Abstract
In early 1980, Irwin A. Rose, Avram Hershko, and Aaron Ciechanover published two papers in PNAS that reported the astounding observation that energy-dependent intracellular proteolysis was far more complicated than the previously accepted models of lysosomal proteolysis or the action of ATP-dependent proteases such as bacterial lon. In fact, it has turned out to be even more complicated than they could have suspected. The general model of covalently attaching a small protein as a targeting signal has proved to be every bit as important to eukaryotic cells as the better understood modifications such as phosphorylation or acetylation. The key player in this modification, a small protein called ubiquitin (APF-1 in these papers), is the founding member of a large family of proteins containing the β-grasp fold and is used as a posttranslational targeting signal to modify the structure, function, and/or localization of other proteins. The story of this discovery is a textbook example of the confluence of intellectual curiosity, unselfish collaboration, chance, luck, and preparation.
It is a truism in science that the first example of any biological phenomenon is the hardest to prove. We rely so much on precedent to formulate our hypothesis that something truly unique and novel is often over-looked for many years. The covalent modification of proteins by the attachment of other proteins is one such example (1–6). As we now know, this modification is a targeting mechanism used to move proteins around in the cell. The ubiquitin family of modifiers (ubiquitin, Nedd8, SUMO, ISG15, etc.) has been implicated in the regulation of proteolysis, nuclear localization, chromatin structure, genetic integrity, protein quality control, and signaling (6). The prototypical example of this modification is the covalent attachment of ubiquitin to proteins to target them for degradation by the proteasome and was first reported in two PNAS papers published early in 1980 by the 2004 Nobel laureates in chemistry, Avram Hershko, Aaron Ciechanover, and Irwin A. Rose (Fig. 1), and their collaborators (1, 7). These two papers outlined the essentials of the system and were amazingly prescient in their interpretations and predictions based on simple biochemical analysis.
Fig. 1.
The laureates at the Karolinska Institute after their Nobel addresses. Shown are (left to right) Aaron Ciechanover, Irwin Rose, and Avram Hershko.
The authors set out to explain a simple biological curiosity: the fact that intracellular proteolysis in mammalian cells requires energy. Melvin Simpson first showed this in his 1953 studies with isotopic labeling of cellular proteins (8), and for the next 25 years there were few insights into the mechanisms or metabolic logic of this observation. The hydrolysis of the peptide bond is exergonic, and there is no thermodynamic reason to use energy. The apparent requirement for energy could mean only that there was something we didn't understand. Part of the answer began to become apparent in the mid 1970s when Goldberg's group showed that damaged or abnormal proteins were rapidly cleared from the cell (9–11). He and Schimke (11) pointed out that enzymes that catalyzed rate-limiting steps in metabolic pathways were generally short-lived and that their amounts were responsive to metabolic conditions. Thus, by the late 1970s, we began to suspect that the energy dependence of intracellular proteolysis reflected some energy-dependent regulation of proteolytic systems.
The collaboration of Ciechanover, Hershko, and Rose was uniquely positioned and qualified to define this regulation. Avram Hershko (M.D. 1965 and Ph.D. 1969 from Hebrew University-Hadassah Medical School) did postdoctoral work in the laboratory of Gordon Tompkins at the University of California at San Francisco where he first became interested in protein degradation. His early studies were on tyrosine amino transferase and on the rates of bulk protein turnover in bacteria and mammalian cells. Hershko then established his own laboratory at the Technion-Israel Institute of Technology in Haifa and continued to collaborate with Tompkins until his untimely death in 1975. Aaron Ciechanover (M.D. 1974 and Ph.D. 1981 from Hebrew University-Hadassah Medical School) completed his military service before joining Hershko as a graduate student at the Technion-Israel Institute of Technology. Irwin A. “Ernie” Rose (Ph.D. in 1952 from the University of Chicago) did postdoctoral studies with Charles Carter at Case Western Reserve University and Severo Ochoa at New York University. He joined the Department of Pharmacology at Yale University in 1954 and moved to the Institute for Cancer Research at the Fox Chase Cancer Center in 1963. As a mechanistic enzymologist, he gained fame for his studies on proton transfer reactions and the use of isotopic labeling to examine the chemical mechanisms used by enzymes. Rose's interest in protein degradation dated back to the observations of Simpson, his colleague at Yale, who had demonstrated the ATP dependence of proteolysis in 1953. They talked often about this biochemical curiosity, and Rose would come back to this question periodically, but made little progress. Hershko and Rose first met at a Fogarty Foundation meeting in 1977 where they discovered their mutual interests in ATP-dependent proteolysis. Rose invited Hershko to do a sabbatical in his laboratory at the Institute for Cancer Research in Philadelphia. This began a 10-year collaboration that saw Rose hosting the Israeli group every summer. Rose was a patron and intellectual contributor far beyond what might be indicated by his authorship on the papers of that era. Thus, these two talented investigators entered into the collaboration that would define one of the several mechanisms of ATP-dependent protein degradation and frame a new means of viewing cellular regulation. [See Ciechanover (12) and Goldberg (13) for a more complete discussion of the various ATP-dependent processes.]
By 1979, Hershko and Ciechanover had exploited the seminal observations of Etlinger and Goldberg that reticulocyte lysates (which lack lysosomes) exhibited ATP-dependent proteolysis of denatured proteins (14) and would be amenable to biochemical fractionation. Hershko and Ciechanover first showed the system could be separated into two fractions (I and II) that had to be recombined to generate ATP-dependent proteolysis (15). Fraction I contained a single required component, a small, heat-stable protein they termed APF-1 (ATP-dependent proteolysis factor 1 because it was the first factor to be characterized). They then went on to further analyze fraction II and discovered a high molecular weight fraction (APF-2) that was stabilized by ATP and required for reconstitution of the ATP-dependent proteolysis (16). In retrospect, APF-1 was ubiquitin and APF-2 was probably the active protease, the 26S proteasome. At the time, however, Hershko, Ciechanover, and Rose considered that APF-2 might contain a kinase domain that phosphorylated APF-1 or an ATP-dependent binding protein that interacted with APF-1. Thus, the work reported in the cited PNAS papers (1, 7) began as an attempt to see whether there was an ATP-dependent association of APF-1 with other components of the system.
In the first paper (1), Ciechanover et al. showed that 125I-labeled APF-1 was promoted to a high molecular weight form upon incubation with fraction II and ATP. This association required low concentrations of ATP and was reversed upon removal of the ATP. At this time, a postdoctoral fellow in Rose's laboratory, Art Haas, began to characterize this association, and he found that the complex survived high pH. To everyone's surprise, the association of 125I-labeled APF-1 with proteins in fraction II was covalent! They went on to show that the bond was stable to NaOH treatment and that APF-1 was bound to many different proteins as judged by SDS/PAGE. The authors concluded that it was likely that conjugation was required for proteolysis; the nucleotide and metal ion requirements were similar for conjugation and proteolysis, as were the amounts of ATP and fraction II necessary to maximally stimulate. The covalent attachment of APF-1 to cellular proteins also explained why some investigators had so much difficulty demonstrating a requirement for APF-1 in ATP dependent proteolysis. When fraction II was prepared from reticulocytes without first depleting the ATP, most of the APF-1 was initially present in high molecular weight conjugates that were subsequently found in fraction II. These conjugates were rapidly disassembled by amidases in fraction II, thereby liberating free APF-1. Thus, there was sufficient APF-1 in fraction II to support proteolysis. If one first depleted the ATP, APF-1 was liberated before the chromatographic preparation of fraction II and APF-1 had to be added back to obtain maximal rates of proteolysis. This series of rather simple experiments convincingly demonstrated that APF-1 was covalently linked to multiple proteins in fraction II and that the linkage was reversible, although they did not demonstrate that this reaction was required for proteolysis. It was also not clear whether the modified proteins were enzymes of the system or substrates destined for degradation.
This paper was profoundly important to me, as well as to many others. At the time this work was being conducted, I was a postdoctoral fellow in Rose's laboratory and was being recruited to identify APF-1. One evening at a local establishment, Haas and I discussed these results with Michael Urban, a postdoctoral fellow from the next laboratory. He pointed out that this covalent attachment of two proteins was unusual, but not without precedent. Goldknopf and Busch (2) had shown that histone H2a was covalently modified by the attachment of a small protein called ubiquitin. Gideon Goldstein first discovered ubiquitin in his search for thymopoietin (17), and he generously shared authentic samples with me. Intrigued by this similarity, Urban, Haas, and I went on to show that APF-1 was the previously known protein called ubiquitin (18). Although it was known that ubiquitin was widely distributed, its physiological role was unclear until the 1980 PNAS papers suggested its role in ATP-dependent proteolysis (1, 7).
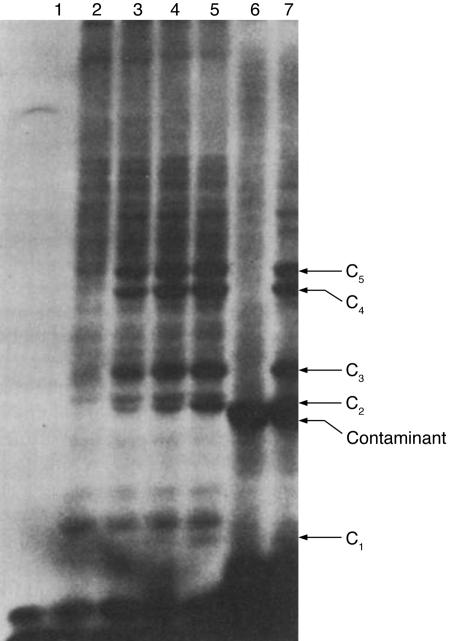
To ask whether this covalent bond formation was related to proteolysis, Hershko et al. (7) next went on to show that authentic substrates of the system were heavily modified and that multiple molecules of APF-1 were attached to each molecule of substrate (Fig. 2). These experiments demonstrated many elements of the system. The conjugation seemed to be enzyme-catalyzed, demonstrating for the first time the activity of ubiquitin ligases (19–22). The ligase activity was processive, preferring to add additional ubiquitin molecules to existing conjugates even in the presence of excess free substrate. Recent proteomics analyses suggest that there are hundreds of such ligases of at least two different types (21, 22). Thus, it seems likely that multiple ligases were active in fraction II and that this might explain why so many different proteins were ubiquitinated. Nearly 10 years later, Chau et al. (23) showed that substrates for proteolysis were polyubiquitinated, forming a chain linked through K48 of one ubiquitin and the C terminus of the next. Pioneering work out of the Finley and the Ellison laboratories later showed that other types of polyubiquitin chains also exist and that they are required in nonproteolytic pathways (24, 25).
Fig. 2.
Formation of covalent compounds between APF-1 and lysozyme in an ATP-dependent reaction. Tracks 1–5, compound formation with 125I-APF-1; track 1, without ATP; track 2, with ATP; tracks 3–5, with ATP and 5, 10, or 25 mg of unlabeled lysozyme, respectively; and tracks 6 and 7, compound formation with 125I-lysozyme. Conditions were as detailed in Methods of ref. 7, except that 5 μgof 125I-lysozyme (40,000 cpm) and 3 μgof unlabeled APF-1 were used. Track 6, ATP omitted; track 7, with 2 mM ATP. Contamination in 125I-labeled lysozyme is indicated. [Reproduced with permission from ref. 7 (Copyright 1980).]
Hershko et al. (7) then followed up on the observation that conjugation was reversed upon removal of ATP by demonstrating an enzyme-catalyzed disassembly of conjugates and liberation of intact ubiquitin that could be used for another round of conjugation (see figure 5 in ref. 7). Thus, they demonstrated the presence of specific amidases, or deubiquitinating enzymes as we now know them (26, 27), that accurately reversed conjugation. They pointed out the possibility that these could be “correction enzymes,” whose role we now understand as similar to the role of phosphatases in a kinase/phosphatase cycle. I shamelessly appropriated this idea upon taking my first job at Emory University and developed an assay for these amidases (deubiquitinating enzymes). We purified and cloned the first of these important regulatory enzymes from mammals (28), whereas Miller et al. (29) cloned Yuh1, the homologous protein from yeast. The elegant work of Varshavsky and his colleagues (30) in the yeast system soon followed, and in the ensuing years we have learned that there are >80 of these enzymes in at least six distinct gene families that regulate vital aspects of ubiquitination.
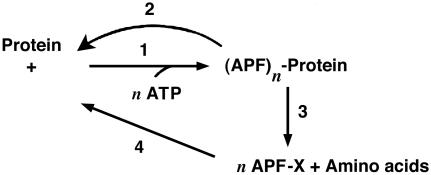
Thus, this second PNAS paper (7) concluded with a simple scheme outlining important aspects of ubiquitin-dependent proteolysis (Fig. 3). Ligases, deubiquitinating enzymes, and a specific protease were all predicted based on these biochemical assays. This has been an amazingly durable representation of the system, requiring only elaboration as details emerge. At its simplest, it pointed out that covalent attachment of ubiquitin targets proteins for delivery to a protease, thus resulting in the degradation of the target protein and release of free ubiquitin for another catalytic cycle. The protease turned out to be the proteasome, to be ATP-dependent, and to produce peptides and not amino acids. But it also explained the chemistry of other ubiquitin-like modifications (1–6). Modification of proteins by a single ubiquitin targets proteins in the endocytic pathway and in chromatin remodeling. Modification by SUMO is involved in altering the enzymatic activities of modified proteins or in targeting proteins to specific locations within the nucleus. Modification of cullins by Nedd8 helps to assemble active enzyme complexes. We need only to change the identity of the ubiquitin-like protein in step 1 and the consequences of the modification in step 3 to make this scheme completely general. There are even amidases that reverse the conjugation of ubiquitin-like proteins, accounting for steps 2 and 4 (31–33).
Fig. 3.
Proposed sequence of events in ATP-dependent protein breakdown (see the text). 1, APF-1-protein amide synthetase (acting on lysine ε-NH2 groups). 2, Amidase that allows correction when n = 1 or 2. 3, Peptidases that act strongly on (APF-1)n derivatives, when n > 1 or 2. 4, Amidase for APF-1-X; X is lysine or a small peptide. [Reproduced with permission from ref. 7 (Copyright 1980).]
These papers drew powerful conclusions from fairly simple biochemical experiments, but at the time there was considerable skepticism of this new paradigm. As Hershko subsequently showed, these conclusions were largely correct (34) and both of these PNAS contributions were extremely influential, being in the top ten (based on citations) of Hershko's original research publications. Their predictions and models have withstood the test of time and are a tribute to the imagination and clarity that Hershko, Ciechanover, and Rose have brought to the field of ubiquitin-dependent metabolism.
Author contributions: K.D.W. wrote the paper.
References
- 1.Ciechanover, A., Heller, H., Elias, S., Haas, A. L. & Hershko, A. (1980) Proc. Natl. Acad. Sci. USA 77, 1365-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldknopf, I. L. & Busch, H. (1977) Proc. Natl. Acad. Sci. USA 74, 864-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahajan, R., Delphin, C., Guan, T., Gerace, L. & Melchior, F. (1997) Cell 88, 97-107. [DOI] [PubMed] [Google Scholar]
- 4.Kamitani, T., Kito, K., Nguyen, H. P. & Yeh, E. T. (1997) J. Biol. Chem. 272, 28557-28562. [DOI] [PubMed] [Google Scholar]
- 5.Ohsumi, Y. (2001) Nat. Rev. Mol. Cell Biol. 2, 211-216. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz, D. C. & Hochstrasser, M. (2003) Trends Biochem. Sci. 28, 321-328. [DOI] [PubMed] [Google Scholar]
- 7.Hershko, A., Ciechanover, A., Heller, H., Haas, A. L. & Rose, I. A. (1980) Proc. Natl. Acad. Sci. USA 77, 1783-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson, M. V. (1953) J. Biol. Chem. 201, 143-154. [PubMed] [Google Scholar]
- 9.Goldberg, A. L. & Dice, J. F. (1974) Annu. Rev. Biochem. 43, 835-869. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg, A. L. & St. John, A. C. (1976) Annu. Rev. Biochem. 45, 747-803. [DOI] [PubMed] [Google Scholar]
- 11.Schimke, R. T. (1976) Circ. Res. 38, I131-7. [PubMed] [Google Scholar]
- 12.Ciechanover, A. (2005) Nat. Rev. Mol. Cell Biol. 6, 79-87. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg, A. L. (2005) Neuron 45, 339-344. [DOI] [PubMed] [Google Scholar]
- 14.Etlinger, J. D. & Goldberg, A. L. (1977) Proc. Natl. Acad. Sci. USA 74, 54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciehanover, A., Hod, Y. & Hershko, A. (1978) Biochem. Biophys. Res. Commun. 81, 1100-1105. [DOI] [PubMed] [Google Scholar]
- 16.Hershko, A., Ciechanover, A. & Rose, I. A. (1979) Proc. Natl. Acad. Sci. USA 76, 3107-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein, G., Scheid, M., Hammerling, U., Schlesinger, D. H., Niall, H. D. & Boyse, E. A. (1975) Proc. Natl. Acad. Sci. USA 72, 11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson, K. D., Urban, M. K. & Haas, A. L. (1980) J. Biol. Chem. 255, 7529-7532. [PubMed] [Google Scholar]
- 19.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka, K., Suzuki, T. & Chiba, T. (1998) Mol. Cells 8, 503-512. [PubMed] [Google Scholar]
- 21.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503-533. [DOI] [PubMed] [Google Scholar]
- 22.Petroski, M. D. & Deshaies, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 9-20. [DOI] [PubMed] [Google Scholar]
- 23.Chau, V., Tobias, J. W., Bachmair, A., Marriott, D., Ecker, D. J., Gonda, D. K. & Varshavsky, A. (1989) Science 243, 1576-1583. [DOI] [PubMed] [Google Scholar]
- 24.Spence, J., Sadis, S., Haas, A. L. & Finley, D. (1995) Mol. Cell. Biol. 15, 1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnason, T. & Ellison, M. J. (1994) Mol. Cell. Biol. 14, 7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amerik, A. Y. & Hochstrasser, M. (2004) Biochim. Biophys. Acta 1695, 189-207. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson, K. D. (1997) FASEB J. 11, 1245-1256. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson, K. D., Lee, K. M., Deshpande, S., Duerksen-Hughes, P., Boss, J. M. & Pohl, J. (1989) Science 246, 670-673. [DOI] [PubMed] [Google Scholar]
- 29.Miller, H. I., Henzel, W. J., Ridgeway, J. B., Kuang, W., Chisholm, V. & Liu, C. (1989) Biotechnology 7, 698-704. [Google Scholar]
- 30.Baker, R. T., Tobias, J. W. & Varshavsky, A. (1992) J. Biol. Chem. 267, 23364-23375. [PubMed] [Google Scholar]
- 31.Li, S. J. & Hochstrasser, M. (1999) Nature 398, 246-251. [DOI] [PubMed] [Google Scholar]
- 32.Gan-Erdene, T., Nagamalleswari, K., Yin, L., Wu, K., Pan, Z. Q. & Wilkinson, K. D. (2003) J. Biol. Chem. 278, 28892-28900. [DOI] [PubMed] [Google Scholar]
- 33.Malakhov, M. P., Malakhova, O. A., Kim, K. I., Ritchie, K. J. & Zhang, D. E. (2002) J. Biol. Chem. 277, 9976-9981. [DOI] [PubMed] [Google Scholar]
- 34.Hershko, A., Heller, H., Elias, S. & Ciechanover, A. (1983) J. Biol. Chem. 258, 8206-8214. [PubMed] [Google Scholar]