–3). Coexpression of ectopic p65/RelA completely blocked E1A-induced sensitivity to TNF-α (Fig. 3B, bar 5 vs. bar 4), whereas coexpression of p50/KBF1 did not rescue cells from E1A-induced sensitivity to TNF-α (not shown). p65/RelA overexpression did not affect E1A protein expression in cotransfected cells.
Figure 3.
NF-κB p65/RelA blockade of E1A-induced repression of the NF-κB activation and apoptotic responses to TNF-α in NIH 3T3 cells. (A) κB-LUC assays. Cells were divided into sham-transfected (empty vector, stippled bar), E1A transfected, or E1A+NF-κB subunit-transfected populations. All cells were treated with TNF-α. TNF-α-induced κB-LUC responses are presented as the percentages of the response of the sham-transfected control. Low dose and high dose p65/RelA significantly blocked E1A repression of κB-LUC responses (P < 0.05), whereas p50/KBF1 and c-Rel did not. (B) For apoptosis assays, cells were divided into sham-transfected (stipple bar) or E1A-transfected populations. All cells were cotransfected with β-gal marker. After treatment with TNF-α (24 h), dishes were counted for surviving/transfected (blue) cells. Cell survival is presented as the percentage of sham-transfected cells not treated with TNF-α (n = 4). Apoptotic death was confirmed by microscopic examination of nuclear morphologies of control and TNF-α-treated cells (e.g., cells in box).
Lack of a Requirement for p300/CBP-Binding Domains in the N Terminus and Conserved Region 1 of E1A for Either Repression of the NF-κB Activation Response or E1A-Induced Sensitivity to TNF-α.
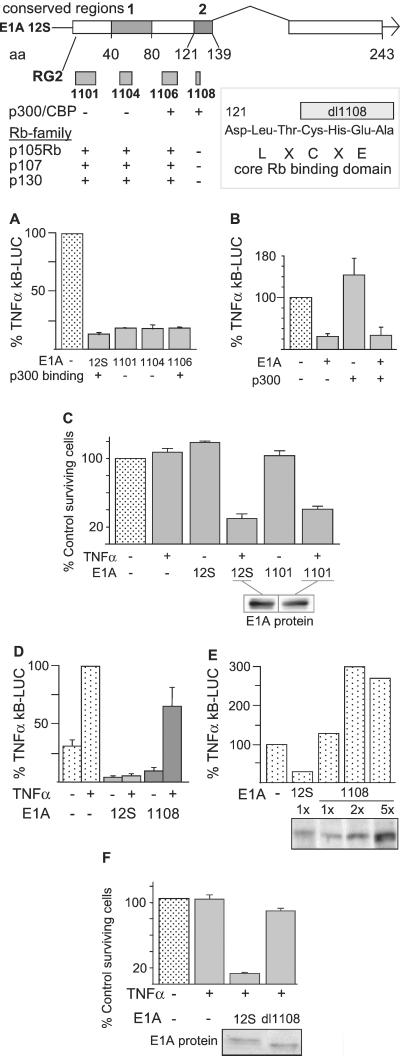
E1A mutant genes—12S.RG2, 12S/dl1101, 12S/dl1104 (see map in Fig. 4), and 12SΔ2–36 (not shown)—whose proteins either do not bind or bind poorly to p300/CBP were compared with nonmutant E1A (12S) for repression of the NF-κB response to TNF-α (Fig. 4A). E1A 12S/dl1106, whose deletion does not affect E1A binding to either p300/CBP or Rb, was used as a mutation control. p300-nonbinding E1A blocked NF-κB activation equally compared with nonmutant E1A. E1A expression was monitored by Western blotting to ensure that comparisons were done by using comparable expression of mutant and nonmutant E1A proteins. CMV promoter concentrations were kept constant.
Figure 4.
Effect of E1A first exon mutations on repression of the NF-κB activation and apoptotic responses to TNF-α. Transcription map of the E1A 12S cDNA. Amino acid (aa) boundaries of conserved regions 1 and 2 are shown below the bar diagram. E1A deletion mutations are represented as shaded boxes above mutation designations. Mutation effects on E1A binding to p300/CBP or Rb family proteins. +, binding; −, elimination/reduction of binding. Box, blow-up of Rb-binding domain and four amino acid deletion in this region for the mutant 12S/dl1108. All data are presented as the mean ± SEM of n different experiments. (A) Effect of deletions that eliminate E1A binding to p300 on repression of κB-LUC activation in transiently transfected NIH 3T3 cells (n = 2–4). (B) Effect of p300 overexpression on E1A repression of κB-LUC activation in transiently transfected U2OS cells (n = 3). (C) Effect of dl1101 deletion (eliminates p300 binding) on E1A-induced cellular sensitivity to TNF-α (n = 4). (D and E) Effect of dl1108 deletion (eliminates binding to Rb family proteins) upon E1A repression of κB-LUC activation in transiently transfected NIH 3T3 cells (D, n = 3; E, n = 1). (E) Sister cultures were tested by Western analysis for E1A protein expression (identical amounts of protein loaded per lane; E1A expression quantitated by PhosphorImager). (F) Effect of dl1108 deletion on E1A-induced cellular sensitivity to TNF-α (n = 4). Sister cultures from matched killing assays were tested for E1A protein expression by Western analysis as shown.
One mechanism of E1A-induced transcription repression in other cell and transcription factor systems involves E1A-binding-dependent “depletion” of limiting p300/CBP coactivator, as evidenced by relief of repression by coactivator overexpression (e.g., see refs. 45 and 46). p300 overexpression experiments were done to test this possibility (Fig. 4B). Overexpression of p300 alone in U2OS cells could slightly enhance the TNF-α-induced NF-κB response (third bar). However, p300 coexpression did not block E1A repression of this response (fourth bar). CBP overexpression also failed to relieve E1A repression (not shown).
The E1A mutant, 12S/dl1101, was used to test whether p300-nonbinding mutant E1A proteins could sensitize cells to TNF-α-induced apoptosis in this transient transfection cell system (Fig. 4C). E1A 12S/dl1101 and nonmutant E1A 12S proteins could be expressed equally after transfection and were not toxic to NIH 3T3 cells in the absence of TNF-α treatment (Fig. 4C, bars 5 and 3, respectively). However, cells expressing equal amounts of E1A 12S/dl1101 protein were just as sensitive to TNF-α-induced apoptosis as were cells expressing nonmutant E1A protein (Fig. 4C, bar 6 vs. bar 4, respectively, contrasted with TNF-treatment control, bar 2; P < 0.05).
Requirement for the E1A-Rb-Binding Domain for Repression of the NF-κB Activation Response and Sensitization to TNF-α.
The four amino acid deletion in E1A 12S/dl1108 eliminates E1A binding to all Rb-family proteins but does not prevent E1A binding to p300/CBP (map in Fig. 4; ref. 47). Like nonmutant E1A 12S, dl1108 protein repressed basal (no TNF-α treatment) κB-LUC activity (Fig. 4D, bars 3 and 5, respectively, vs. bar 1; P < 0.05). However, unlike nonmutant E1A 12S, dl1108 did not eliminate the TNF-α-induced κB-LUC signal (Fig. 4D, bars 4 and 6, respectively, vs. bar 2; 12S, P < 0.05, dl1108, NS). In fact, the relative difference between the basal and the TNF-α-induced signals was increased in dl1108-expressing contrasted with E1A-negative cells. This difference was most clearly seen in dose-response studies of dl1108 expression, where increasing mutant E1A protein expression had the net effect of widening the fold amplification of the TNF-α signal (representative experiment shown in Fig. 4E, bars 3–5 vs. bar 1). The prediction that deletion of the E1A-Rb-binding domain would reduce E1A-induced repression of the NF-κB defense against TNF-α was tested in apoptosis assays (Fig. 4F). Cells expressing comparable levels of 12S/dl1108 protein were significantly less sensitive to TNF-α than cells expressing nonmutant E1A 12S protein (Fig. 4F, bar 4 vs. bar 3, P < 0.01).
Discussion
Reports from several laboratories have shown that E1A oncoprotein expression in cells of different types and from different species induces conversion from a TNF-α-resistant to a TNF-α-susceptible phenotype (1, 2, 6, 22, 48–51). Previous studies of cells stably transfected with E1A or E1A plus activated ras or of adenovirus infected cells expressing E1A suggested that this effect involves E1A binding to either p300/CBP or Rb-family proteins but could not distinguish between these E1A-cell protein-binding activities (5, 20–22). Among the possible explanations for differences in these reported data are clonal variations of stably transfected cells, E1A-independent effects of viral infection on the cellular NF-κB response that might affect TNF-α sensitivity and inadequate E1A protein expression in some tested cell populations. These problems were eliminated in the present studies by using unselected populations of mouse or human cells adjusted to express equal amounts of nonmutant or mutant E1A proteins using transient transfection. The data show that the Rb-binding domain encoded by E1A-conserved region 2 (Fig. 4) can function independently of p300/CBP-binding domains in the N terminus and conserved region 1 to repress the NF-κB-dependent transcription response and cellular defense against TNF-α-induced apoptosis. This E1A-induced transcription repression was independent of any detectable abnormalities in the upstream signaling pathway that have been reported for other cell systems (Fig. 2 A and B; refs. 22 and 52).
E1A repression of the TNF-induced, NF-κB response was observed across a wide range of E1A protein expression (Fig. 1A). We have reported that E1A expression levels similar to those observed during viral infection and virus-induced cellular immortalization are required to sensitize cells to immune-mediated apoptosis induced by TNF-α or cytolytic lymphocytes (2, 14). In contrast, lower E1A expression levels that are sufficient for cellular immortalization are not sufficient to sensitize cells to injury-induced apoptosis (9). These observations suggest that a threshold level of E1A repression of one or more NF-κB-dependent cellular defenses against proapoptotic activities triggered by TNF-α is required to convert cells to the cytolytic-susceptible phenotype. The E1A repressive effect was selective for κB-dependent transcription (Fig. 1D) and was specifically reversed by p65/RelA (Fig. 3A). In these respects, E1A-positive cells are similar to p65/RelA knockout cells that lack an NF-κB-dependent defense against TNF-α (23). In addition to rendering cells functionally deficient for p65/RelA activity, it is also possible that E1A has other proapoptotic effects in cells (11, 53, 54).
These data identify repression of the NF-κB activation response and cellular defense against TNF-α as an activity of the Rb-binding domain of E1A (map and box in Fig. 4). Specifically, the results show that E1A-Rb-binding blocks the TNF-α-induced activation response but does not prevent E1A repression of basal, κB-dependent transcription (Fig. 4 D and E). This finding is consistent with the results of studies by Parker et al. (55) on the repressive effects of E1A on the HIV promoter (LTR) that contains two κB sites (56). They reported that an E1A deletion including E1A CR2 resulted in retention of E1A repression of basal transcription from the HIV LTR but eliminated E1A-induced repression of TNF-α-activated transcription.
There are at least three mechanisms by which this E1A effect might be mediated: (i) E1A-Rb interactions might repress NF-κB activation by increasing E2F activity; (ii) E1A might also block Rb activities unrelated to E2F that enhance NF-κB activation; or (iii) the Rb-binding domain of E1A might possess NF-κB-repressive activities in addition to Rb-binding-dependent effects.
The best characterized effect of the E1A-Rb-binding domain (box in Fig. 4) is its relief of Rb repression of E2F-family transcription factors (reviewed in ref. 57). One question is whether this E1A-induced activation of E2F could explain repression of the NF-κB response to TNF-α. E2F-1 blocks an upstream step in TNF-α signaling by inducing degradation of TRAF-2, a step leading to IκB turnover (58). This E2F-1 effect does not explain the present results, because IκB turnover was not blocked by E1A (Fig. 2A). Furthermore, we have observed that E1A represses NF-κB activation by phorbol myristate acetate that does not signal through TRAF-2 (unpublished data; ref. 59). E2F-1 can also repress NF-κB-dependent transcription by binding directly to the NF-κB p50/KBF1 subunit (60). Direct binding of E2F-1 to NF-κB p50 probably does not explain our results either, because E1A repression was selective for NF-κB p65/RelA activity and was not relieved by overexpression of p50/KBF1 (Fig. 3A). Increased E2F activity could compete with NF-κB p65/RelA for limiting p300/CBP (29, 61). It would be expected that this E2F effect would be relieved by overexpression of p300/CBP, which was not the case in our studies (Fig. 4B). Furthermore, specific repression of E2F-dependent transcription using a dominant negative mutant does not relieve E1A repression of the NF-κB response to TNF-α (unpublished observations). Therefore, it is unlikely that E2F transcription factor activation is the key factor in E1A repression of the NF-κB activation response to TNF-α.
Rb-family proteins alter the function of other transcription factors—in some cases increasing transcription—but there are no reports of direct Rb-mediated activation of NF-κB (e.g., refs. 62 and 63). The emerging model of Rb-mediated transcription control is that Rb-associated histone deacetylases cause transcription repression (64–70). Therefore, it is possible that Rb could indirectly enhance NF-κB activation by repressing another transcriptional repressor. Theoretical examples of this mechanism include Rb-family protein enhancement of SP-1 activity that could, in turn, synergistically increase the p65/RelA response (63, 71, 72) and Rb relief of YY1 repression of p65/RelA function (73, 74). Further studies will be needed to test these and other possibilities.
There is limited evidence for E1A CR2 activities in addition to Rb binding. E1A binding to p300/CBP is usually associated with E1A domains encoded by the N terminus and conserved region 1 (Fig. 4, map) that interact with the CH3 domain of p300/CBP (75, 76). Chakravarti et al. (77) have shown that there are E1A interactions with the histone acetyltransferase (HAT) domain of p300/CBP that is adjacent to the CH3 domain and that may involve E1A CR2 (78). Therefore, it is possible that the E1A repression of NF-κB activation observed in our studies could involve complex interactions between E1A CR2, Rb, and p300/CBP. The recent linkage between HAT activity and the functional modulation of transcription factors, including NF-κB p65/RelA, provides a basis for new hypotheses in this area of investigation (79–81).
The observations reported here will be useful for studies of the consequences of E1A-induced sensitivity to TNF-α for tumor cell survival at the interface with the host cellular immune response. These E1A gene mapping data can be used to test the relevance of TNF-α-induced cell killing of E1A-positive cells by tumoricidal macrophages and cytolytic lymphocytes in vitro and to evaluate the importance of this cytokine for rejection of E1A-positive tumor cells by immunocompetent animals (17). These studies also might lead to a better understanding of Rb-dependent mechanisms that modify NF-κB-dependent effects on cell cycling, viral gene expression, and the host inflammatory response.
Acknowledgments
We thank Kelley Colvin, Barbara Routes, Jim Nichol, Colleen Platt, Nishath Quadar, and Dimple Damani for technical assistance, Drs. Robert Scheinman, Stanley Bayley, Josehp Mymryk, and Betty Moran for providing the indicated reagents, and Drs. Jack Routes, Robert Scheinman, Lauren Sompayrac, David Ucker, and Tanya Miura for critical reviews of the manuscript. This work was supported by Public Health Service Grant CA86727.
Abbreviations
- TNF-α
tumor necrosis factor-α
- IκB
NF-κB inhibitor
- EMSA
electrophoretic mobility-shift assay
- CBP
cyclic AMP response element-binding protein
- Rb
retinoblastoma
- κB-LUC
κ B-luciferase
- EMSA
electrophoretic mobility-shift assays
- CMV
cytomegalovirus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chen M J, Holskin B, Strickler J, Gorniak J, Clark M A, Johnson P J, Mitcho M, Shalloway D. Nature (London) 1987;330:581–583. doi: 10.1038/330581a0. [DOI] [PubMed] [Google Scholar]
- 2.Cook J L, May D L, Wilson B A, Holskin B, Chen M J, Shalloway D, Walker T A. J Immunol. 1989;142:4527–4534. [PubMed] [Google Scholar]
- 3.Lowe S W, Ruley H E, Jacks T, Housman D E. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Prieto R, Quintanilla M, Cano A, Leonart M L, Martin P, Anaya A, Ramon y Cajal S. Oncogene. 1996;13:1083–1092. [PubMed] [Google Scholar]
- 5.Shisler J, Duerksen-Hughes P, Hermiston T M, Wold W S, Gooding L R. J Virol. 1996;70:68–77. doi: 10.1128/jvi.70.1.68-77.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brader K R, Wolf J K, Hung M C, Yu D, Crispens M A, van Golen K L, Price J E. Clin Cancer Res. 1997;3:2017–2024. [PubMed] [Google Scholar]
- 7.Samuelson A V, Lowe S W. Proc Natl Acad Sci USA. 1997;94:12094–12099. doi: 10.1073/pnas.94.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao R, Karunagaran D, Zhou B P, Li K, Lo S S, Deng J, Chiao P, Hung M C. J Biol Chem. 1997;272:32739–32742. doi: 10.1074/jbc.272.52.32739. [DOI] [PubMed] [Google Scholar]
- 9.Cook J, Routes B, Walker T, Colvin K, Routes J. Exp Cell Res. 1999;251:414–423. doi: 10.1006/excr.1999.4597. [DOI] [PubMed] [Google Scholar]
- 10.Stiewe T, Parssanedjad K, Esche H, Opalka B, Putzer B M. Cancer Res. 2000;60:3957–3964. [PubMed] [Google Scholar]
- 11.Ueno N T, Bartholomeusz C, Herrmann J L, Estrov Z, Shao R, Andreeff M, Price J, Paul R W, Anklesaria P, Yu D, Hung M C. Clin Cancer Res. 2000;6:250–259. [PubMed] [Google Scholar]
- 12.Sawada Y, Fohring B, Shenk T E, Raska K., Jr Virology. 1985;147:413–421. doi: 10.1016/0042-6822(85)90143-6. [DOI] [PubMed] [Google Scholar]
- 13.Cook J L, May D L, Lewis A M, Jr, Walker T A. J Virol. 1987;61:3510–3520. doi: 10.1128/jvi.61.11.3510-3520.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook J L, Wilson B A, Wolf L A, Walker T A. Oncogene. 1993;8:625–635. [PubMed] [Google Scholar]
- 15.Walker T A, Wilson B A, Lewis A M, Jr, Cook J L. Proc Natl Acad Sci USA. 1991;88:6491–6495. doi: 10.1073/pnas.88.15.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Routes J M, Ryan S. J Virol. 1995;69:7639–7647. doi: 10.1128/jvi.69.12.7639-7647.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Routes J M, Ryan S, Li H, Steinke J, Cook J L. Virology. 2000;277:48–57. doi: 10.1006/viro.2000.0571. [DOI] [PubMed] [Google Scholar]
- 18.Cook J, Routes B, Leu C, Walker T, Colvin K. Exp Cell Res. 1999;252:199–210. doi: 10.1006/excr.1999.4617. [DOI] [PubMed] [Google Scholar]
- 19.Cook J, Krantz C, Routes B. Proc Natl Acad Sci USA. 1996;93:13985–13990. doi: 10.1073/pnas.93.24.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ames R S, Holskin B, Mitcho M, Shalloway D, Chen M J. J Virol. 1990;64:4115–4122. doi: 10.1128/jvi.64.9.4115-4122.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duerksen-Hughes P J, Hermiston T W, Wold W S, Gooding L R. J Virol. 1991;65:1236–1244. doi: 10.1128/jvi.65.3.1236-1244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao R, Hu M C, Zhou B P, Lin S Y, Chiao P J, von Lindern R H, Spohn B, Hung M C. J Biol Chem. 1999;274:21495–21498. doi: 10.1074/jbc.274.31.21495. [DOI] [PubMed] [Google Scholar]
- 23.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 24.Wang C-Y, Mayo M, Baldwin A., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 25.Van Antwerp D, Martin S, Kafri T, Green D, Verma I. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 26.Rensing-Ehl A, Hess S, Ziegler-Heitbrock H W, Riethmuller G, Engelmann H. J Inflamm. 1995;45:161–174. [PubMed] [Google Scholar]
- 27.Ponton A, Clement M V, Stamenkovic I. J Biol Chem. 1996;271:8991–8995. doi: 10.1074/jbc.271.15.8991. [DOI] [PubMed] [Google Scholar]
- 28.Dudley E, Hornung F, Zheng L, Scherer D, Ballard D, Lenardo M. Eur J Immunol. 1999;29:878–886. doi: 10.1002/(SICI)1521-4141(199903)29:03<878::AID-IMMU878>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 31.Hottiger M O, Felzien L K, Nabel G J. EMBO J. 1998;17:3124–3134. doi: 10.1093/emboj/17.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merika M, Williams A J, Chen G, Collins T, Thanos D. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhong H, Voll R E, Ghosh S. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 34.Brockmann D, Putzer B M, Lipinski K S, Schmucker U, Esche H. Gene Expression. 1999;8:1–18. [PMC free article] [PubMed] [Google Scholar]
- 35.Vanden Berghe W, De Bosscher K, Boone E, Plaisance S, Haegeman G. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 36.Tamami M, Lindholm P F, Brady J N. J Biol Chem. 1996;271:24551–24556. doi: 10.1074/jbc.271.40.24551. [DOI] [PubMed] [Google Scholar]
- 37.Janaswami P M, Kalvakolanu D V, Zhang Y, Sen G C. J Biol Chem. 1992;267:24886–24891. [PubMed] [Google Scholar]
- 38.Scheinman R I, Beg A A, Baldwin A S., Jr Mol Cell Biol. 1993;13:6089–6101. doi: 10.1128/mcb.13.10.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H G, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelsma T N, Howe J A, Evelegh C M, Cunniff N F, Skiadopoulos M H, Floroff M R, Denman J E, Bayley S T. Virol. 1988;163:494–502. doi: 10.1016/0042-6822(88)90290-5. [DOI] [PubMed] [Google Scholar]
- 41.Routes J M, Cook J L. Virology. 1995;210:421–428. doi: 10.1006/viro.1995.1358. [DOI] [PubMed] [Google Scholar]
- 42.Schindler U, Baichwal V R. Mol Cell Biol. 1994;14:5820–5831. doi: 10.1128/mcb.14.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duke R, Cohen J. Curr Prot Immunol. 1992;3:3.17.1–3.17.16. [Google Scholar]
- 44.Baldwin A S., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 45.Smits P H, de Wit L, van der Eb A J, Zantema A. Oncogene. 1996;12:1529–1535. [PubMed] [Google Scholar]
- 46.Ghosh A K, Yuan W, Mori Y, Varga J. Oncogene. 2000;19:3546–3555. doi: 10.1038/sj.onc.1203693. [DOI] [PubMed] [Google Scholar]
- 47.Barbeau D, Charbonneau R, Whalen S G, Bayley S T, Branton P E. Oncogene. 1994;9:359–373. [PubMed] [Google Scholar]
- 48.Chen M-J, Earl C Q, Holskin B, Anzano M, Shalloway D, Cook J L. In: Monokines and Other Non-Lymphocytic Cytokines. Powanda M C, Oppenheim J J, Kluger M J, Dinarello C A, editors. New York: Liss; 1988. pp. 243–248. [Google Scholar]
- 49.Duerksen-Hughes P, Wold W S, Gooding L R. J Immunol. 1989;143:4193–4200. [PubMed] [Google Scholar]
- 50.Rodrigues M, Dion P, Sircar S, Weber J M. Virus Res. 1990;15:231–236. doi: 10.1016/0168-1702(90)90030-f. [DOI] [PubMed] [Google Scholar]
- 51.Vanhaesebroeck B, Timmers H T, Pronk G J, van Roy F, Van der Eb A J, Fiers W. Virology. 1990;176:362–368. doi: 10.1016/0042-6822(90)90006-d. [DOI] [PubMed] [Google Scholar]
- 52.Shao R, Tsai E M, Wei K, von Lindern R, Chen Y H, Makino K, Hung M C. Cancer Res. 2001;61:7413–7416. [PubMed] [Google Scholar]
- 53.Fearnhead H O, McCurrach M E, O'Neill J, Zhang K, Lowe S W, Lazebnik Y A. Genes Dev. 1997;11:1266–1276. doi: 10.1101/gad.11.10.1266. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen M, Branton P E, Roy S, Nicholson D W, Alnemri E S, Yeh W C, Mak T W, Shore G C. J Biol Chem. 1998;273:33099–33102. doi: 10.1074/jbc.273.50.33099. [DOI] [PubMed] [Google Scholar]
- 55.Parker S F, Felzien L K, Perkins N D, Imperiale M J, Nabel G J. J Virol. 1997;71:2004–2012. doi: 10.1128/jvi.71.3.2004-2012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nabel G, Baltimore D. Nature (London) 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 57.Cress W D, Nevins J R. Curr Top Microbiol Immunol. 1996;208:63–78. doi: 10.1007/978-3-642-79910-5_3. [DOI] [PubMed] [Google Scholar]
- 58.Phillips A C, Ernst M K, Bates S, Rice N R, Vousden K H. Mol Cell. 1999;4:771–781. doi: 10.1016/s1097-2765(00)80387-1. [DOI] [PubMed] [Google Scholar]
- 59.Vertegaal A C, Kuiperij H B, Yamaoka S, Courtois G, van der Eb A J, Zantema A. Cell Signalling. 2000;12:759–768. doi: 10.1016/s0898-6568(00)00133-9. [DOI] [PubMed] [Google Scholar]
- 60.Kundu M, Guermah M, Roeder R G, Amini S, Khalili K. J Biol Chem. 1997;272:29468–29474. doi: 10.1074/jbc.272.47.29468. [DOI] [PubMed] [Google Scholar]
- 61.Lee C W, Sorensen T S, Shikama N, La Thangue N B. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- 62.Klemm D J, Colton L A, Ryan S, Routes J M. J Biol Chem. 1996;271:8082–8088. doi: 10.1074/jbc.271.14.8082. [DOI] [PubMed] [Google Scholar]
- 63.Johnson-Pais T, Degnin C, Thayer M J. Proc Natl Acad Sci USA. 2001;98:2211–2216. doi: 10.1073/pnas.051415898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 65.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 66.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 67.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai A, Lee J M, Yang W M, DeCaprio J A, Kaelin W G, Jr, Seto E, Branton P E. Mol Cell Biol. 1999;19:6632–6641. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meloni A R, Smith E J, Nevins J R. Proc Natl Acad Sci USA. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- 71.Udvadia A J, Rogers K T, Higgins P D, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanceau J, Kaisho T, Hirano T, Wietzerbin J. J Biol Chem. 1995;270:27920–27931. doi: 10.1074/jbc.270.46.27920. [DOI] [PubMed] [Google Scholar]
- 73.Petkova V, Romanowski M J, Sulijoadikusumo I, Rohne D, Kang P, Shenk T, Usheva A. J Biol Chem. 2001;276:7932–7936. doi: 10.1074/jbc.M007411200. [DOI] [PubMed] [Google Scholar]
- 74.Lu S Y, Rodriguez M, Liao W S. Mol Cell Biol. 1994;14:6253–6263. doi: 10.1128/mcb.14.9.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 76.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 77.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 78.Goodman R H, Smolik S. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 79.Ashburner B P, Westerheide S D, Baldwin A S., Jr Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen L, Fischle W, Verdin E, Greene W C. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 81.Furia B, Deng L, Wu K, Baylor S, Kehn K, Li H, Donnelly R, Coleman T, Kashanchi F. J Biol Chem. 2002;277:4973–4980. doi: 10.1074/jbc.M107848200. [DOI] [PubMed] [Google Scholar]