Abstract
Placozoans, the simplest free-living animals, have never been observed to reproduce sexually. Here, we describe molecular evidence for sexual reproduction within one clade of the Placozoa. In a population sample of 10 individuals, within-individual and overall nucleotide diversity were similar to each other and consistent with levels observed in sexually reproducing species. Intergenic recombination as well as the sharing of alleles between heterozygous and homozygous individuals was also observed. These hallmarks of sexual reproduction establish that sex is indeed present in this phylum.
Keywords: obligate asexual, sexual reproduction, Trichoplax, basal metazoan, Belize
Members of the Phylum Placozoa are microscopic marine invertebrates possessing the smallest genomes of any known metazoan (1) as well as one of the simplest body plans of any free-living animal (2). They occupy a basal position in the metazoan phylogeny, along with Cnidaria, Ctenophora, and Porifera, but the phylogenetic ordering among these phyla has remained unresolved (3–6). These characteristics, along with simple conditions for laboratory culture, make placozoans an attractive, and arguably essential, model system for studies of early animal evolution and development. Many aspects of placozoan biology, however, are unknown. Most critically, whether placozoans have a sexual phase in their life cycle has remained unresolved ever since their discovery over a century ago (7). In this paper, we show that at least one taxon within the Phylum Placozoa can indeed reproduce sexually. This finding resolves a long-standing question in placozoan biology and recommends further development of the Placozoa as a model system.
Placozoans were first discovered crawling on the wall of a seawater aquarium by F. E. Schulze in 1883. They have since been found throughout the tropic and subtropic latitudes (8–11) where, in their largely benthic existence, they feed on detrital matter by extracellular digestion. Histologically, they have only four somatic cell types, none of which is found in other animals (2). These cells are organized into two layers, both lacking a basement membrane. There is no axis of symmetry; placozoans can assume a variety of shapes and folds, but a definite dorsal and ventral orientation exists.
Placozoan life history has been studied primarily in laboratory cultures, where they readily reproduce asexually by either binary fission or budding of pelagic “swarmers” (smaller protrusions of the animal) from the upper cell layer (2, 12–14). Asexual division occurs at a rate of once per day, thus a Petri dish containing only a few animals quickly becomes densely populated. Under high animal density, we consistently observe the formation of egg-like structures, as originally reported 30 years ago (15–18). Although the formation of these putative eggs is suggestive of sexual reproduction, sperm, fertilization, and complete development have never been observed. Despite the failure to observe male gametes, cleaving embryos are observed (16). These putative embryos, however, eventually exhibit a runaway genome amplification (18) and cease to develop past the 64-cell stage. Could this failure to observe complete development be due to improper laboratory conditions, or could the putative early embryos be the remnants of a past sexual phase in a presently obligate asexual organism? Traditional observational methods have failed to answer these questions and could have easily overlooked rare or cryptic sex.
Molecular genetics offers a sensitive and efficient approach to answering the question of sexuality in the Placozoa. An organism's mode of reproduction shapes the passage of genetic information from one generation to the next, resulting in distinct patterns of nucleotide variation at the population level. In particular, the absence of recombination and genetic exchange in apomictic obligate asexual diploids produces a genealogical history that essentially contains two parallel lineages, one for each formerly allelic copy of the genome. The formerly allelic gene copies within an obligate asexual individual therefore become increasingly divergent over time (19–21). This characteristic molecular signal of obligate asexual reproduction has been used as evidence for the ancient obligate asexuality of bdelloid rotifers (20). By contrast, in a sexual population, there is on average no difference between the genealogical history of a pair of homologous genes found in the same individual and the history of a pair that includes copies from separate individuals. Because of genetic drift, recombination, and genetic exchange, both pairs will have the same expected time to a common coalescent ancestor and, therefore, the same expected degree of nucleotide variation. Furthermore, the decay of linkage disequilibrium over physical distance, because of recombination, and the occurrence of heterozygotes and homozygotes, because of segregation and assortment, are distinguishing features of sexual reproduction. Frequent inbreeding or automixis can lead to even more reduced levels of nucleotide variation and increased levels of homozygosity.
In this study, we used molecular patterns in DNA sequences from a population sample to resolve the long-standing question of sexuality in the Placozoa. We show that one clade within the Placozoa carries the molecular signatures of sexual reproduction: low nucleotide polymorphism, intergenic or interchromosomal recombination, and sharing of alleles between heterozygotes and homozygotes.
Materials and Methods
Animal Material and DNA Extraction. Placozoans were collected from Twin Cays, Belize, during the months of August 2003 and June 2004. Single animal isolates were fixed on a 96-well CloneSaver FTA card (Whatman), and live animals were transported in seawater and cultured in the laboratory at Yale University. Clonal cultures were established from single animal isolates in glass Petri dishes filled with ≈250 ml of filter-sterilized (0.2 μm) artificial seawater (37 parts per thousand) and supplemented with 250 μl of Micro Algae Grow (Florida Aqua Farms, Dade City, FL) micronutrients and 2 ml of a stationary culture of cryptomonad algae, Pyrenomonas salina. At weekly intervals, one-third of the medium was exchanged with fresh supplemented seawater. Animals were periodically subcultured by seeding new dishes containing supplemented medium and fresh algae.
Live animals were harvested for DNA extraction by performing several washes with unsupplemented seawater followed by a 2-day period in total darkness and several additional washes with fresh seawater. Animals were transferred to 1.5-ml tubes and pelleted by centrifugation for 5 sec in a minicentrifuge (Thermo Electron, Milford, MA). The seawater was replaced with 500 μl of urea extraction buffer (22), extracted twice with an equal volume of phenol:chloroform (1:1), and precipitated with 0.7 vol of isopropanol. The resulting pellet was washed with 70% ethanol, air-dried, and resuspended in 100 μl of 10 mM Tris/1 mM EDTA, pH 8.0. DNA was purified from fixed animals by taking a 1.25-mm core from the FTA card; this core was cleaned using the manufacturer's protocol and amplified in a 20-μl reaction using the GenomiPhi amplification enzyme (General Electric).
Genotyping. The Placozoa was, until recently, described as a phylum containing only a single species, Trichoplax adhaerens. Recent work by Voigt et al. (11) has demonstrated that this phylum is actually composed of at least five divergent mitochondrial clades. Although these clades are molecularly very different, morphologically they appear indistinguishable. To ensure that our study included only members of a single clade, we sequenced each of our 31 isolates at an ≈450-bp region of the mitochondrial 16S ribosomal DNA locus. The 16S region was PCR-amplified by using forward 5′-CGAGAAGACCCCATTGAGCTTTACTA-3′ and reverse 5′-TACGCTGTTATCCCCATGGTAACTTT-3′ primers under the following conditions: 95°C denaturation for 2 min; 5 cycles of 95°C for 30 sec, 63°C for 30 sec, and 72°C for 1 min; 5 cycles of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 1 min; 20 cycles of 95°C for 30 sec, 61°C for 30 sec, and 72°C for 1 min; and a 72°C final extension for 10 min. Amplification products were purified by using QIAquick kit (Qiagen) and sequenced in both directions by using each PCR primer.
All of our samples fell into three of the five previously described mitochondrial clades. The clade identified by strain H8 in Voigt et al. (11) contained the largest number of our sampled individuals and was, for this reason, selected for further analysis. There were a total of 10 samples: BZ10101, BZ264, BZ413, BZ46, BZ384, BZ2115, BZC2, BZC6, BZE8, and BZF1. Strains BZ384, BZE8, BZC2, and BZC6 were obtained from single isolates deposited on the FTA card.
Full-Length cDNA Library. Total RNA was extracted from the cultured BZ10101 strain using RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The full-length cDNA (fl-cDNA) library was constructed by using the GeneRacer kit and SuperScript II reverse transcription (Invitrogen) with RT-PCR amplification carried out using both the 3′ and 5′ manufacturer's primers and PfuUltra high-fidelity enzyme (Stratagene). The cDNA fraction between 1 and 3 kb in length was selected on a 0.9% agarose gel, purified with QIAquick gel purification kit (Qiagen), cloned into pCR4Blunt-TOPO vector, and transformed into TOP10 chemically competent cells according to the manufacturer's instructions (Invitrogen). By using the M13 forward and reverse primers (Invitrogen), 96 random cDNA clones were sequenced.
PCR and Sequencing. Primers designed using the program primer3 (23) to the 5′ and 3′ untranslated regions (UTRs) and internal regions of the cDNAs are listed in Table 1. Genomic DNA was amplified by PCR with primer combinations using Taq enzyme (Qiagen), and conditions were 95°C denaturation for 2 min; 30 cycles of 95°C for 30 sec, 58°C for 30 sec, and 72°C for 4 min; and a 72°C final extension for 10 min. P. salina DNA was used as a negative control for each PCR. PCR amplification products were purified by using PEG precipitation (24) and sequenced with both forward and reverse PCR primers and with the internal primers listed in Table 1. In select cases, specific haplotypes in heterozygous DNA samples were analyzed by cloning PCR products into pCR4Blunt-TOPO vector (Invitrogen) according to the manufacturer's protocol, transformed into TOP10 chemically competent cells, and sequenced with corresponding forward and reverse PCR primers. All DNA sequences were generated by the W. M. Keck Facility (Yale University) or Genaissance Pharmaceuticals (New Haven, CT) by TaqFS dyeterminator cycle-sequencing reactions using PRISM 3730 DNA sequencers (Applied Biosystems).
Table 1. PCR and sequencing primers, in the 5′ to 3′ direction, for the seven loci used in this study.
| Gene | Forward | Reverse |
|---|---|---|
| Signal peptidase | TTGCCATTTTATGCTGAGCTGAAT | TTTTGGAGCTTCAAGGCTTTATTT |
| CCGCTTGTATAGTAGCTGCCATTG | CAACACACAATTAGAACCGTTTTCG | |
| Unknown P1439 | GCCTCAAAGAGTGAAAGAGGGTTT | ATCCAGAGGTAGCAAACGGCTTAT |
| TCACAGTCCTTTAGCCTGATACTACC | ||
| Clathrin light chain | GCCGTGAGGGTCTTTGGTTAAG | CGGTATATAGTATGGGCCTTGGTTC |
| TCCTCCATGATAAATGACATGCAA | CAGGAATAGTGCTATCGCGTTCAT | |
| PTTGIIP | TACTTGATCACAGTTCACGCCAAA | CCTATACAGCGCTGATTGCTGATG |
| Phospholipase A21-precursor | CAGCACGTACTGTGTATCGAAAGAA | AAAAGCAATTTGGAACAAAGTATGAA |
| TCATCTGTCGCAGGTACGTATGAA | ACTTGTATCTGGCGAAGCATTCAG | |
| TTTTGTGCGCGCTAAAACTATCTT | ||
| CTGGTAGCTGTGACCGTGCTAACT | ||
| Unknown P1500 | ATCCCGCGTTCTCCTACTATTTTT | AATTTTATGATCGCTTCCTGCAAT |
| TCATTCGTCGTGTGGCTGTTACTA | GCACAAGTATTAACCACACCGTCA | |
| GCGCCTTGGTTTCTACTGTTTTGT | ||
| Glutathione S-transferase | GGTGTTGCCTTGTGTTCACTTTT | GGGATGCAAAACTAATAATTACCAAA |
| TGGTCAAATTCGTCAACTTTACGTTC | TTTCTTAGATTTGGCTGGAAAGAC |
The first primer pair for each gene was designed to the 5′ and 3′ UTR; subsequent primers listed below these spanning primers are internal primers. All PCRs were carried out by using the 5′ and 3′ UTR primers and sequencing was performed using all primers listed.
Sequence Analyses. Applied Biosystems chromatogram files and DNA sequences were analyzed by the lasergene software package (DNASTAR, Madison, WI). Sequences were assembled by using the SeqMan module and aligned using clustalw (25). Genomic sequences were queried through tblastx (26) with a cutoff expect value, e, of 0.001. In directly sequenced PCR products, heterozygous sites were identified by using the program polyphred as part of the phred, phrap, and consed package (27–30) and confirmed by manually examining chromatograms. Haplotype phase was inferred by using phase 2.1.1 (31, 32) with 1,000 replicates under the no-recombination model. The two measures of nucleotide variation used in this paper, πW and  , are distinguished by the former being a within-individual measure defined as
, are distinguished by the former being a within-individual measure defined as
 |
where Si is the number of heterozygous sites in individual i and L is the gene length in base pairs, and the latter, 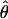 , being proportional to the total number of segregating sites in the sample as defined by Watterson (33).
, being proportional to the total number of segregating sites in the sample as defined by Watterson (33).
Results
Nucleotide Variation. We analyzed seven genes for overall levels of nucleotide variation,  , and within-individual levels of variation, πW. All sequences generated by this study were deposited in GenBank. Two of the seven genes analyzed returned no blast hits. The Unknown P1439 gene was found to contain two motifs similar to the thrombospondin type I (TSP1) repeat and a transmembrane segment at the 3′ end of the gene. The Unknown P1500 contained a motif similar to CD20 and three transmembrane segments.
, and within-individual levels of variation, πW. All sequences generated by this study were deposited in GenBank. Two of the seven genes analyzed returned no blast hits. The Unknown P1439 gene was found to contain two motifs similar to the thrombospondin type I (TSP1) repeat and a transmembrane segment at the 3′ end of the gene. The Unknown P1500 contained a motif similar to CD20 and three transmembrane segments.
Among the 10 individuals included in this study, 2 were found to be identical at every locus (BZ46 and BZ413). This identity is not unexpected because (i) these two individuals were collected from nearby sites on the island fringe, and (ii) placozoans possess an asexual dispersal stage (i.e., swarmers) (14). BZ46 and BZ413 are, for these reasons, most likely clonal copies. Table 2 provides a summary of the nucleotide diversity among the >10,000 nucleotides of sequence from these seven genes. The number of segregating sites per gene, S, ranged from 3 to 61, and corresponding estimates of 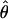 varied from 0.00091 to 0.00869. The within-individual divergence estimates, πW, ranged from 0.00069 to 0.01333. These estimates of nucleotide variation fall well within the range of variation found in invertebrate and vertebrate sexually reproducing organisms (34–39).
varied from 0.00091 to 0.00869. The within-individual divergence estimates, πW, ranged from 0.00069 to 0.01333. These estimates of nucleotide variation fall well within the range of variation found in invertebrate and vertebrate sexually reproducing organisms (34–39).
Table 2. Nucleotide sequence variation.
| Gene | Length, nt | No. of individuals sequenced | πW* | No. of segregating sites |
 † †
|
|---|---|---|---|---|---|
| Signal peptidase | 1,449 | 6 | 0.00069 | 4 | 0.00091 |
| Unknown P1439 | 2,042 | 9 | 0.01333 | 61 | 0.00869 |
| Clathrin light chain | 1,604 | 4 | 0.00156 | 6 | 0.00144 |
| PTTGIIP | 956 | 6 | 0.00157 | 3 | 0.00104 |
| Phospholipase A21 precursor | 1,444 | 6 | 0.00242 | 18 | 0.00413 |
| Unknown P1500 | 1,482 | 8 | 0.00464 | 24 | 0.00488 |
| Glutathione S-transferase | 1,297 | 5 | 0.00709 | 23 | 0.00627 |
Data excludes all gapped and insertion/deletion sites, and gene lengths include concatenated sequences.
The average per site no. of within-individual differences
The average per site no. of segregating sites
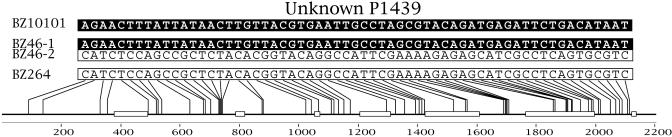
Heterozygous and Homozygous Individuals. Sexual reproduction leads to the sharing of alleles between heterozygous and homozygous individuals because of genetic exchange and the subsequent union of gametes during fertilization. At the Unknown P1439 locus, we observe individuals that are homozygous for two different alleles (BZ10101 and BZ264 in Fig. 1) and individuals that are heterozygous for the same two alleles (BZ46 in Fig. 1, for example). The occurrence of this pattern in an obligate asexual would be extremely unlikely, requiring a set of 61 nucleotide positions in the Unknown P1439 locus to mutate in exactly the same way along at least two separate lineages. The sharing of alleles between heterozygous and homozygous individuals was also observed in all other loci.
Fig. 1.
Heterozygous and homozygous states in three placozoan strains. Bars located at the bottom represent exons, and the connecting lines are introns. Numbers below the exons indicate nucleotide positions along the gene. Alleles were determined by the polymorphic nucleotide sites shown. Heterozygous individual BZ46 shares one identical allele with homozygous individual BZ10101 and one allele with homozygous individual BZ264. The homozygous states were inferred from the absence of another allele in their sequences.
All variable sites observed in the data are biallelic. These sites fell into two categories: (i) those in which all three possible genotypes exist and (ii) those in which only one homozygous and one heterozygous genotype exist. Sites in which all individuals are heterozygous or homozygous were not detected.
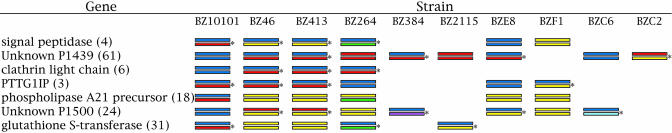
Intergenic Recombination. Obligate asexual reproduction enforces complete linkage across all regions of the genome because recombination and genetic exchange are not present to break up these associations. The inferred and directly cloned haplotype phases in Fig. 2 show no pattern of complete linkage between the loci we examined (except for individuals BZ46 and BZ413, which were found to be identical everywhere). Consider, for example, individuals BZ264 and BZ46 in Fig. 2. At the clathrin light-chain locus, these two individuals were inferred to be heterozygous and identical. If placozoans were obligate asexuals, then at any other variable loci examined, these two individuals should also be identical or very similar to each other. At the Unknown P1439 locus, individual BZ46 was found to be heterozygous with 61 heterozygous sites, whereas individual BZ264 was homozygous for one of these alleles. This pattern can be seen throughout Fig. 2. Intergenic or interchromosomal recombination is the most parsimonious explanation for such patterns.
Fig. 2.
Summary of all haplotypes observed in the seven loci. The values in parentheses after the gene names indicate the number of polymorphic sites found in these regions. Each column represents an individual or strain, and bars indicate color-coded haplotypes. Pairs of bars designate genotypes. Inferred haplotype phase obtained through the software phase (31, 32) are indicated by an asterisk to the right of the genotype. Genotypes with two of the same colored haplotypes are homozygous, and those with differing colors are heterozygous. Intergenic or interchromosomal recombination is inferred from the genotypic pattern across all individuals and loci.
Discussion
The overall population level of variation,  , and the average within-individual level of variation, πW, are expected to, on average, be equal in a sexually reproducing population. In contrast, πW is expected to be much larger than
, and the average within-individual level of variation, πW, are expected to, on average, be equal in a sexually reproducing population. In contrast, πW is expected to be much larger than 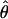 in an ancient obligate asexual population. This difference arises from the parallel inheritance (Fig. 3) of the two haploid genome copies in the genealogy of a diploid obligate asexual population (19). If we consider homologous gene copies chosen from two different individuals, because of genetic drift their lineages may coalesce much more recently at an asexual ancestor. In fact, of the four between-individual gene pairs that can be formed from two diploids, two will have their most recent common ancestor at the last sexual event, which may be quite ancient, and the other two will share a more recent asexual ancestor. Consequently, obligate asexual populations will have higher within-individual divergence compared with the overall population level of variation. On the other hand, genes of sexually reproducing animals are constantly recombining, and, as a result, two gene copies within a diploid individual are expected to have the same level of divergence as two gene copies sampled from different individuals. The observed values of πW and
in an ancient obligate asexual population. This difference arises from the parallel inheritance (Fig. 3) of the two haploid genome copies in the genealogy of a diploid obligate asexual population (19). If we consider homologous gene copies chosen from two different individuals, because of genetic drift their lineages may coalesce much more recently at an asexual ancestor. In fact, of the four between-individual gene pairs that can be formed from two diploids, two will have their most recent common ancestor at the last sexual event, which may be quite ancient, and the other two will share a more recent asexual ancestor. Consequently, obligate asexual populations will have higher within-individual divergence compared with the overall population level of variation. On the other hand, genes of sexually reproducing animals are constantly recombining, and, as a result, two gene copies within a diploid individual are expected to have the same level of divergence as two gene copies sampled from different individuals. The observed values of πW and 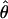 were not only low but also generally similar to each other at each locus analyzed (Table 2), supporting the sexual model. Specifically, the per site nucleotide difference (S/L) ranged from 0.3% to 3%, which is similar to the observed range of 0.75%–4% polymorphism reported for sexually reproducing invertebrates and vertebrates (34–39).
were not only low but also generally similar to each other at each locus analyzed (Table 2), supporting the sexual model. Specifically, the per site nucleotide difference (S/L) ranged from 0.3% to 3%, which is similar to the observed range of 0.75%–4% polymorphism reported for sexually reproducing invertebrates and vertebrates (34–39).
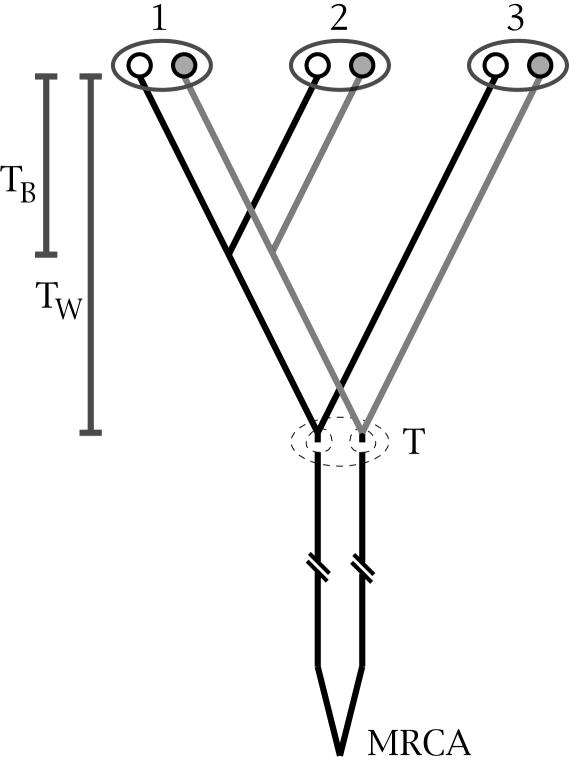
Fig. 3.
A gene tree of an obligate asexual population depicting six gene copies from three diploid individuals. Each ellipse represents an individual, and the two circles within each ellipse are homologous gene regions. TB, divergence time between individuals 1 and 2; TW, divergence time within individual 3; T, point in time when obligate asexual reproduction began; MRCA, most recent common ancestor of all gene copies sampled.
The absence of nucleotide sites that were heterozygous for the same allele pair in all individuals provides further evidence against ancient asexuality. A large fraction (64%) of the polymorphic sites across all loci were observed in all three possible genotypic pairings. This pattern would not be surprising in a sexual population, but in an obligate asexual population, these sites would each require at least two mutations in their coalescent genealogies and should therefore constitute only a small fraction of the segregating sites. This pattern is therefore grossly inconsistent with ancient obligate asexual reproduction.
Frequent inbreeding or automixis tends to increase homozygosity. Our data, however, did not contain any one segregating site in which all genotypes were homozygous. Moreover, alleles at each of three genes, signal peptidase, Unknown P1500, and glutatione S-transferase, occur in three distinct heterozygous genotypes that share only one allele in common (Fig. 2). If placozoans were obligate asexual organisms with automixis, all three heterozygous genotypes for each gene should be derivable through recombination and limited mutation from only two parental haplotypes, because they all share a common allele that presumably arose only once. Such a derivation is not possible for these three genes, leaving outbreeding sexual reproduction as the more parsimonious explanation.
Two additional lines of evidence in support of a sexual mode of reproduction in the placozoan life cycle were also uncovered from the population genetic data generated in this study. The sharing of alleles between heterozygous and homozygous individuals (e.g., Fig. 1) can only take place if sex is possible. The seven loci we examined behaved much as if they were unlinked to one another (Fig. 2), and this behavior is inconsistent with the expected total linkage between loci in obligate asexual genomes. Together with the low levels of polymorphism found both within individuals and overall in the population, these two additional observations led us to conclude that placozoans possess, or at least have possessed in their very recent history, the ability to reproduce sexually. This article is a conclusive report of the existence of a sexual phase in the Phylum Placozoa.
As the entire placozoan genome is slated for sequencing this year, the low levels of nucleotide variation revealed by this study predict that assembling the genome should proceed without the major problems that would be expected in an obligate asexual taxon. Moreover, if placozoans are indeed sexually reproducing, other questions regarding their life cycle are raised. How frequent is sexual reproduction? Is there a parthenogenic phase, and does it include a resting stage? Are the observed egg-like structures part of a complex life cycle that alternates between parthenogenesis, apomixis, and sex? The fact that placozoans possess molecular signatures for sexual reproduction raises the prospect that protocols might be discovered that would allow the life cycle of this animal to be completed in the laboratory, providing a look at embryogenesis in this basal metazoan phylum. Successful sexual reproduction in the laboratory would also open this system to the power of classical genetic analysis. With their simple body plan, basal phylogenetic position, rapid generation time, and ease of culture, along with having the smallest genome of any animal completely sequenced, placozoans make excellent candidate model systems for the study of early animal evolution and development.
Acknowledgments
We thank Maria Moreno and James Signorovitch for helpful discussions and critical reading of the manuscript and Klaus Rützler for making field research in Belize possible. Zack Snable, Chad Kritzberger, and Aaron Thier helped with animal care and Rafael Rosengarten helped with field work. Casey Dunn kindly provided the BZ10101 strain. This work was supported in part by the Caribbean Coral Reef Ecosystems Program Award (contribution no. 726) and National Institutes of Health Genetics Training Grant 5 T32 GM07499-28 (to A.Y.S.) and National Science Foundation Grant EF-0319076 (to L.W.B.).
Author contributions: A.Y.S., S.L.D., and L.W.B. designed research; A.Y.S. performed research; S.L.D. and L.W.B. contributed new reagents/analytic tools; A.Y.S. analyzed data; and A.Y.S., S.L.D., and L.W.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. DQ012104–DQ012115 (signal peptidase); DQ012116–DQ012125 (Unknown P1439); DQ012126–DQ012137 (clathrin light chain); DQ012098–DQ012103 (PTTG1IP); DQ012138–DQ012145 (phospholipase A21 precursor); DQ019421–DQ019444 (Unknown P1500); and DQ012146–DQ012155 (glutathione S-transferase)].
References
- 1.Ruthmann, A. (1977) Cytobiologie 15, 58-64. [Google Scholar]
- 2.Grell, K. G. & Ruthmann, A. (1991) in Placozoa, Porifera, Cnidaria, and Ctenophora, Microscopic Anatomy of Invertebrates, eds. Harrison, F. W. & Westfall, J. A. (Wiley–Liss, New York), Vol. 2, pp. 13-27. [Google Scholar]
- 3.Christen, R., Ratto, A., Baroin, A., Perasso, R., Grell, K. G. & Adoutte, A. (1991) EMBO J. 10, 499-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, A. G. (1998) Proc. Natl. Acad. Sci. USA 95, 15458-15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zrzavy, J., Mihulka, S., Kepka, P., Bezdek, A. & Tietz, D. (1998) Cladistics 14, 249-285. [DOI] [PubMed] [Google Scholar]
- 6.Medina, M., Collins, A. G., Silberman, J. D. & Sogin, M. L. (2001) Proc. Natl. Acad. Sci. USA 98, 9707-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulze, F. E. (1883) Zool. Anz. 6, 92-97. [Google Scholar]
- 8.Grell, K. G. (1987) Ann. Limnol. 14, 255-256. [Google Scholar]
- 9.Pearse, V. B., Vehara, T. & Miller, R. L. (1994) Trans. Am. Microscopical Soc. 113, 385-389. [Google Scholar]
- 10.Pearse, V. B. (1989) Pacific Sci. 43, 117-121. [Google Scholar]
- 11.Voigt, O., Collins, A. G., Pearse, V. B., Pearse, J. S., Ender, A., Hadrys, H. & Schierwater, B. (2004) Curr. Biol. 14, R944-R945. [DOI] [PubMed] [Google Scholar]
- 12.Thiemann, M. & Ruthmann, A. (1988) Z. Naturforsch. C Biosci. 43, 955-957. [Google Scholar]
- 13.Thiemann, M. & Ruthmann, A. (1990) Zoomorphology 110, 37-45. [Google Scholar]
- 14.Thiemann, M. & Ruthmann, A. (1991) Zoomorphology 110, 165-174. [Google Scholar]
- 15.Grell, K. G. (1971) Naturwissenschaften 58, 570. [Google Scholar]
- 16.Grell, K. G. (1972) Z. Morphol. Tiere 73, 297-314. [Google Scholar]
- 17.Grell, K. G. & Benwitz, G. (1974) Z. Morphol. Tiere 79, 295-310. [Google Scholar]
- 18.Ruthmann, A., Grell, K. G. & Benwitz, G. (1981) Z. Naturforsch. C Biosci. 36, 564-567. [Google Scholar]
- 19.Birky, C. W. (1996) Genetics 144, 427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mark Welch, D. B. & Meselson, M. (2000) Science 288, 1211-1215. [DOI] [PubMed] [Google Scholar]
- 21.Normark, B. B., Judson, O. P. & Moran, N. A. (2003) Biol. J. Linn. Soc. 79, 69-84. [Google Scholar]
- 22.Chen, J. & Dellaporta, S. (1994) in The Maize Handbook, ed. Walbot, V. (Springer, New York), pp. 526-527.
- 23.Rozen, S. & Skaletsky, H. (2000) Methods Mol. Biol. 132, 365-386. [DOI] [PubMed] [Google Scholar]
- 24.Ausebel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (2001) Current Protocols in Molecular Biology, ed. Janssen, K. (Wiley, New York), Vol. 3, pp. 15.2.1-15.2.13. [Google Scholar]
- 25.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 27.Nickerson, D. A., Tobe, V. O. & Taylor, S. L. (1997) Nucleic Acids Res. 25, 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 29.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 30.Ewing, B., Hillier, L., Wendl, M. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 31.Stephens, M. & Donnelly, P. (2003) Am. J. Hum. Genet. 73, 1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens, M., Smith, N. J. & Donnelly, P. (2001) Am. J. Hum. Genet. 69, 912-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watterson, G. A. (1975) Theor. Popul. Biol. 7, 256-276. [DOI] [PubMed] [Google Scholar]
- 34.Palumbi, S. R. & Metz, E. C. (1991) Mol. Biol. Evol. 8, 227-239. [DOI] [PubMed] [Google Scholar]
- 35.Wang, R. L., Wakeley, J. & Hey, J. (1997) Genetics 147, 1091-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindblad-Toh, K., Winchester, E., Daly, M. J., Wang, D. G., Hirschhorn, J. N., Laviolette, J. P., Ardlie, K., Reich, D. E., Robinson, E., Sklar, P., et al. (2000) Nat. Genet. 24, 381-386. [DOI] [PubMed] [Google Scholar]
- 37.Sachidanandam, R., Weissman, D., Schmidt, S. C., Kakol, J. M., Stein, L. D., Marth, G., Sherry, S., Mullikin, J. C., Mortimore, B. J., Willey, D. L., et al. (2001) Nature 409, 928-933. [DOI] [PubMed] [Google Scholar]
- 38.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298, 2157-2167. [DOI] [PubMed] [Google Scholar]
- 39.Hillier, L. W., Miller, W., Birney, E., Warren, W., Hardison, R. C., Ponting, C. P., Bork, P., Burt, D. W., Groenen, M. A., Delany, M. E., et al. (2004) Nature 432, 695-716.15592404 [Google Scholar]