Abstract
The peptide pheromone cCF10 of Enterococcus faecalis is an intercellular signal for induction of conjugative transfer of plasmid pCF10 from donor cells to recipient cells. When a donor cell is exposed to recipient-produced cCF10, expression of the pCF10-encoded aggregation substance of pCF10 (Asc10) and other conjugation gene products is activated. Asc10 also increases enterococcal virulence in several models, and when donor cells are grown in animals or in plasma, Asc10 expression is induced by means of the cCF10-sensing machinery. Plasmid pCF10 carries two genes that function to prevent self-induction by endogenous cCF10 in donor cells. The membrane protein PrgY reduces endogenous pheromone activity in donor cells, and the inhibitor peptide iCF10 neutralizes the residual endogenous cCF10 that escapes PrgY. In the current study, we found that E. faecalis strains with allelic replacements abolishing active cCF10 production showed reduced ability to acquire pCF10 by conjugation; prgY-null mutations had no phenotype in the cCF10-negative strains. We observed that expression of the mRNA for iCF10 was reduced in this background and that these mutations also blocked plasma induction of Asc10 expression. These findings support a model in which plasma induction in wild-type donors results from iCF10 inactivation by a plasma component, causing disruption of a precisely maintained balance of iCF10 to cCF10 activity and allowing subsequent induction by endogenous cCF10. Although cCF10 has traditionally been viewed as an intercellular signal, these results show that pCF10 has also adapted cCF10 as an autocrine signal that activates expression of virulence and conjugation functions.
Keywords: cell–cell signaling, Enterococcus, horizontal gene transfer, pCF10
Expression of the conjugation functions encoded by Enterococcus faecalis plasmid pCF10 is induced by the heptapeptide mating pheromone cCF10, which is produced by plasmid-free cells. This communication between donor and recipient cells results in highly efficient transfer of pCF10 and its associated antibiotic resistance and virulence properties through E. faecalis populations. Although intercellular communication systems in Gram-positive bacteria typically employ peptides as signal molecules (1), the mating pheromone system has unique features that distinguish it from most quorum-sensing systems that control expression of multicellular behavior in these bacteria. Quorum-sensing systems involve a single cell type whose population density in a particular niche is sensed by using the signal molecule. In many cases, the genes encoding critical components of both the sensing machinery and production of the signal are genetically linked (2). In contrast, the pCF10 pheromone system involves two distinct populations, donors and plasmid-free recipients. Donors sense cCF10 pheromone produced by nearby recipient cells and induce plasmid transfer to these recipients. In addition, the pheromone-sensing machinery of pCF10 is encoded by one genetically autonomous entity (the plasmid), whereas the ability to produce the pheromone is encoded by a second genetic entity, the chromosome of the host bacterium.
The pheromone is produced by proteolytic processing of a predicted secreted lipoprotein encoded by the ccfA gene in the E. faecalis chromosome (3). A putative integral membrane protease called Eep is involved in processing the CcfA-signal peptide to active cCF10 pheromone (4). After processing in the membrane, mature cCF10 can be found both in the culture medium and associated with the cell wall fraction of recipient cells (5). The pheromone response in donor cells entails binding of the cCF10 peptide by the pCF10-encoded PrgZ protein, followed by import into the cytoplasm by a chromosomal oligopeptide permease ABC transporter (6). The internalized pheromone binds to PrgX and abolishes PrgX negative-regulatory functions, leading to increased expression of conjugation ability (7, 8). The primary target for PrgX regulation is the promoter region of the prgQ operon (7, 9). This operon encodes production of many, if not all, of the pCF10 conjugation factors, including the aggregation substance of pCF10 (Asc10), a surface protein that mediates initial donor/recipient attachment. Pure liquid cultures of wild-type donor cells induced with cCF10 aggregate as visible clumps, and this clumping is used as a simple phenotypic screen for induction. The prgQ promoter is active in uninduced cells, but transcripts in these cells, termed Qs RNAs, extend only ≈400 nt, which is not sufficient for production of conjugation proteins such as Asc10 (10). Pheromone binding to PrgX inactivates the weak repressor function of PrgX, leading to increased initiation of prgQ transcripts and ultimately to extension of these transcripts past a predicted inverted repeat terminator, termed IRS1, and into the distal portion of the operon (7). Extension of transcription past IRS1 likely results from titration of a constitutively expressed terminating antisense RNA Qa (7).
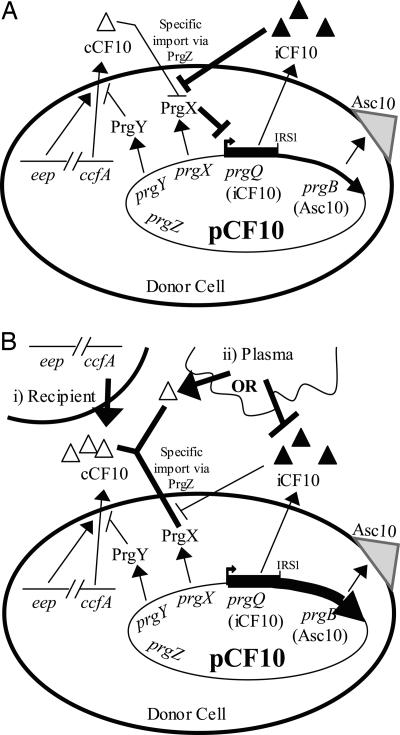
A significant evolutionary problem faced by the plasmid is the ability to enable donor cells to detect the pheromone produced by recipients in close proximity while avoiding energetically wasteful self-induction of conjugation functions by their own endogenous pheromone. Two pCF10 gene products are required to block self-induction, PrgY and iCF10. PrgY is a membrane protein that is essential to block self-induction by pCF10-carrying donors. PrgY has been shown to reduce the amount of both excreted and cell-associated pheromone in donor cells, possibly by binding or degrading cCF10 as it is released from the outer surface of the membrane (5, 11). Control of the residual donor pheromone activity that is not blocked by PrgY is mediated by an inhibitor peptide, iCF10. Interestingly, the iCF10 peptide is encoded within a 66-bp ORF located at the 5′ end of the prgQ operon within Qs (12). Previous analyses indicated that the culture fluids of donor cells contain a mixture of pheromone and inhibitor peptides, with iCF10 being present in excess at an ≈80:1 molar ratio (13). This ratio apparently controls self-induction in donor cells while still allowing a high level of sensitivity to exogenous pheromone produced by nearby recipient cells. Our current view of the operative regulatory circuits in pure donor cultures and in donor/recipient mating mixtures is shown in Fig. 1.
Fig. 1.
Model of regulation of pheromone and pheromone response in a donor cell. (A) Donors in pure culture. PrgY reduces endogenous cCF10, and iCF10 neutralizes remaining cCF10. PrgX represses Asc10 expression so that Asc10 is not induced. (B) Donors induced by cCF10 from a nearby recipient cell (i) or growth in plasma (ii). In both cases, PrgX repression is abolished by binding of imported cCF10 and Asc10 is expressed. Induction in plasma (ii) occurs through two possible mechanisms: a small host molecule (probably a peptide) with cCF10 activity acts as a direct signal to induce Asc10 expression, or a host factor (possibly albumin; see text) sequesters iCF10, altering the ratio of peptides and allowing endogenous cCF10 to act as an autocrine inducer.
The medical significance of the enterococcal pheromone systems is increased by the fact that expression of the conjugation machinery not only enhances the spread of antibiotic resistance determinants but also enhances virulence of the host bacteria. For example, the pheromone-induced Asc10 protein (and the corresponding homologous proteins encoded by other pheromone plasmids) has been shown to augment the ability of E. faecalis to attach and invade mammalian epithelial cells (14, 15), to resist killing by phagocytes (16), and to produce large cardiac vegetations in experimental endocarditis (16, 17). Interestingly, expression of Asc10 and other pCF10 conjugation proteins is induced in the absence of recipient cells when E. faecalis harboring pCF10 is grown in vivo or in human or rabbit plasma; this induction depends on the pheromone-sensing system (17). We could not discern from previous data whether in vivo induction was due to a host-encoded factor with direct pheromone activity or due to inactivation of iCF10, resulting in a shift in the balance between endogenously produced cCF10 and iCF10 peptides in favor of cCF10 (Fig. 1B).
Here, we report the construction and characterization of E. faecalis strains with null mutations in the ccfA gene that abolish cCF10 production. Analysis of these mutants confirmed the important role of pheromone production by recipients in plasmid acquisition and showed that the sole function of the prgY gene is to prevent self-induction of donor cells by reducing endogenous pheromone production. We also found that induction of plasmid transfer in human plasma is abolished in mutant strains carrying pCF10. This result suggests that, in addition to using the chromosomally encoded bacterial cCF10 peptide to promote its dissemination, pCF10 has also taken advantage of this pheromone to up-regulate expression of Asc10 in the mammalian bloodstream in the absence of bacterial recipient cells, where it can function to enhance survival and virulence of the bacterial host. Further, we report results demonstrating that the inhibitor peptide iCF10 is regulated in response to endogenous pheromone levels, which provides evidence for precise plasmid regulation of the iCF10:cCF10 balance. This balance facilitates both a sensitive response to recipient cells and the ability to induce virulence expression in vivo.
Materials and Methods
Bacterial Strains, Culture Conditions, and Plasmids. E. faecalis was grown at 37°C in Todd–Hewitt broth (THB) (Difco) or M9-YE medium (27), a semidefined M9-based medium supplemented with 0.3% yeast extract, 1% casamino acids, 0.1% glucose, 1 mM MgSO4, and 0.1 mM CaCl2, supplemented with antibiotics at the following concentrations: tetracycline, 10 μg/ml; erythromycin, 10 μg/ml; chloramphenicol, 20 μg/ml; rifampicin, 200 μg/ml; streptomycin, 1,000 μg/ml; and spectinomycin, 1,000 μg/ml. X-Gal was used at 250 μg/ml, and 5-fluorouracil was used at 130 μg/ml. Human plasma was used fresh and was acquired by drawing blood from healthy human volunteers and by adding heparin to a final concentration of 100 units/ml, followed by centrifugation for 10 min at 2,500 rpm in a standard clinical centrifuge. All plasmids were expressed in E. faecalis strains OG1RF (18), OG1SSp (18), or OG1Sp (C. J. Kristich, personal communication) or in the OG1RF-derived strains CK104, JRC101, or JRC102. CK104 encodes a chromosomal deletion of the upp2 gene encoding sensitivity to the base analog 5-fluorouracil (19). CK104 was used in this study to make JRC101 and JRC102 by allelic exchange according to Kristich et al. (19). Plasmids pJRC101 or pJRC102, containing the ccfA mutations ccfA1 and ccfA2, respectively (Table 1), were transformed into E. faecalis strain CK104 carrying pVE6007 (20). Upon raising the temperature to 37°C, plasmid pVE6007 was lost and chromosomal integrants of pJRC101 or pJRC102 were selected. These strains were grown for ≈20 generations at 37°C, then 5-fluorouracil was used to select for excision of the integrated plasmid. These strains were screened for the desired ccfA mutations by their pCF10-recipient ability, and mutations were confirmed by sequencing.
Table 1. Mutations made in ccfA.
| Strain | Mutation | CcfA amino acid sequence* |
|---|---|---|
| OG1RF† | Wild type | MKKYKRLLMAGLVTLVFVLSACG... |
| JRC101 | ccfA1 | MKKYKRLLMAGLATLVFVLSACG...‡ |
| JRC102 | ccfA2 | MKKYKRLLMAG stop stop§ |
Active cCF10 sequence is in boldface.
Strain CK104 also has this sequence.
GTG encoding valine at the second position of cCF10 was changed to GCG encoding alanine (underlined).
TTA GTG encoding leucine and valine at the first and second positions of cCF10 were changed to TAA TAA encoding two consecutive stop signals.
Plasmid Transfer. Overnight cultures (15 hr) were grown at 37°C in M9-YE (for plasma or cCF10-induction measurement) or THB. These were diluted 1:10 into human plasma, M9-YE or THB, and donors were induced with cCF10 or left uninduced. These cultures were incubated for 1 hr, and then donors and recipients were combined for mating and incubated at 37°C for the indicated time. In the case of plasma or cCF10 induction, donors and recipients were incubated for 2 hr in the presence of cCF10, plasma, or M9 medium (as a control), followed by a 10-min incubation of mating mixtures. In both cases, donors and recipients were combined in a 1:9 ratio, respectively. Transconjugants were enumerated by serial dilution onto THB agar, and either transconjugants (recipient + plasmid) or donors were selected for to determine the transconjugants per donor. Measurements reported represent means from duplicate assays, and error bars represent 1 SD from the mean. Assays were repeated at least two times in duplicate; representative results from one experiment performed in duplicate are shown.
Affinity Chromatography. C-terminal tyrosinated iCF10 (iCF10Y, Microchemical Facility, University of Minnesota) was coupled to a Sepharose 6B column (Amersham Pharmacia) according to the manufacturer's recommendations. Fresh human plasma was diluted 1:100 in PBS (10 mM sodium phosphate, pH 7.4/0.9% NaCl), and 500 μl was incubated with 3.5 ml of the column material for 30 min under gentle rotation at room temperature. The material was washed with 50 ml of PBS and eluted with 5 ml of acetonitrile. The eluate was aliquoted, lyophilized, and stored at –20°C until it was used for further experiments. For gel electrophoresis, 50% of a lyophilized sample eluate was used.
Protein Preparation, SDS/PAGE, Silver Stain, and Western Analysis. For Asc10, cell extracts were prepared and analyzed as described (11) except that cells were grown overnight in M9-YE medium, then diluted 1:10 into fresh M9, plasma, or M9 + 1ng/ml cCF10 and grown for 2 hr at 37°C before harvesting. SDS/PAGE and Western blot analyses were performed as described in ref. 21 or with use of the ECL system (Pierce) according to manufacturer recommendations. Anti-human albumin antibodies (Sigma) were used in a 1:1,000 dilution. Silver staining of SDS/PAGE gels was performed as described in ref. 22.
RNA Preparation and Northern Blot Analysis. Purification of total RNA and Northern hybridization analysis were performed as described in ref. 23 except that cells were grown in M9-YE overnight (15 hr), diluted 1:5, and grown for an additional 20 min. RNA was isolated by using the BIO101-FastRNA RE kit (Qbiogene), and RNA samples were separated on a 2% agarose gel containing 2% formaldehyde and 1× Mops (20 mM morpholinopropanesulfonic acid). RNA was transferred to a nylon membrane (Roche Applied Science, Indianapolis) and probed by digoxigenin-labeled anti-Qs RNA, which was generated by in vitro transcription as described (24). The chemiluminescent substrate CDP-STAR (Roche Applied Science, Indianapolis) was used for detection of the digoxigenin-labeled probe. The x-ray films were then analyzed by using the Bio-Rad model GS-700 Imaging Densitometer (2400 dpi) at the University of Minnesota Biomedical Image Processing Laboratory and the Bio-Rad molecular analyst (version 21) software. The profile area of each band was analyzed and depicted as OD·mm.
Results
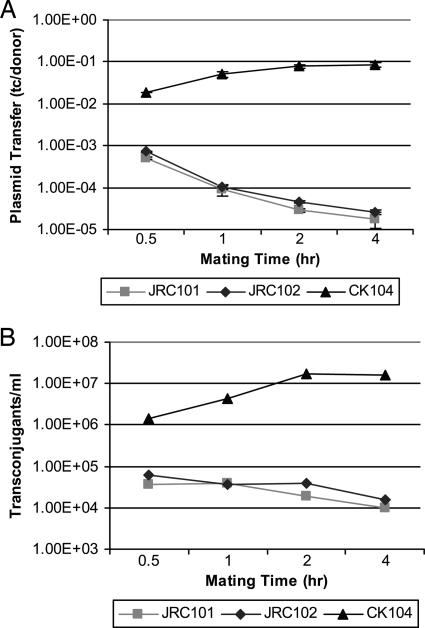
Mutations in ccfA That Abolish cCF10 Activity Reduce pCF10-Recipient Ability. Previously, cCF10 was found to be encoded within ccfA in the chromosome of E. faecalis (3). At the time, deletion or disruption of ccfA within the chromosome could not be achieved. Recently, our group developed a more effective method of markerless allelic exchange in E. faecalis (19). We used this method to create two isogenic strains, JRC101 and JRC102, carrying chromosomal mutations in ccfA (see Materials and Methods). The first mutation, ccfA1, was a single base change in the valine codon at the second position of the cCF10 coding sequence, resulting in a V to A transition. The resulting peptide, LATLVFV, was previously shown to have no measurable cCF10 activity (3). The second mutation, ccfA2, replaced the leucine and valine codons at the first and second positions of cCF10 with two stop codons (TAA TAA), so that neither the cCF10 peptide nor processed CcfA protein would be translated (Table 1). The growth rate of JRC101 and JRC102 in both THB and M9 medium were found to be similar to that of the parental strain CK104 (data not shown). These strains were also tested in a conjugation assay for their pCF10-recipient ability. Transfer of pCF10 into both JRC101 and JRC102 was reduced as compared with the parental strain CK104 (Fig. 2). These results confirm that the ccfA1 and ccfA2 mutations abolish production of functional cCF10. Surprisingly, the total number of transconjugants generated from pCF10 transfer into JRC101 and JRC102, as well as the frequency of pCF10 transfer into these strains, showed a marked decrease as the length of mating increased (Fig. 2). This trend was opposite that of the wild type, CK104. The total donors and recipients in the mating mixtures at each time point were enumerated, and there were no significant differences among the three mating assays at any time point (data not shown). This finding indicates that the decreased number of transconjugants obtained after longer periods of mating is not due to a significant die-off of the mutant recipient strains. In addition, several independent JRC102(pCF10) transconjugants stably maintained the pCF10 plasmid after six subsequent transfers in THB medium without antibiotics (data not shown), indicating the plasmid is stably maintained in cCF10-negative strains once it has been acquired.
Fig. 2.
Mutations in the ccfA gene, which encodes cCF10 pheromone (JRC101 and JRC102), decrease pCF10 recipient ability compared with wild type (CK104). OG1SSp(pCF10) was used as a donor strain. Overnight cultures were diluted 1:10 in THB and were grown for 1 hr before donors and recipients were combined to begin mating. Mating was allowed to proceed for the time indicated before plating. (A) Plasmid transfer depicted as transconjugants per donor cell. Experiments were done in duplicate; error bars represent one standard deviation of the mean. (B) Total numbers of transconjugants per ml.
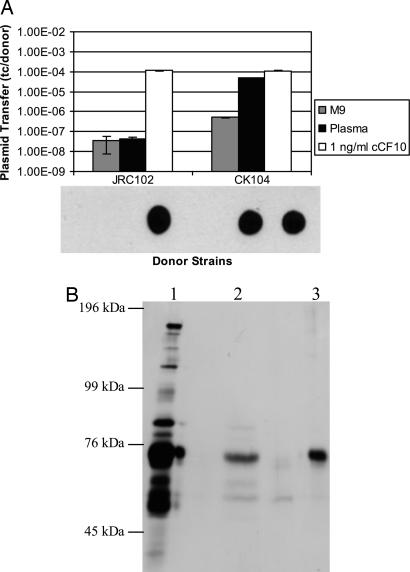
pCF10 Transfer out of JRC102 Is Not Inducible by Human Plasma. Until now, it has been difficult to distinguish between two models for induction by human plasma; either plasma contains a cCF10-like inducing agent or the balance of inhibitor to pheromone activities produced by the donor cells is shifted in favor of pheromone through sequestration or suppression of inhibitor peptide (Fig. 1B). We sought to distinguish between these two possibilities by using strain JRC102. Plasma induced pCF10 to transfer out of strain CK104, but it did not induce pCF10 transfer from strain JRC102 (Fig. 3A). This trend was also seen when the recipient strain and source of human plasma were varied (data not shown). Transfer of pCF10 out of both strains was elevated by the addition of 1 ng/ml synthetic pheromone (Fig. 3A). In addition, induction of pCF10 conjugation by plasma was restored when the plasmid was transferred back to the CK104 strain from JRC102 (data not shown). Asc10 expression was similarly shown to be induced by plasma in CK104 but not in JRC102 (Fig. 3A). These results indicate that plasma induces pCF10 transfer and Asc10 expression by causing endogenous pheromone induction probably through sequestering or suppressing iCF10 activity and disrupting the iCF10 to cCF10 ratio.
Fig. 3.
Endogenous pheromone is required for induction by plasma. (A) pCF10 transfer and Asc10 expression of donor strains JRC102(pCF10) or CK104(pCF10). pCF10 transfer and Asc10 expression are induced by plasma in strain CK104 (wild-type) but not in the cCF10-negative strain JRC102. Strain OG1Sp was used as a recipient for mating. For quantification of plasmid transfer, overnight cultures of recipients and donors were diluted 1:10 in M9 medium (with or without 1 ng/ml cCF10) or in plasma and grown for 2 hr before donors and recipients were combined to begin mating. Mating mixtures were incubated for 10 min before plating. Plasmid transfer is depicted as transconjugants per donor. Experiments were done in duplicate; error bars represent 1 SD about the mean. Immunoblot analysis for Asc10 by using a polyclonal anti-Asc10 antibody is depicted below the mating results; samples are in the same order. As for the mating assay, overnight cultures were diluted 1:10 in M9 medium (with or without 1 ng/ml cCF10) or in plasma. Cell extracts were washed twice before harvesting. An equivalent amount of protein was spotted for each sample. (B) Retention of albumin by fractionation of plasma on an iCF10 affinity column. The iCF10 peptide was coupled with the carboxyl terminus to Sepharose 6B. The matrix material was incubated with human plasma and eluted with acetonitrile (see Supporting Text, which is published as supporting information on the PNAS web site). Eluate was separated on an SDS/7.5% PAGE gel and silver stained. Lane 1, human plasma 1:1000; lane 2, acetonitrile eluate from the affinity column; lane 3, purified (commercially obtained) human serum albumin (100 ng).
iCF10 Affinity Columns Specifically Retain Albumin from Human Plasma. According to our model, plasma could influence the balance of the cCF10 and iCF10 peptides if it contained a factor that preferentially interacted with iCF10. An affinity column with iCF10 coupled to Sepharose 6B was used to identify plasma components with iCF10-binding acitivity. After passing plasma over the column, a 60-kDa protein was retained (Fig. 3B), which was subsequently identified as albumin by Western blot analysis (Fig. 5, which is published as supporting information on the PNAS web site). Independent confirmation that albumin acted as an inducer was also obtained by fractionating plasma over a gel filtration column and determining that the active fractions corresponded to those containing albumin (data not shown). Further data implicating albumin/lipid complexes as the plasma components responsible for alteration of the iCF10/cCF10 balance are presented as Figs. 6 and 7, which are published as supporting information on the PNAS web site.
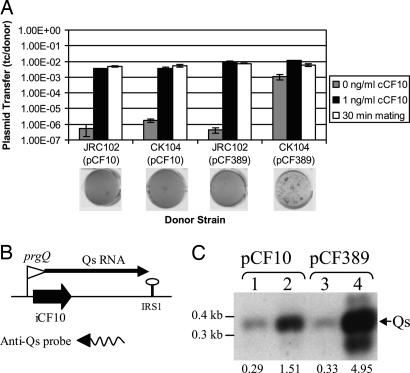
The Ratio of iCF10 to cCF10 Activity in a Donor Cell Is Regulated by Both PrgY and Endogenous Pheromone. By the model proposed above for plasmid induction in vivo, the ratio of endogenous iCF10 to cCF10 must be maintained in a precise balance when in noninducing conditions so that disruption of this balance results in a sensitive response in vivo. The balance between iCF10 and endogenous cCF10 has long been believed to be important also for donor sensing of pheromone from recipient cells, but until now the mechanism for controlling this balance has not been explored. It appears that a necessary requirement for controlling this balance is the reduction of endogenous cCF10 activity by PrgY (5, 11), but previous data did not elucidate whether PrgY played additional roles in regulation, such as affecting the response of donor cells to exogenous pheromone. To address these possibilities, a PrgY-null plasmid, pCF389, was used. In wild-type CK104, pCF389 is self-induced, as can be seen by elevated plasmid transfer levels in uninduced conditions (Fig. 4A). In addition, CK104(pCF389) visibly clumps in broth culture due to constitutive Asc10 expression (Fig. 4A, clumping is depicted as aggregation of cell culture as seen from above). In the cCF10-negative strain JRC102, pCF389 did not have elevated transfer levels or a clumpy phenotype and resembled the phenotype of CK104(pCF10) and JRC102(pCF10) (Fig. 4A). These results indicate that the cells carrying the prgY-null plasmid pCF389 do not express conjugation genes constitutively in the absence of endogenous pheromone. Upon addition of exogenous pheromone, pCF389 had the same response as that of pCF10 in both strains, and similar transfer frequencies of both plasmids were also seen when the mating time was increased to allow donor induction by pheromone synthesized by the recipient cells during the mating (Fig. 4A). These results indicate that the sole function of PrgY is to reduce endogenous pheromone, and that it is not involved in regulating response to exogenous pheromone.
Fig. 4.
Regulation of plasmid transfer and Qs (encoding iCF10) expression by PrgY and by endogenous pheromone. (A) Transfer of pCF10 and pCF389 (prgY-null) out of CK104 and JRC102 donor strains. OG1Sp was used as a recipient strain. Pheromone induction (0 and 1 ng/ml cCF10 samples) experiments were carried out by a protocol different from that with the longer mating time (30-min sample), with the data displayed side-by-side for comparison. For 0 and 1 ng/ml cCF10, donors and recipients were grown overnight in M9 medium and diluted 1:10, then grown an additional 2 hr in the presence or absence of 1 ng/ml cCF10 and combined for a 10-min mating before plating. For the 30-min samples, donor and recipient strains were grown overnight in THB, diluted 1:10, and grown for 1 hr in the absence of cCF10, then they were combined for 30 min to allow induction of donors by recipient pheromone before plating. Plasmid transfer is depicted as transconjugants per donor. Experiments were done in duplicate; error bars represent one SD about the mean. Clumping assay results for each strain are shown below. Strains induced for plasmid transfer express Asc10 and clump in liquid culture (see text), which causes uneven settling at the bottom of the well (lane 4) of a 96-well plate. Overnight cultures were grown in THB without pheromone and were shaken for 2 hr before a picture was taken. (B) Location of the Qs probe within the regulatory region of pCF10 and pCF389. The digoxigenin-labeled RNA probes were generated by in vitro transcription. The 3′ end of the probes is indicated by an arrowhead. (C) Results of Northern blotting analysis of Qs RNA levels. For Northern analysis, 1 μg of total RNA was separated on a 2% agarose gel, transferred onto nylon membrane, and hybridized with the Qs probe shown in B. The arrow indicates Qs. Strains depicted are as follows: JRC102 (lanes 1 and 3) and CK104 (lanes 2 and 4). These strains harbor the plasmid pCF10 (lanes 1 and 2) or pCF389 (lanes 3 and 4). Relative amounts of Qs as determined by densitometry are displayed below each lane. These data were obtained by determining the profile area with the Bio-Rad molecular analyst (Version 21) software.
Endogenous pheromone activity is not abolished by PrgY (5, 11), so in an uninduced donor cell the plasmid-produced inhibitor peptide iCF10 must be at a level that just sufficiently neutralizes the remaining endogenous pheromone activity. For the plasmid to ensure an exact balance of iCF10 and cCF10 activities, we hypothesized that iCF10 production is regulated by the endogenous cCF10 in donor cells. To explore this hypothesis, the abundance of Qs RNA (encoding iCF10) in the cCF10-negative strain JRC102 was analyzed by Northern hybridization using a Qs RNA probe (Fig. 4B). The results showed that pCF10 Qs RNA was 5-fold less abundant in JRC102 than in wild-type CK104 (Fig. 4C, lanes 1 and 2), strongly supporting the notion that endogenous pheromone regulates Qs RNA levels. CK104 carrying the prgY-null plasmid pCF389 had high Qs RNA levels as compared with CK104(pCF10) (Fig. 4C, lanes 3 and 4) because of higher levels of endogenous pheromone in this strain. Qs RNA from pCF389 was low in the cCF10-negative strain JRC102 (Fig. 4C, compare lanes 1 and 3), further supporting the notion that the level of Qs RNA depends on the level of endogenous pheromone. These results show that in wild-type donor cultures endogenous cCF10 levels are reduced by PrgY, and the remaining cCF10 acts as a signal for regulation of iCF10 production. This signaling maintains a threshold balance of iCF10 and cCF10, which was previously believed to be important for a sensitive response to pheromone from recipient cells and has now also been shown to be important for plasmid induction in vivo.
Discussion
In this study, we report the construction and characterization of two E. faecalis strains with abolished production of cCF10 pheromone and the use of one of these strains to elucidate a model for the induction of pCF10 transfer and virulence factor expression in vivo. Both mutant strains exhibited normal growth characteristics, but pCF10 transfer into these strains was reduced compared with wild type. In 30-min matings, transfer into the mutant strains was reduced to ≈10% of the wild-type level. This low level of pheromone-independent plasmid transfer was previously observed in 10-min matings (25) and may be due to a low level of transient self-induction in donor cells. As the length of mating time increased, the total numbers of transconjugants and the transfer frequencies decreased substantially (Fig. 2) in contrast with the temporal increase in transconjugants observed with wild-type recipients. This decrease may be explained by the fact that cCF10 interacts with the PrgW replication protein (6) and may play a role in plasmid maintenance. We believe that wild-type pCF10 may not be stably maintained in cCF10-negative strains and is rapidly lost before selective plating on antibiotics (mating mixtures are incubated in the absence of antibiotic selection; see Materials and Methods). If newly acquired pCF10 plasmid is lost by transconjugants in the mating mixture, and the numbers of new transfer events decrease with time as nutrients are depleted in the growth medium (25), then there would be a temporal decrease in transconjugants, as seen in Fig. 2. In the presence of antibiotic selection, mutations that support pheromone-independent plasmid maintenance could be selected for before the unstable plasmid is segregated. Mutations of this nature would promote stable maintenance of pCF10 in cCF10-negative strains without affecting pheromone-inducible plasmid transfer, because transfer of pCF10 from the JRC102 background when the donors were induced with exogenously supplied cCF10 was identical to wild-type donors (Figs. 2 and 4). In addition, normal donor transfer properties were retained through multiple rounds of transfer of pCF10 between JRC102 and wild-type strains (data not shown).
We previously reported that expression of pCF10-encoded Asc10 and conjugative transfer functions is induced by growth in vivo or in plasma in the absence of bacterial recipient cells (17). This induction depended on the pCF10 PrgZ pheromone-binding protein, suggesting that cCF10 or a closely related peptide was the active inducing molecule. It was also observed that incubation of mixtures of iCF10 and cCF10 with plasma affected the biological activities of these mixtures, shifting the ratio of pheromone activity to inhibitor activity in favor of pheromone (17). This shift suggested that one or more plasma components could preferentially sequester or degrade iCF10, leading to self-induction by cCF10 of endogenous origin, but the alternative possibility that a host factor could have a direct inducing activity was not eliminated. The most significant result was the observation that a mutation in the chromosomal ccfA gene that abolished pheromone production also prevented induction of expression of the conjugation system by pCF10-carrying E. faecalis strains grown in human plasma. The response of these strains to exogenously supplied cCF10 pheromone was not impaired by the ccfA mutations.
The present results argue strongly against the existence of a pheromone-like inducing factor of host origin in human plasma and imply instead that a plasma host factor(s) reduces iCF10 activity relative to cCF10. Our results indicate that albumin in human plasma interacts with iCF10 (Fig. 3B). As described in the supporting information, we also found that pure albumin/oleic acid complexes have essentially the same biological activities as whole plasma in terms of inducing Asc10 expression (Fig. 6) and in altering the ratios of iCF10 to cCF10 activity in solutions containing mixtures of the two peptides (Fig. 7). Thus, it is possible that interaction of certain albumin/lipid complexes with the bacterial peptides in vivo leads to induction by shifting the effective ratios of endogenous cCF10 to iCF10 (Fig. 1B, ii). This shift in ratios could result either from preferential binding of iCF10 in solution or from a higher rate of iCF10 inactivation due to the propensity of cCF10 to remain cell-associated (5). The present results clearly demonstrate a requirement for endogenous cCF10 in plasma induction, but the available data on the interactions of the bacterial peptides with albumin complexes are not yet sufficient to prove that these complexes are the critical host factors.
The finding that iCF10 expression is regulated in response to endogenous pheromone provides a mechanistic explanation for the precise balance of pheromone and inhibitor activities previously observed in cultures of wild-type donor cells (Fig. 4). The cumulative results reported here indicate that endogenous pheromone acts as a signal for production of an appropriate level of iCF10, and that it also acts as the inducing agent for expression of conjugation genes when iCF10 activity is decreased by plasma. The results reported here increase our knowledge of the complexity of this system and of the remarkable extent of coevolution of the plasmid with its host so that endogenous pheromone is neutralized with the minimal amount of iCF10 required to block a mating response and no more.
Regulation of expression of pCF10 transfer and virulence in vivo appears to be similar to the mechanism of induction of cytolysin expression in E. faecalis recently reported by Coburn et al. (26). The inhibitor of transcription of the cytolysin operon was hypothesized to be sequestered by erythrocytes, thereby disrupting the ratio of inducer to inhibitor and causing induction of cytolysin transcription. The cytolysin inducer and inhibitor are also short peptides but are completely unrelated to iCF10 and cCF10. It is striking that these two systems, which probably evolved separately, both appear to use an interaction of a host component with one member of a two-peptide signaling system to control virulence factor expression in vivo. These two signaling systems require no physical contact between the eukaryotic and bacterial cells, nor do they rely on a direct signal produced by the infected host. Instead, they both sense a balance of two antagonizing peptides and respond to the disruption of that balance that occurs upon invasion of a mammalian host. The mechanism of pCF10 induction in vivo demonstrates additional complexity in that this balance is the result of a symbiotic relationship in which the inducing peptide is produced by the host bacterial cell and the neutralizing peptide is produced by the plasmid, and the result is advantageous to both entities.
Supplementary Material
Acknowledgments
We thank O. Chuang, B. Kozlowicz, I. Vlasova, A. Pragman, and P. Schlievert for technical assistance with plasma acquisition and mating experiments, and C. Waters for comments on the manuscript. This research was supported by National Institutes of Health Grants HL51987 and GM49530 (to G.M.D.). J.R.C. is a predoctoral trainee under National Institutes of Health Grant T32DE07288.
Author contributions: J.R.C., H.H., and G.M.D. designed research; J.R.C. and H.H. performed research; J.R.C., H.H., and G.M.D. analyzed data; and J.R.C. and G.M.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: Asc10, aggregation substance of pCF10.
References
- 1.Lyon, G. J. & Novick, R. P. (2004) Peptides 25, 1389–1403. [DOI] [PubMed] [Google Scholar]
- 2.Dunny, G. M. & Leonard, B. A. (1997) Annu. Rev. Microbiol. 51, 527–564. [DOI] [PubMed] [Google Scholar]
- 3.Antiporta, M. H. & Dunny, G. M. (2002) J. Bacteriol. 184, 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, F. Y., Sulavik, M. C. & Clewell, D. B. (1999) J. Bacteriol. 181, 5915–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buttaro, B. A., Antiporta, M. H. & Dunny, G. M. (2000) J. Bacteriol. 182, 4926–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonard, B. A., Podbielski, A., Hedberg, P. J. & Dunny, G. M. (1996) Proc. Natl. Acad. Sci. USA 93, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae, T., Kozlowicz, B. K. & Dunny, G. M. (2004) Mol. Microbiol. 51, 271–281. [DOI] [PubMed] [Google Scholar]
- 8.Kozlowicz, B. K., Bae, T. & Dunny, G. M. (2004) Mol. Microbiol. 54, 520–532. [DOI] [PubMed] [Google Scholar]
- 9.Bae, T., Kozlowicz, B. & Dunny, G. M. (2002) J. Mol. Biol. 315, 995–1007. [DOI] [PubMed] [Google Scholar]
- 10.Chung, J. W. & Dunny, G. M. (1995) J. Bacteriol. 177, 2118–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler, J. R., Flynn, A. R., Bryan, E. M. & Dunny, G. M. (2005) J. Bacteriol. 187, 4830–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama, J., Ruhfel, R. E., Dunny, G. M., Isogai, A. & Suzuki, A. (1994) J. Bacteriol. 176, 7405–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama, J., Dunny, G. M., Clewell, D. B. & Suzuki, A. (1995) Dev. Biol. Stand. 85, 35–38. [PubMed] [Google Scholar]
- 14.Kreft, B., Marre, R., Schramm, U. & Wirth, R. (1992) Infect. Immun. 60, 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waters, C. M., Wells, C. L. & Dunny, G. M. (2003) Infect. Immun. 71, 5682–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlievert, P. M., Gahr, P. J., Assimacopoulos, A. P., Dinges, M. M., Stoehr, J. A., Harmala, J. W., Hirt, H. & Dunny, G. M. (1998) Infect. Immun. 66, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirt, H., Schlievert, P. M. & Dunny, G. M. (2002) Infect. Immun. 70, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunny, G. M., Brown, B. L. & Clewell, D. B. (1978) Proc. Natl. Acad. Sci. USA 75, 3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristich, C. J., Manias, D. A. & Dunny, G. M. (2005) Appl. Environ. Microbiol. 71, 5837–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguin, E., Duwat, P., Hege, T., Ehrlich, D. & Gruss, A. (1992) J. Bacteriol. 174, 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tortorello, M. L. & Dunny, G. M. (1985) J. Bacteriol. 162, 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wray, W., Boulikas, T., Wray, V. P. & Hancock, R. (1981) Anal. Biochem. 118, 197–203. [DOI] [PubMed] [Google Scholar]
- 23.Bae, T., Clerc-Bardin, S. & Dunny, G. M. (2000) J. Mol. Biol. 297, 861–875. [DOI] [PubMed] [Google Scholar]
- 24.Bae, T. & Dunny, G. M. (2001) Mol. Microbiol. 39, 1307–1320. [DOI] [PubMed] [Google Scholar]
- 25.Dunny, G., Yuhasz, M. & Ehrenfeld, E. (1982) J. Bacteriol. 151, 855–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coburn, P. S., Pillar, C. M., Jett, B. D., Haas, W. & Gilmore, M. S. (2004) Science 306, 2270–2272. [DOI] [PubMed] [Google Scholar]
- 27.Dunny, G. M. & Clewell, D. B. (1975) J. Bacteriol. 124, 784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.