Abstract
The Rel/NF-κB transcription factor Relish performs a central role in the acute-phase response to microbial challenge by activating immune antibacterial peptides. We cloned and molecularly characterized the gene homologous to Drosophila Relish from the mosquito Aedes aegypti. Unlike Drosophila Relish, Aedes Relish has three alternatively spliced transcripts encoding different proteins. First, the predominant Aedes Relish transcript of 3.9 kb contains both the Rel-homology domains and the inhibitor κB (IκB)-like domain, which is similar to Drosophila Relish and to the mammalian p105 and p100 Rel/NF-κB transcription factors. Second, Aedes Relish transcript contains Rel-homology domains identical to those of the major transcript but it completely lacks the IκB-like domain-coding region, which has been replaced by a unique 3′-untranslated region sequence. In the third transcript, a deletion replaces most of the N-terminal sequence and Rel-homology domains; however, the IκB-like domain is intact. All three Aedes Relish transcripts were induced by bacterial injection but not by blood feeding. In vitro-translated protein from the Rel-only construct specifically binds to the κB motif from Drosophila cecropin A1 and Aedes defensin genes. PCR and Southern blot hybridization analyses show that these three transcripts originated from the same large inducible mRNA encoded by a single Relish gene.
Activation of innate immune factors in both mammals and insects shares a conserved pathway in which Rel/NF-κB transcription factors are chief regulators (1). In Drosophila, three Rel/NF-κB molecules are involved in two distinct pathways: the antifungal immune response, mediated by dorsal and Dif factors, and the antibacterial immune response, regulated by Relish (2–4). Dorsal, the dorsal/ventral morphogen (5), and Dif are activated by way of the Toll-signaling pathway and stimulate the production of antifungal factors such as Drosomycin (6). The antibacterial immune response against Gram-negative bacteria is mediated by another signaling pathway termed IMD, after immunodeficient mutation (imd). The IMD pathway requires another Drosophila Rel/NF-κB factor, Relish, which is transcriptionally up-regulated in response to microbial infection (4). In Drosophila relish mutants, the induction of immune defense is severely reduced, and insects are extremely sensitive to bacterial and fungal infection (7). Known immune effector genes under the regulation of Relish include Cecropin, Diptericin, Attacin, Defensin, and Metchnikowin (7, 8). Similar to the mammalian immune transcription factors p105 and p100, Relish consists of an N-terminal Rel/NF-κB homology domain and a C-terminal inhibitor κB (IκB)-like domain with ankyrin repeats (4). The Rel/NF-κB domain functions in dimerization and DNA binding, and the IκB domain interacts to control the subcellular localization of NF-κB (1). Relish processing is distinctly different from that of p105, cleavage of which is proteasome-dependent and the C-terminal domain of which is completely degraded (9, 10). In contrast, Relish is activated by endoproteolytic cleavage in response to bacterial infection, leading to the production of N-terminal Rel-homology domains that translocate to the nucleus and of a stable C-terminal ankyrin domain that remains in the cytoplasm (11). The Drosophila IKK complex regulates Relish activity (12), like the mammalian IKK complex that function in the IL1-R and tumor necrosis factor (TNF)-R pathways. Dredd, which encodes a protease related to mammalian Caspase 8 (13), functions downstream of Drosophila IKK complex (14) and may be directly involved in Relish cleavage and activation (8, 11). The immune deficiency gene (imd) and dTAK1, a homologue of mammalian MAPKKK kinase, have been suggested to function upstream of Relish, Dredd and the IKK complex, constituting the IMD pathway for antibacterial defense (14, 15). The imd gene encodes a protein with a death domain similar to that of a mammalian receptor interacting protein, a protein that plays a role in both NF-κB activation and apoptosis (16).
The study of the inducible immune genes and their regulatory mechanisms in model insect species, in particular, the fruit fly Drosophila, has provided powerful tools to elucidate the insect innate immune response (17). The basic knowledge of invertebrate immunity has been applied to mosquitoes, dipteran insects of medical importance (18). Initial studies of humoral immunity led to the purification of Defensin and the cloning of Defensin-encoding cDNAs in the yellow fever mosquito, Aedes aegypti (19–21), and later in the major African vector of the malaria parasite, Anopheles gambiae (22). Cecropins and other immune factors have been characterized from the cell lines of three mosquito species, Aedes albopictus, A. aegypti, and A. gambiae (for review, see ref. 23). Recently, gambicin from A. gambiae has been reported to be a novel antimicrobial peptide induced by immune challenge (24).
Some of these inducible components of the mosquito immune system may play a role in limiting the development of parasites that cause diseases such as malaria and lymphatic filariasis. Defensins have been shown to have effects on certain stages of Plasmodium either in vitro or when injected into the hemolymph of infected A. aegypti (25). The large losses of the parasite during invasion of epithelial tissues and translocation to the salivary glands are correlated with transcriptional activation of immune genes by malaria infection (26). This transcriptional activation in mosquitoes may be regulated by the NF-κB factor in a similar manner to Drosophila and vertebrates.
Little is known about the pathways regulating the immune responses in mosquitoes despite the enormous importance of such knowledge for our understanding of the immune system of these vectors of many devastating human diseases. We report herein the molecular cloning and characterization of mosquito homologue to Drosophila Relish from A. aegypti. Cloning of Aedes Relish provides evidence that the regulatory mechanism against bacterial challenge shown in Drosophila is generally conserved. As in Drosophila, Aedes Relish is a compound protein consisting of both NF-κB and IκB domains. Similarly, the Aedes Relish gene is induced by bacterial challenge, and Relish protein binds to the κB motif. However, characteristics of Aedes Relish revealed in this work provide insights in the regulatory mechanisms of antibacterial immune genes in the mosquitoes.
Materials and Methods
Isolation of cDNA Clones.
A PCR product was obtained from genomic DNA by degenerate primers based on the conserved region of the RHD from Drosophila Relish and mammalian Rel/NF-κB proteins. The following primers were used: AaRELF1 (5′-CTGCGGATCGT(T/G)GAGCA (A/G)CC-3′) and AaRELR1 (5′-CGAATATGTA(T/C)TT(T/C)TTGGCG-3′). This PCR fragment was subcloned by using a TA cloning kit (Novagen) and then was used as a probe to screen the λ ZAPII cDNA library prepared from previtellogenic female A. aegypti mosquitoes. A cDNA clone (C8) was isolated and then used as a probe to rescreen the cDNA library. In total, 12 clones were obtained and sequenced from both 5′ and 3′ ends. On the basis of sequencing and restriction-mapping analyses, the 12 clones were subdivided into three groups. The longest representative of each cDNA group, R6, R7, and C8 cDNA clones were fully sequenced from both strands.
Northern Hybridization, Reverse Transcription (RT)-PCR, and Rapid Amplification of cDNA 3′ Ends (3′-RACE).
Adult 2- or 3-day-old A. aegypti females were injected with a stationary-phase culture of Enterobacter cloacae. For the developmental study, adult males, females, third instar larvae and pupae were collected without any treatment and/or 5 h after bacterial challenge. The vitellogenic mosquitoes were collected 1 and 2 days after blood feeding. The unfertilized eggs were obtained by dissecting mosquitoes 3 days after blood feeding, whereas fertilized eggs were collected 1 day after laying eggs. Total RNA was prepared by the Trizol technique (GIBCO/BRL). Samples of 10 μg of total RNA were separated on a formaldehyde gel, blotted, and hybridized with a corresponding DNA probe. RT-PCR was performed by using the Titan one-step RT-PCR kit (Roche Molecular Biochemicals) with samples of 0.2 μg of total RNA as templates. Tubes containing RNA and RNase inhibitor (1 unit/μl, Roche Molecular Biochemicals) were incubated for 30 min at 50°C for RT reaction. Amplification conditions included rapid heating to 94°C for 2 min followed by 25–30 cycles of 55°C for 1 min, 72°C for 3 min, and 94°C for 45 s. Exact 3′ end of C8 clone with poly(A) was identified by 3′-RACE system (GIBCO/BRL).
Genomic DNA Isolation and Southern Blot.
Genomic DNA from ≈10 mosquitoes was purified by using the DNeasy tissue kit (Qiagen, Chatsworth, CA). Two-microgram aliquots of DNA were digested with the corresponding endonucleases; the DNA fragments were separated by electrophoresis in a 0.8% agarose gel, transferred to nitrocellulose filters, and hybridized with a DNA probe. DNA from individual mosquitoes, purified by the method of Bender et al. (27), was digested with EcoRI, and treated by the same method.
Electrophoretic Gel Mobility-Shift Assay.
The Rel-only protein was synthesized by a coupled in vitro transcription-translation system (Promega). The entire Rel-type transcript C8 cDNA clone was subcloned into pcDNA3.1/Zeo(+) (Invitrogen). The in vitro transcription-translation reactions programmed by the circular plasmid DNA used the T7 promoter. To confirm the synthesis of proteins with expected size, control transcription-translation reactions of both luciferase and the Rel-only protein were performed in the presence of [35S]methionine, and the resulting reactions were analyzed by SDS/PAGE and autoradiography.
The annealed deoxyoligonucleotide 5′-tcgagacacGGGGATTTTTgcac of Drosophila cecropin A1 κB motif and 5′-cacgctttcGGTGATTTACacag of A. aegypti Defensin κB motif were purified from 3.5% agarose gel (high-resolution agarose, Sigma) by electroelution. Labeling of double-stranded oligonucleotides and electrophoretic gel mobility-shift assay was performed with a gel-shift assay system (Promega). The protein-DNA complex was separated on 5% TBE Criterion Precast Gel (Bio-Rad) and visualized by autoradiography. For supershift tests we used α-RHD antibody against Drosophila Relish, which were a kind gift of D. Hultmark (Umeå University, Umeå, Sweden) (11).
Results and Discussion
Cloning of Three Types of Relish cDNAs from A. aegypti.
Several degenerate primer sets were designed and synthesized on the basis of the nucleotide sequence and amino acid sequence comparisons of Rel-homology domains (RHD and IPT domain) between Drosophila Relish and other NF-κB proteins. PCRs were performed with genomic DNA or reverse transcribed total RNA as templates. A 338-bp-long DNA fragment was obtained by using the genomic DNA and a set of a sense and an antisense primers based on the sequence of RHD. The deduced amino acid sequence of this PCR fragment indicated that it was highly similar to Drosophila Relish (55% identity) compared with other NF-κB proteins (less than 30% identity with Dif, dorsal, Gambif1, and NF-κB factors from vertebrates).
We used this PCR fragment as a probe to screen the cDNA library prepared from previtellogenic A. aegypti female mosquitoes. A cDNA clone, designated C8, was isolated and sequenced. The nucleotide sequence of C8 showed that it was a 5′-truncated cDNA clone containing 2,140 bp with a stop codon and 3′-untranslated region (UTR) without a poly(A) at the 3′ end. The deduced amino acid sequences from C8 clone exhibited high similarity with Drosophila Relish in the Rel-homology domains, but it had no IκB-like domain with ankyrin repeats.
To find out whether a true Relish homologue existed that contained an IκB-like domain in the mosquito, the C8 clone was used as a probe to rescreen the cDNA library. Eleven additional clones were isolated and sequenced from both 5′ and 3′ ends. Their nucleotide sequence showed that all 11 clones had the same 3′-UTR sequence different from that of C8 clone, although some of them were truncated and did not have poly(A) sequences. Further sequencing and restriction mapping analyses demonstrated that the 12 cDNA clones could be divided into three groups, with the longest representatives designated as R6, R7, and C8 clones.
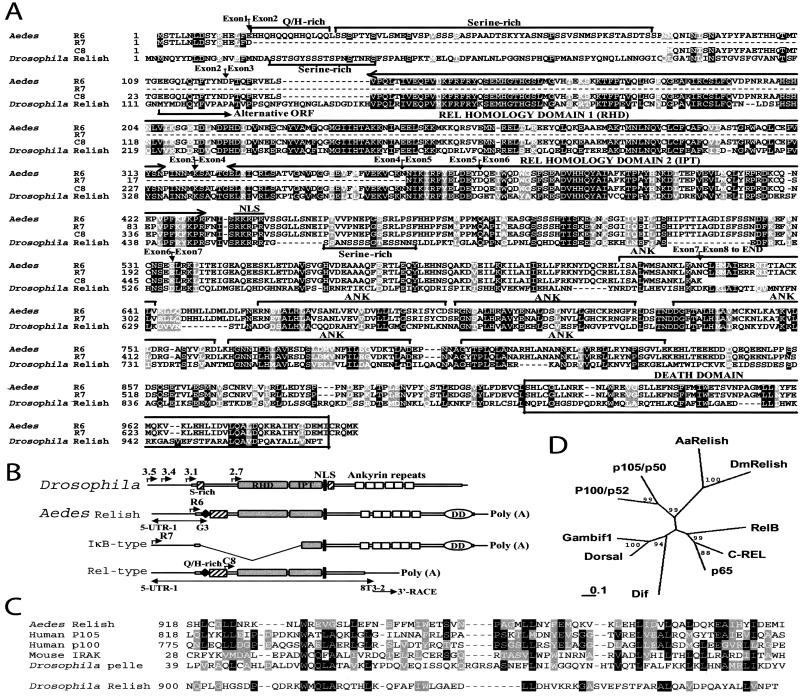
R6 group cDNAs consisted of the R6 clone that contains the full ORF and the other seven cDNA clones with different truncations in the 5′-region. The conceptual protein translated from the 3,280-bp sequence of the longest cDNA clone R6 contained both Rel-homology domains and an IκB-like domain with six ankyrin repeats (Fig. 1 a and b). Rel-homology domains (RHD and IPT), the location of nucleus localization signal, and overall structure of the protein was highly similar to Drosophila Relish, suggesting that it was a mosquito homologue of Drosophila Relish. Both proteins had an unusually long region to the N terminus of the Rel-homology domains compared with other insect Rel proteins. A RHD, an IPT, a nucleus localization signal, and six ankyrin repeats were found with the same arrangements. In Drosophila Relish, two serine-rich stretches were at the N-terminal region (S22–S45) and just downstream of the nucleus localization signal (S460–S475), whereas in the mosquito homologue, a long serine-rich stretch was found in the N-terminal region (S31–S84) but not downstream of the nucleus localization signal.
Figure 1.
A comparison between Aedes Relish and Drosophila Relish. (A) Comparison of amino acid sequences. Alignments were done by CLUSTALW 1.8 and manually adjusted. The accession number for Drosophila Relish is U62005. Exon–exon junctions confirmed from the genomic sequence are indicated with arrows. (B) Domain structure of Aedes and Drosophila Relish factors. The four alternative start sites of Drosophila Relish are indicated with the size of corresponding transcripts (kb). The three transcripts of Aedes Relish are reconstructed from three types of clones, R6, R7, and C8, according to RT-PCR and 3′-RACE results. (C) Comparison of death domains. Accession numbers of Swiss-Prot for other sequences shown are: human p105, P19838; human p100, Q00653; mouse IRAK, Q9QY63; Drosophila pelle, Q05652. (D) Phylogenetic comparison of Aedes Relish with other Rel proteins. The distance matrix analysis is based on the Rel- homology domains only. Numbers indicate the percentage of bootstrap replications that support each branch.
The unique features of Aedes Relish primary sequence compared with Drosophila were the presence of a short His/Gln-rich stretch at the N terminus and of a death domain at the C terminus (Fig. 1 A–C). Glutamine-rich domains constitute one of the three main classes of transcriptional activation domains in transcription factors; they are often associated with histidine-rich stretches. These domains promote protein–protein interactions that facilitate the recruitment of transcription initiation complexes (28). A much longer His/Gln stretch is found in the transactivation domain of Drosophila Dorsal (2, 29).
The Pfam profile search with Aedes Relish indicated the presence of a death domain located at the extreme C terminus. On the basis on the alignment and followed phylogenetic analysis, the death domain found in the mosquito Relish was related to those of p105 and p100, the vertebrate homologue to Relish, to tube and pelle, factors involved in Drosophila development, and several other proteins, including IRAK (Fig. 1C). In contrast, the C terminus of Drosophila Relish showed a low homology to the death domains of other proteins.
The death domain (FAS/TNF cytosolic interaction domain) has been described as a region in the cytoplasmic tail of the 75-kDa TNF receptor (TNFR-1) which is involved in TNF-mediated cell death signaling (30). Several proteins contain regions with significant similarity to the death domain. In most of these proteins, the death domain is located at the extreme C terminus (31). In Drosophila, the connection between the transduction of cell-death signals and the induction of the antimicrobial response has recently been reported by the dual function of Dredd. This protease of the caspase family, involved both in apoptosis during Drosophila development (32) and in mediating antibacterial resistance (8, 11), specifically associates with dFADD (33) that in turn interacts with dMyD88 (34). The cleavage and following activation of Drosophila Relish during the immune activation by bacterial challenge is effected by an IKK complex and Dredd, which may be directly involved in the Relish cleavage. Further studies are required to understand the role of the death domain in Aedes Relish processing, which may be more reminiscent of p105 processing than of Drosophila Relish.
A phylogenetic study with Rel-homology domains demonstrated that both insect Relish factors were clustered to the same subgroup, distinguished from other Rel family proteins (Fig. 1d). This Relish subgroup could be grouped with p105 and p100, indicating that these Rel/IκB compound proteins might have branched off at an early evolutionary stage from other Rel family proteins including a group of three insect Rel proteins, Dorsal, Dif and Gambif1, and a group of mammalian Rel proteins like p65, RelB, and c-rel. The phylogenetic relationship of IκB domains was not clear; each IκB domain did not cluster with those from other proteins with more than 90% of bootstrap replication (data not shown).
Interesting features were found in R7 clone, the longest representative of a cDNA group consisting of three clones. R7 clone had a deletion of the coding region including most N-terminal sequence and Rel-homology domains (whole of RHD and N-terminal of IPT) (Fig. 1 A and B), thus it was called IκB-type. It was unlikely a truncated cDNA because R7 clone showed longer 5′-UTR sequence compared with R6 clone, and shared the same sequence of 5′ UTR and N-terminal coding region (M1-D16) with R6 clone.
Only a single C8 cDNA clone, so-called Rel-type, had a different 3′ region sequence compared with other two type cDNA clones. The stop codon, TGA, in the ORF was found a base after the merger of the different sequences between C8 and other clones, followed by the 3′ UTR sequence specific to the C8 clone (Fig. 1A).
Aedes Relish Binds Specifically to Insect κB Motifs.
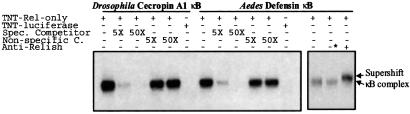
The Drosophila Cecropin A1 gene promoter has a functional κB site (35) and is stimulated by overexpression of Relish after cotransfection of the mbn-2 blood cell line (4). The sequences present in the Rel-only construct without IκB-like domain are sufficient for this effect. Drosophila Relish protein is cleaved into two parts after immune challenge, and the RHD-containing part is translocated to the nucleus, where it binds to the κB motif of the Cecropin A1 promoter (11). To test whether or not the cloned Aedes Relish homologue binds to insect κB motifs, we expressed the Rel-domain region and its downstream without IκB-like domain (M81-A623) by using an in vitro coupled transcription-translation assay. In the electrophoretic gel mobility shift assay, this in vitro expressed protein bound to both κB motifs of Drosophila Cecropin A1 and Aedes Defensin promoters (Fig. 2). In both cases, the addition of a 50-fold excess of the unlabeled specific oligonucleotide effectively completed binding to the labeled probe, whereas the addition of nonspecific competitor, AP1, did not effect the binding. This band was not present in the control reaction with luciferase gene instead of the Rel-domain region, showing that the proteins in the transcription-translation expression system were not involved in this binding. Binding depended on the presence of the κB-like sequences, because the change of the last 2 bp in Cecropin A1 κB motif, from TT to AC, almost abolished the binding (data not shown). The addition of the antibodies against Drosophila Relish RHD peptide caused a supershift of the κB-binding complex. This effect was specific and was not observed with the preimmune serum (Fig. 2).
Figure 2.
Mobility-shift assay for the binding of Relish to insect κB motifs. In vitro-translated protein of Rel-type C8 cDNA clone specifically binds to the κB motif of Drosophila cecropin A1 and Aedes Defensin promoters. The binding is completely abolished by the addition of 50-fold of unlabeled specific competitor, but not by the nonspecific probe, AP1. Supershift of κB-binding complex by addition of antibody against Drosophila Relish is indicated (Left). *, preimmune serum; TNT, transcription-translation.
Three Inducible Relish Transcripts Corresponding to Three Types of cDNA Clones Are Expressed in A. aegypti.
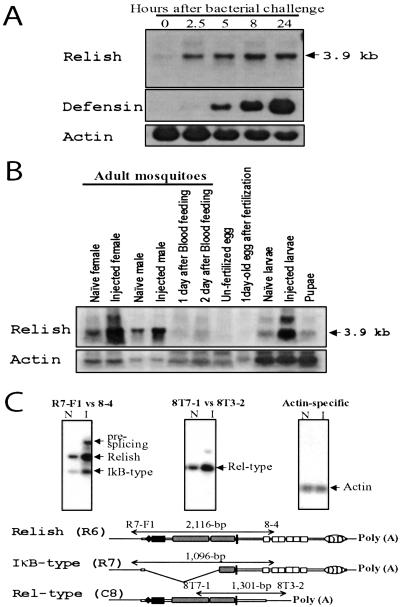
Northern blot analyses were performed to examine the expression of Relish transcripts. Utilization of the total mRNA revealed a 3.9-kb transcript that was constitutively expressed at a low level during the normal unchallenged state of the naïve adult females (Fig. 3 A and B). Probes from both RHD (D81–I330 region of R6) and IκB domain (V677 end of 3′-UTR of R6) hybridized to the same 3.9-kb transcript, suggesting that it represented the major Relish mRNA (data not shown). It was highly induced less than 2.5 h after injection with a stationary-phase culture of Enterobacter cloacae. The level of induced 3.9-kb transcript was constant for the next 24 h. This expression profile was different from that of the Aedes Defensin transcript. The latter was undetectable in naïve mosquitoes, and its induction was weak at 2.5 h after infection. However, the level of Defensin transcript rose exponentially during the next 24 h (Fig. 3A). Additional Northern blot analysis with poly(+) RNA from challenged females and probes from Iκ-B-type (R7) and Rel-type (C8) cDNAs revealed minor additional bands of 2.8 and 3.5 kb, respectively (data not shown).
Figure 3.
Expression of Aedes Relish transcripts during infection and development. (A and B) Northern blot analysis; (C) RT-PCR analysis. (A) Induction of Relish and Defensin mRNA after bacterial challenge in female mosquitoes. Lane numbering indicates the number of hours after injection. The same blot was used for both Relish and Defensin Northern blot hybridizations. (B) The developmental expression of Relish mRNA. (C) Inducible expression of three alternately spliced transcripts shown by Southern analysis of RT-PCR products. The total RNA from naïve (N) and challenged (I) female mosquitoes was reverse-transcribed and amplified with the indicated primer sets. Aedes actin gene was used as RNA-loading control. The 3.2-kb R6 cDNA and actin cDNA were used as probes for hybridization, respectively.
Expression of Relish was examined at several stages of mosquito development. Relish transcript was below the detection level in both unfertilized and fertilized eggs. However, the 3.9-kb transcript was present at similar and relatively high levels in naïve third instar larvae, midpupae, males and previtellogenic females 3–5 days after eclosion, which indicated that Relish was constitutively expressed at most stages of mosquito development. In larvae and adult males examined 5 h after bacterial challenge, the 3.9-kb transcript exhibited inducible expression similar to that observed in previtellogenic females (Fig. 3B). In females, expression of Aedes Relish was reduced during vitellogenesis after its activation by blood feeding (Fig. 3B).
Expression of putative transcripts corresponding to three Relish cDNA clones, Relish (R6), IκB-type (R7), and Rel-type (C8), was investigated further by using RT-PCR followed by hybridization analysis (Fig. 3C). To ascertain whether the deletion in the IκB-type transcript, located between the N terminus and RHD, represented an alternative RNA splicing of the precursor RNA as the 3.9-kb transcript, a primer set of R7-F1 and 8–4 spanning the deletion region was used. Because the sequence corresponding to the 8–4 primer was present in the IκB domain, two bands, corresponding to Relish and IκB-type transcripts, were expected to be amplified from the RNA pool of naïve female mosquitoes. Indeed, two bands of the predicted sizes, 2,116 and 1,096 bp, were the products of the PCR with a primer set of R7-F1 and 8–4. Testing of infected mosquitoes 5 h after injection with the same primer set showed that IκB-type transcript was inducible similarly to Relish transcript. In this reaction, a larger transcript was also detected which likely represented prespliced RNA. A similar large transcript was also observed in Northern blot analyses of infected mosquitoes (Fig. 3 A and B).
Similar tests were performed to detect a transcript corresponding to the Rel-type (C8) cDNA clone. RT-PCR was first performed by using RNA from naïve females with a primer set of 8T7–1 and 8T3–2, specific to 3′-UTR sequence of the C8 clone. This reaction revealed the presence of a 1,301-bp transcript that matched the expected size of the C8 cDNA. The Rel-type transcript was also inducible after a challenge with infection (Fig. 3C). The effect of blood feeding on the expression of Rel-type and IκB-type transcripts was also investigated by using RT-PCR and hybridization analysis. Similar to the pattern of Relish shown by Northern analyses, Rel-type and IκB-type transcripts exhibited reduced levels of expression after blood feeding and during vitellogenesis (data not shown). Expression of Relish after blood feeding tested by RT-PCR as a control was in agreement with the data obtained by the Northern blot analyses.
Alternative Splicing and Polyadenylation Are Involved in Generation of Three A. aegypti Relish Transcripts from a Single Gene.
We used RT-PCR analysis to investigate whether the three transcripts have the same 5′-UTR regions (Fig. 1B). The 3,280-bp R6 cDNA clone was smaller than the major 3.9-kb Relish transcript, even though both contained Rel-homology and an IκB-like domain. Thus, it seemed that R6 clone was truncated at its 5′-UTR region. This suggestion was proven by RT-PCR analysis with a primer set of 5-UTR-1 and G3, which could not generate a PCR product from the IκB-type transcript. Sequencing of the resulted 739-bp PCR fragment showed the presence of the same 5′-UTR sequence as in Rel-type transcript. It was only a base pair different from R7 5′-UTR sequence. The 739-bp PCR fragment was used to screen cDNA library, however, attempts to clone full-length Relish with a complete 5′-UTR region had failed.
A 2.5-kb PCR fragment from the RT-PCR experiment with 5-UTR-1 and 8T3–2, a specific primer to 3′-UTR of C8 clone, showed same nucleotide sequence of 5′-UTR and N terminus as the Relish-type transcript, suggesting that all three transcripts shared the same 5′-UTR sequence and most likely originated from the same prespliced RNA. 3′-RACE confirmed that in the Rel-type transcript and C8 cDNA clone, the polyadenylation sequence was present immediately after the 3′-UTR sequence.
Two overlapping genomic clones spanning ≈25 kb were isolated and partially sequenced. This genomic region contained the central portion of Relish, except for 5′-UTR, short stretch of the N terminus (M1–D16), and the C-terminal IκB region (N624 end). Partial sequencing and PCR analyses, focused on the exon–intron structure of the region, revealed that at least eight exons were in the Aedes Relish gene (Figs. 1A and 4A). The exon–intron structure of the gene exhibited strong correlation with the proposed alternative splicing of Aedes Relish RNA (Fig. 4A). The deleted region shown at IκB-type cDNAs coincided with the 5′ start of the second exon and ended at the 3′ end of the fourth exon (Fig. 4A). The 3′-UTR sequence unique to the C8 clone was only a base different from the sequence after the seventh exon, indicating that utilization of alternative polyadenylation was involved in generation of the Rel-type transcript (Fig. 4B).
Figure 4.
The genomic structure of Aedes Relish gene. (A) Schematic diagram of the Aedes relish gene. About 15 kb of genomic DNA containing the second exon to seventh exon has been sequenced (underlined by a double-headed arrow). The 5′-UTR region common to all three transcripts is likely present in the first exon. *, ≈1-kb region flanking the fifth exon was not confirmed by sequencing analysis because of the interference caused by the strong secondary structure. (B) Comparison between genomic sequence of seventh exon and the 3′-UTR sequence of Rel-type transcript indicates the utilization of alternative polyadenylation. A base difference between the 3′-UTR of C8 cDNA and genomic sequences is indicated by shading. The putative polyadenylation site is boxed.
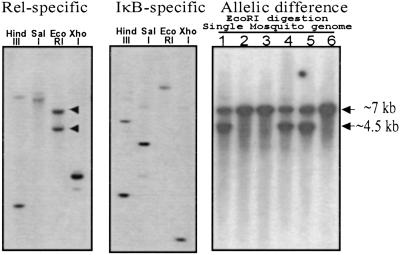
Southern blot analysis was performed by using genomic DNA from 10 mosquitoes and two probes, one expanding RHD (D81–I330 region of R6) and another the IκB domain (V677 end of 3′-UTR of R6). The hybridization results with either probe showed two or three bands of irregular density in a digest with different restriction enzymes (Fig. 5). The banding patterns that resulted from hybridization with each probe were very different, indicating that both Relish domains were apart from each other, separated by large introns.
Figure 5.
Genomic Southern analysis showing the allelic difference and single genomic locus of Aedes Relish. (Left and Center) The genomic DNA from 10 mosquitoes was digested with the indicated restriction enzyme. The same blot was hybridized by Rel-specific (Left) and IκB-specific (Center) probes. (Right) Genomic DNA from individual mosquitoes was digested with EcoRI, blotted, and hybridized with a Rel-specific probe.
To test whether these multiple bands originated from the allelic polymorphism of a single Relish gene or resulted from multiple genomic copies, Southern blot analysis was performed with genomic DNA from six individual mosquitoes. For this analysis, we selected EcoRI digestion and a probe from RHD, which generated two bands of 7 and 4.5 kb. This pattern was present in three of six individuals, whereas three others had only a 7-kb band (Fig. 5). The latter analysis suggested that a single genomic copy of Relish gene existed in A. aegypti, and the two bands observed in the Southern blots of individual mosquitoes originated from allelic difference. The presence of allelic polymorphism was also in agreement with sequence variation found in the nucleotide sequence of cDNA clones. The heterogeneity was between 0 and 2% in the nucleotide sequences of each of the 12 clones, and two amino acid substitutions occurred between the R6 and R7 clones.
Among insect members of the Rel family of immune transcription factors, Relish is unique, possessing both Rel/NF-κB domains and an inhibitory IκB domain. In this respect, it is similar more to the mammalian p100 and p105 factors than to other insect Rel proteins. Previously, only Drosophila Relish has been investigated. In this work, we report characterization of Relish from the mosquito A. aegypti. Significantly, we found three alternatively spliced transcripts of the Aedes Relish gene encoding dramatically different proteins, Relish, Rel-type, and IκB-type. In contrast, the Drosophila Relish gene encodes four transcripts, which originate from alternative start sites and make proteins differently truncated at their N termini.
Mammalian p105 and p100 are processed to release their N-terminal activation domains, p50 and p52, respectively (1). However, the same NF-κB1 gene that encodes p105 generates the NF-κB inhibitor IκBγ by alternative splicing. The formation of Relish and IκB-type transcripts from a single gene in the mosquito seems to occur by the same mechanism. Recruitment of similar products from both mosquito Relish and mammalian NF-κB1 genes during activation by infection suggests an even closer evolutionary link between these two Rel/IκB genes. This evolutionary link further indicates a high level of conservation between regulatory mechanisms of innate immunity pathways.
Acknowledgments
We thank Dan Hultmark and Svenja Stöven (Umeå University, Umeå, Sweden) for the kind gift of the antibodies and Geoff Attardo for critical reading of the manuscript. This work was supported by Grants AI24716 and AI45123 from National Institutes of Health.
Abbreviations
- IκB
inhibitor κB
- 3′-RACE
rapid amplification of cDNA 3′ ends
- UTR
untranslated region
- RT
reverse transcription
- TNF
tumor necrosis factor
Footnotes
References
- 1.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Steward R. Science. 1987;238:692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- 3.Ip Y T, Reach M, Engstrom Y, Kadalayil L, Cai H, Gonzales-Crespo S, Tatei K, Levine M. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 4.Dushay M S, Åsling B, Hultmark D. Proc Natl Acad Sci USA. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govind S, Steward R. Trends Genet. 1991;7:119–125. doi: 10.1016/0168-9525(91)90456-z. [DOI] [PubMed] [Google Scholar]
- 6.Lemaitre B, Reichhart J, Hoffmann J. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedengren M, Asling B, Dushay M S, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 8.Leulier F, Rodriguez A, Khush R S, Chen P, Abrams J M, Lemaitre B. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin L, DeMartino G N, Greene W C. EMBO J. 2000;19:4712–4722. doi: 10.1093/emboj/19.17.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orian A, Gonen H, Bercovich B, Fajerman I, Eytan E, Israel A, Mercurio F, Iwai K, Schwartz A L, Ciechanover A. EMBO J. 2000;19:2580–2591. doi: 10.1093/emboj/19.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman N, Zhou J, Stöven S, Pandey N, Hultmark D, Maniatis T. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams J M. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 14.Vidal S, Khush R S, Leulier F, Tzou P, Nakamura M, Lemaitre B. Genes Dev. 2001;15:1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J, Hoffmann J. Proc Natl Acad Sci USA. 1995;92:9365–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart J, Hoffmann J A. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann J A, Reichhart J M, Hetru C. Curr Opin Immunol. 1996;8:8–13. doi: 10.1016/s0952-7915(96)80098-7. [DOI] [PubMed] [Google Scholar]
- 18.Richman A, Kafatos F C. Curr Opin Immunol. 1996;8:14–19. doi: 10.1016/s0952-7915(96)80099-9. [DOI] [PubMed] [Google Scholar]
- 19.Chalk R, Townson H, Natori S, Desmond H, Ham P J. Insect Biochem Mol Biol. 1994;24:403–410. doi: 10.1016/0965-1748(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 20.Lowenberger C, Bulet P, Charlet M, Hetru C, Hodgeman B, Christensen B M, Hoffmann J A. Insect Biochem Mol Biol. 1995;25:867–873. doi: 10.1016/0965-1748(95)00043-u. [DOI] [PubMed] [Google Scholar]
- 21.Cho W L, Fu Y C, Chen C C, Ho C M. Insect Biochem Mol Biol. 1996;26:395–402. doi: 10.1016/0965-1748(95)00108-5. [DOI] [PubMed] [Google Scholar]
- 22.Richman A M, Bulet P, Barillas-Mury C, Hoffmann J A, Kafatos F C. Insect Mol Biol. 1996;5:203–210. doi: 10.1111/j.1365-2583.1996.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 23.Fallon A M, Sun D. Insect Biochem Mol Biol. 2001;31:263–278. doi: 10.1016/s0965-1748(00)00146-6. [DOI] [PubMed] [Google Scholar]
- 24.Vizioli J, Bulet P, Hoffmann J A, Kafatos F C, Muller H M, Dimopoulos G. Proc Natl Acad Sci USA. 2001;98:12630–12635. doi: 10.1073/pnas.221466798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shahabuddin M, Fields I, Bulet P, Hoffmann J A, Miller L H. Exp Parasitol. 1998;89:103–112. doi: 10.1006/expr.1998.4212. [DOI] [PubMed] [Google Scholar]
- 26.Dimopoulos G, Seeley D, Wolf A, Kafatos F C. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender W, Spierer P, Hogness D. J Mol Biol. 1983;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- 28.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 29.Isoda K, Roth S, Nusslein-Volhard C. Genes Dev. 1992;8:619–630. doi: 10.1101/gad.6.4.619. [DOI] [PubMed] [Google Scholar]
- 30.Cleveland J, Ihle J N. Cell. 1995;81:479–482. doi: 10.1016/0092-8674(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 31.Feinstein E, Kimchi A, Wallach D, Boldin M, Varfolomeev E. Trends Biochem Sci. 1995;20:342–344. doi: 10.1016/s0968-0004(00)89070-2. [DOI] [PubMed] [Google Scholar]
- 32.Chen P, Rodriguez A, Erskine R, Thach T, Abrams J M. Dev Biol. 1998;201:202–116. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 33.Hu S, Yang X. J Biol Chem. 2000;275:30761–30764. doi: 10.1074/jbc.C000341200. [DOI] [PubMed] [Google Scholar]
- 34.Horng T, Medzhitov R. Proc Natl Acad Sci USA. 2001;98:12654–12658. doi: 10.1073/pnas.231471798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engstrom Y, Kadalayil L, Sun S C, Samakovlis C, Hultmark D, Faye I. J Mol Biol. 1993;232:327–333. doi: 10.1006/jmbi.1993.1392. [DOI] [PubMed] [Google Scholar]