Abstract
Chronic microbial infections are associated with fibrotic and inflammatory reactions known as granulomas showing similarities to wound-healing and tissue repair processes. We have previously mapped three leishmaniasis susceptibility loci, designated lmr1, -2, and -3, which exert their effect independently of T cell immune responses. Here, we show that the wound repair response is critically important for the rapid cure in murine cutaneous leishmaniasis caused by Leishmania major. Mice congenic for leishmaniasis resistance loci, which cured their lesions more rapidly than their susceptible parents, also expressed differentially genes involved in tissue repair, laid down more ordered collagen fibers, and healed punch biopsy wounds more rapidly. Fibroblast monolayers from these mice repaired in vitro wounds faster, and this process was accelerated by supernatants from infected macrophages. Because these effects are independent of T cell-mediated immunity, we conclude that the rate of wound healing is likely to be an important component of innate immunity involved in resistance to cutaneous leishmaniasis.
Keywords: fibroblasts, leishmania, macrophages, resistance, wound healing
Infection by many pathogenic microorganisms results in extensive remodeling of the infected tissue and production of granulomas. Schistosomiasis has been used as a model for the study of granuloma biology, with Schistosoma mansonii eggs stimulating the production of extracellular matrix and the formation of protective granulomas (1, 2). Infection by Mycobacterium tuberculosis results in the production of granulomas that simultaneously inhibit the spread of the organism and offer a safe environment for microbial persistence. Similarly, many Leishmania spp. cause skin granulomas resulting in chronic, indolent ulcers, which upon healing are remodeled into normal structural or scar tissue. In response to infection, host cells extensively remodel tissues by producing collagens and matrix metalloproteinases (3, 4). Fibroblasts migrate into the region, resulting in the destruction of the normal tissue architecture and the laying down of nascent connective tissue matrices (5–7). The severity of disease often depends on the host's ability to form granulomata; in their absence, the organisms spread, resulting in tuberculosis or diffuse cutaneous leishmaniasis.
Resistance to the spread of M. tuberculosis and to the severity of leishmaniasis has been attributed to the adaptive immune response (8, 9). Although it has been suggested that an exaggerated local healing response could result in eradication of the organism from an individual, this response has not been demonstrated (10). However, there is evidence that factors such as tissue repair and remodeling, as well as a vigorous adaptive immune response, are involved (11). We have used the Leishmania major mouse model to explore the hypothesis that the rate of healing of the granulomatous ulcers of cutaneous leishmaniasis contributes significantly to the host's armamentarium of innate resistance. We demonstrate that mice congenic for host response loci controlling the severity of disease differ in their healing response to both infective and surgical lesions, with resistant animals possessing a more robust wound healing response in both cases.
Cutaneous leishmaniasis is caused by the obligatory intracellular protozoan Leishmania and is transmitted by bloodsucking sandflies. Skin macrophages are infected, and a granulomatous ulcer develops at the site of infection. The severity of human cutaneous leishmaniasis ranges from self-healing ulcers to lesions lasting for years (12). Although the importance of host genetics has been recognized, correlates of disease severity have not been identified in humans (13, 14). A murine model of leishmaniasis using the human pathogen L. major has been extensively studied, and loci mediating disease severity have been described (15–18). Like the human disease, resistance equates to a rapid healing of the skin ulcer, whereas susceptibility is characterized by chronicity of disease. Here, we use these mice to show that resistance is closely linked to the ability to heal a skin wound and that lmr1-3 loci control the rate at which wounds heal. Importantly, we show that infection by L. major modulates this response in a host-specific manner, with animals mounting a vigorous wound-healing response also showing resistance to L. major lesion development.
Loci on mouse chromosomes 17 (lmr1), 9 (lmr2), and X (lmr3) modulate functional differences in L. major susceptibility observed between the resistant C57BL/6 and susceptible BALB/c mice. Lines of mice congenic for donor resistance intervals for lmr1 and -2 from C57BL/6 have been bred onto BALB/c and called C.B6-(lmr1.lmr2) (C.lmr1/2), with the symmetrical congenic carrying the BALB/c donor-susceptible intervals called B6.C-(lmr1.lmr2) (B6.lmr1/2), creating two lines of reciprocal congenics, compounded for two donor intervals (19). Both congenic lines retain the appropriate lmr3 locus because the susceptible allele is from C57BL/6 and the resistant from BALB/c (16). These mice differ in susceptibility from their parental lines, exhibiting a statistically significant tendency toward the phenotype of the donor, away from that of the recipient strain (19). Mice congenic for resistance loci lmr1-3 show no role for T cell modulation of the susceptibility phenotype (19, 20), suggesting that T cell responses may act elsewhere in the process (9).
Methods
Mice. BALB/cAn Bradley, C57BL/6J, B6.c-(lmr1, lmr2), and c.B6-(lmr1, lmr2) were bred in the specific pathogen-free facility at The Walter and Eliza Hall Institute. Congenic intervals are on chromosomes 9 and 17. The most proximal and distal markers found to be within the congenic intervals are as follows: B6.c-(lmr1, lmr2), D17Mit57–D17Mit129, D9Mit89–D9Mit71.c.B6-(lmr1, lmr2), D17Mit57–D17Mit39, and D9Mit89–D9Mit329.
Punch Biopsy. Two-month-old female mice were anesthetized with Xylazil/Ketamil (0.013 g/0.1 g per kg body weight), and 4-mm full thickness punch wounds were made. Wound repair was monitored by measuring the diameter of the lesion on days 3, 5, and 7 and by histopathological examination of skin biopsies from two mice from each group after hematoxylin/eosin staining. Masson's trichrome stains were done on day 7 lesions to assess collagen deposition at the wound site. The statistical test used to compare the kinetics of wound closure for both the punch biopsies and L. major-induced lesions is described at http://bioinf.wehi.edu.au/software/russell/perm/help.html.
Parasites and Infections. The virulent cloned line V121, derived from the L. major isolate LRC-L137 (MHOM/IL/67/JerichoII), was used to infect bone marrow-derived macrophages at a ratio of five parasites per cell, as described (21). Mice were infected with L. major as described (19, 20).
Preparation of RNA and Microarray Data Analysis. Cells were collected 24 h after infection by using trypsin, and RNA was prepared by using RNeasy (Qiagen, Valencia, CA). Fifteen micrograms of biotin-labeled cRNA was produced and hybridized to the murine U74Av2 Genechip (Affymetrix, Santa Clara, CA) by using the manufacturer's recommendations. Analysis of the data was carried out by using the robust multiarray analysis (RMA) method (22). Briefly, to assess the differential expression of a gene, a linear model is fitted to the background-corrected, normalized probe level data. There are typically 16–20 probes on the murine U74Av2 GeneChip. This analysis returns an estimate of the log2 expression for each chip as well as an estimated error in the expression. For each comparison, the log ratio (M) is calculated, and the variability in expression measurements is taken into account by constructing a “moderated” T statistic, t* = M̄/SE*, where SE* is a slightly inflated SE (23).
To determine which genes were differentially expressed, quantile-quantile plots, which graph the quantiles of the data against the expectation for the normal distribution, were used. Most of the data lie along a straight line, whereas the genes that are differentially expressed are at the extreme ends of the quantile-quantile plot. The assignment of a threshold is arbitrary, so a compromise is made between detecting as many genes as possible that are truly differentially expressed, and not producing a list so large that any follow-up work becomes impractical. The function of genes was assigned by using the Unigene and National Center for Biotechnology Information Locuslink databases (www.ncbi.nlm.nih.gov).
Isolation of Primary Dermal Fibroblasts. Fibroblasts were obtained from the skin of 4-day-old mice of the parental and congenic mouse strains (24). Dermal fibroblasts were allowed to migrate out into DMEM, 10% FBS, and 50 μg/ml l-ascorbic acid. After 2 weeks at 37°C, 10% CO2 explants were removed, fresh medium was added, and the cells were grown to confluence and replated twice to deplete the residual keratinocytes that do not survive replating. Purity of fibroblast cultures was tested by immunofluorescence with antibodies to potential contaminating keratinocytes and epithelial and endothelial cells, and the cells were used at passages 4–10.
In Vitro Wound Healing. Fibroblasts were seeded in 24-well plates (4 × 105 cells per well) and grown to confluence overnight in DMEM containing 10% FBS and 50 μg/ml l-ascorbic acid. A scratch was made by using a 1-ml pipette tip, cell debris was removed, fresh medium was added, and the wounded monolayers were incubated at 37°C, 10% CO2 (24). Scratch width was measured at various time points. To investigate the contribution to wound healing of fibroblast migration versus proliferation, monolayers of dermal fibroblasts were prepared as above with or without 2 μg/ml mitomycin C (Sigma). In some experiments, the culture supernatants were replaced with supernatants from the infected macrophage cultures described above. All experiments were performed in triplicate.
Results
Microarray Analysis of Infected Macrophages from Congenic Lines. Bone marrow-derived macrophages were isolated from both reciprocal congenic lines (C.lmr1/2 and B6.lmr1/2) and their parental strains (BALB/c and C57BL/6) and infected with L. major parasites in vitro. RNA was extracted from infected and uninfected macrophages of the parental and congenic strains, and cRNA was hybridized to Affymetrix microarray slides according to Affymetrix protocols. Expression levels for each probe set were calculated by using robust multiarray analysis (22). Moderated t statistics, t*, were calculated, and differential expression was assessed by using a normal quantile-quantile plot of the t*. Many genes were differentially expressed between infected and uninfected macrophages, with more genes showing differential expression in C57BL/6 compared with BALB/c mice (Table 1). Over 20 genes were commonly differentially expressed between the reciprocal arms of this experiment (BALB/c vs. C.lmr1/2 and C57BL/6 vs. B6.lmr1/2). Seventeen of these genes showed highly concordant expression patterns (Table 1). Those expressed highly in the susceptible mice were low in the resistant and vice versa, adding confidence in the accuracy of the expression data. Key expression differences were confirmed by real-time PCR (data not shown). Comparisons were made between the congenics and their parental lines and between infected and uninfected cells, and also differences between these differentially expressed genes were noted. An enrichment of genes involved in extracellular matrix deposition emerged from these analyses (Table 2). These genes included a number of molecules involved in the remodeling and composition of the extracellular matrix and include several collagens, a matrix metalloproteinase, and Sparc, a secreted phosphoglycan. Tgfb1 and Ctgf are also differentially expressed, and these are genes involved in the control of wound healing. That these genes play an important role in wound healing and tissue remodeling suggested the possibility that differences in wound healing may play a role in the functional differences in susceptibility between the congenics and their recipient parental lines. After all, the lesion produced by L. major is a chronic ulcer, which is, by definition an aberration in the normal process of wound healing.
Table 1. Genes differentially expressed in infected macrophages between congenic and parental lines, which are also common across the reciprocal congenic lines.
| Gene symbol | Gene name | Chromosome, position | Fold change BALB/c v. C.lmr1/2 | Up or down | Fold change C57BL/6 v. B6.lmr1/2 | Up or down |
|---|---|---|---|---|---|---|
| 1110008E19Rik | RIKEN cDNA | 17, A3.3 | 1.48 | D | 1.48 | U |
| Tmsb10 | Thymosin, β10 | 2, 18.0 cM | 1.32 | U | 1.38 | D |
| Cxcl2 | Chemokine (C-X-C motif) ligand 2 | 5, 51.0 cM | 3.25 | D | 1.57 | U |
| 2010100O12Rik | RIKEN cDNA | 2, H1 | 1.34 | U | 1.34 | D |
| H2-D1 | Histocompatibility 2, D region locus 1 | 17, 19.09 cM | 3.99 | D | 3.83 | U |
| Sdha | Succinate dehydrogenase complex, subunit A, flavoprotein | 13, C1 | 1.44 | U | 1.50 | D |
| Pim1 | Proviral integration site 1 | 17, 16.4 cM | 1.77 | D | 1.50 | U |
| Rpo1-1 | RNA polymerase 1-1 (40-kDa subunit) | 17, B3 | 1.81 | U | 1.85 | D |
| — | IMAGE: 4192122, mRNA | — | 2.39 | D | 1.80 | U |
| Acta2 | Actin, α2, | 19, C3 | 1.97 | U | 1.59 | D |
| Rbbp7 | Retinoblastoma binding protein 7 | X, F4 | 1.47 | U | 1.51 | D |
| H2-Ea | Histocompatibility 2, class II antigen E α | 17, 18.7 cM | 14.34 | D | 6.08 | U |
| Ltb | Lymphotoxin B | 17, 19.06 cM | 1.67 | D | 1.70 | U |
| — | RIKEN clone | — | 1.69 | U | 1.60 | D |
| 5730403E06Rik | RIKEN cDNA | X, A1.1 | 1.49 | U | 1.37 | D |
| qk | Quaking | 17, 5.9 cM | 1.41 | U | 1.78 | D |
| Col1a1 | Procollagen, type I, α1 | 11, 56.0 cM | 1.48 | U | 1.72 | D |
| Fen1 | Flap structure specific endonuclease 1 | 19, A | 1.89 | U | 1.50 | D |
| 2310015N07Rik | RIKEN cDNA | 7, | 1.46 | U | 1.50 | D |
| Ctgf | Connective tissue growth factor | 10, 17.0 cM | 2.17 | U | 1.56 | D |
| 1110008E19Rik | RIKEN cDNA | 17, A3.3 | 1.63 | D | 1.61 | U |
| 2610003J05Rik | RIKEN cDNA | 4, D3 | 1.38 | U | 1.36 | D |
| Pbx3 | Pre-B cell leukemia transcription factor 3 | 2, 22.0 cM | 1.44 | U | 1.45 | D |
| Zfp36 | Zinc finger protein 36 | 7, 10.2 cM | 2.08 | D | 1.43 | D |
| H2-Aa | Histocompatibility 2, class II antigen A, α | 17, 18.65 cM | 1.36 | D | 1.50 | D |
| Mapkapk2 | MAP kinase-activated protein kinase 2 | 1, E4 | 1.4 | D | 1.48 | D |
U, up in congenic; D, down in congenic. All comparisons with infected cells. MAP, mitogen-activated protein.
Table 2. Genes differentially expressed in infected/uninfected macrophages between the congenic strains and their parental lines.
| Symbol | Name | Chromosome | Location | Fold change | Up/down |
|---|---|---|---|---|---|
| C57BL/6 infected v B6.lmr1/2 infected | |||||
| Col1a2 | Procollagen, type I, α2 | 6 | 0.68 cM | 1.46 | D |
| Col18a1 | Procollagen, type XVIII, α1 | 10 | 41.3 cM | 1.43 | U |
| Tgfb1 | Transforming growth factor, β1 | 7 | 6.5 cM | 1.51 | D |
| Postn | Osteoblast specific factor 2 (fasciclin I-like) | 3 | 54.5 Mb | 1.91 | D |
| Ctgf | Connective tissue growth factor | 10 | 17.0 cM | 1.56 | D |
| Col1a1 | Procollagen, type I, α1 | 11 | 56.0 cM | 1.72 | D |
| BALB/c infected v C.lmr1/2 infected | |||||
| Col5a2 | Procollagen, type V, α2 | 1 | 45.8 Mb | 1.50 | U |
| Ctgf | Connective tissue growth factor | 10 | 17.0 cM | 2.17 | U |
| Col1a1 | Procollagen, type I, α1 | 11 | 56.0 cM | 1.48 | U |
| Sparc | Secreted acidic cysteine rich glycoprotein | 11 | 29.9 cM | 1.61 | U |
| C57BL/6 uninfected v B6.lmr1/2 uninfected | |||||
| Col1a2 | Procollagen, type I, α2 | 6 | 0.68 cM | 1.90 | D |
| Col5a2 | Procollagen, type V, α2 | 1 | 45.8 Mb | 2.00 | D |
| Postn | Osteoblast specific factor 2 (fasciclin I-like) | 3 | 54.5 Mb | 2.40 | D |
| Ctgf | Connective tissue growth factor | 10 | 17.0 cM | 2.30 | D |
| Col1a1 | Procollagen, type I, α1 | 11 | 56.0 cM | 2.00 | D |
| Bgn | Biglycan | X | 29.3 cM | 1.80 | D |
| Sparc | Secreted acidic cysteine rich glycoprotein | 11 | 29.9 cM | 2.70 | D |
| BALB/c uninfected v C.lmr1/2 uninfected | |||||
| Col18a1 | Procollagen, type XVIII, α1 | 10 | 41.3 cM | 1.70 | D |
| Ctgf | Connective tissue growth factor | 10 | 17.0 cM | 2.20 | U |
| Col1a1 | Procollagen, type I, α1 | 11 | 56.0 cM | 1.75 | U |
| Sparc | Secreted acidic cysteine rich glycoprotein | 11 | 29.9 cM | 1.95 | U |
D, down in congenic; U, up in congenic; cM, genetic distance from GDB; Mb, physical distance from UCSC May 2004 Assembly.
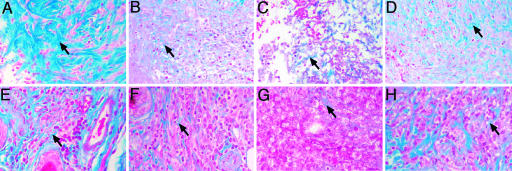
Collagen Bundles Are More Abundant and Ordered in Mice Carrying the Resistance C57BL/6 lmr Loci. We tested the wound-healing hypothesis by histological examination of skin lesions induced by either a 4-mm full thickness punch biopsy or infection by the L. major virulent cloned line V121. Masson's trichrome (MT) stained sections showed obvious strain-specific differences in the deposition of collagen fibrils (Fig. 1). C57BL/6 punch biopsy lesions exhibited an ordered array of collagen deposition (Fig. 1 A) whereas BALB/c mice deposited a sparser and more disordered matrix of collagen (Fig. 1C). The pattern of collagen deposition in the congenic mice was more similar to the donor strain of the congenic interval than the recipient strain, indicating that genes within the congenic intervals controlled this process (Fig. 1 B and D). This strain-specific phenotype was also recapitulated in lesions caused by L. major (Fig. 1 E–H), confirming that normal wound-healing processes with active collagen deposition are present in lesions caused by L. major and that differences between the congenic lines and their parentals are preserved in infective lesions.
Fig. 1.
Histological sections of punch biopsies and L. major lesions stained with Masson's trichrome showing strain-dependent deposition of collagen fibrils. (A–D) Punch biopsy lesions. (E–H) L. major lesions. (A and E) -C57BL/6. (B and F) -B6.lmr1/2. (C and G) -BALB/c. (D and H) –C.lmr1/2.
Wound Healing Is Faster in Mice Carrying the Resistance C57BL/6 lmr Loci. The rate of healing of punch biopsies was used to associate quantitative differences in wound repair with the histological and microarray observations. Kinetics of healing of punch biopsies performed on the congenic and parental lines demonstrated that the presence of the resistance C57BL/6 haplotype at the donor intervals resulted in faster healing. The diameters of the lesions were measured every other day, and the rate of wound closure was calculated (Fig. 2A). There were statistically significant differences between C57BL/6 and BALB/c (P = 0.0001), BALB/c and C.lmr1/2 (P = 0.006), and C57BL/6 and B6.lmr1/2 (P = 0.025). The order of fastest healer to slowest is C57BL/6, B6.lmr1/2, C.lmr1/2, and BALB/c, indicating that the presence of the donor interval significantly affects the speed of wound closure in both congenic directions. The susceptible BALB/c lmr loci slow down wound closure on a C57BL/6 background, and the resistant C57BL/6 loci accelerate closure on a BALB/c background, implicating the presence of controlling genes within the congenic intervals. These differences in the rate of wound healing are consistent with this being a factor in the differential host response to infection by L. major in these mouse strains.
Fig. 2.
Closure kinetics of skin punch biopsy lesions and of scratches in fibroblast monolayers showing strain dependent closure times. (A) Closure kinetics of surgically induced skin lesions from C57BL/6, B6.lmr1/2, C.lmr1/2, and BALB/c. (B and C) Kinetics of closure of scratches in fibroblast monolayer.
lmr Loci Control Fibroblast Motility in an in Vitro Model of Wound Healing. We used an in vitro wound-healing model measuring the mobility of fibroblasts across a scratch, to determine whether these cells are functionally important in the above-mentioned observations in the mouse (24). Fibroblasts were isolated from the skin of parental and congenic mice, plated, and grown to confluence. Interestingly, it was consistently more difficult to obtain fibroblast cultures from the B6.lmr1/2 line due to slower growth of these fibroblasts. Closure times were measured in the presence and absence of mitomycin C, which prevents cell division, but does not affect cell motility. Closure of scratches in fibroblast monolayers showed highly reproducible strain-specific kinetics (Fig. 2 B and C). In concert with the in vivo healing data, cells from C57BL/6 mice closed the scratch faster than BALB/c (P = 0.007). This phenotype was controlled by genes on the resistance intervals because C.lmr1/2 closed the wound more rapidly than the recipient parent, BALB/c(P = 0.031) (Figs. 2B and 3), and they displayed a rate of closure similar to C57BL/6 (P = 0.117) with which they share only the resistance interval. The presence of the susceptible BALB/c congenic donor intervals significantly slowed closure on a C57BL/6 background (Fig. 2C). The addition of mitomycin to the culture did not affect the rate of closure of the scratch (results not shown), indicating that the differences in closure rates were due to differences in cell motility and not proliferation.
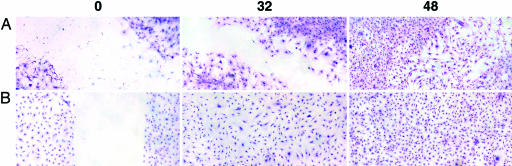
Fig. 3.
Giemsa-stained skin fibroblast monolayer showing the differences in scratch closure between BALB/c(A) and C.lmr1/2 fibroblasts (B) 32 and 48 h after wounding (0).
Supernatant from C57BL/6-Infected Macrophages Accelerates Kinetics of Scratch Closure. We wished to examine the potential contribution of the parasite and the macrophage to the process of wound healing. Supernatant from infected or uninfected C57BL/6 and BALB/c bone marrow-derived macrophages was used as substitute culture medium in the scratch-wound model, and kinetics of scratch closure were compared between fibroblasts grown in infected and uninfected conditioned media. Surprisingly, supernatant from infected, but not uninfected, C57BL/6 macrophages was able to accelerate the closure of scratches in C.lmr1/2 fibroblast monolayers (P = 0.007) (Fig. 4). Although there was no significant overall effect of the C57BL/6-infected macrophage supernatant on C57BL/6 fibroblasts, at the 24-h time point, it seemed to accelerate closure (Fig. 4). There was no difference in closure rates if the fibroblasts were isolated from the susceptible BALB/c or the B6.lmr1/2 mice carrying the congenic interval from BALB/c (Fig. 4). There was also no difference in closure kinetics caused by supernatant from infected or uninfected BALB/c macrophages on any of the cells (data not shown), indicating that the ability of fibroblasts to respond to infected macrophages is also dependent on the congenic intervals.
Fig. 4.
Kinetics of scratch closure in fibroblasts from C57BL/6, BALB/c, C.lmr1/2, and B6.lmr1/2 grown in medium conditioned by infected (filled circles), uninfected (open circles and dashed line) C57BL/6 macrophages, or control medium (gray circles) expressed as a percentage of the original scratch width.
Discussion
Our earlier work demonstrated that the lmr loci mediate susceptibility to L. major infection in the mouse (15, 16, 19, 20) and showed that this effect was not due to T cell-mediated responses, suggesting that they must act elsewhere in the host response pathway. The data presented in this paper show that these same congenic intervals also modulate a differential wound-healing response. This response was first suspected when tissue repair genes were found to be differentially expressed in infected and uninfected macrophages isolated from the congenic lines. Histological examination of either surgically induced lesions or infective lesions showed that the amount and structure of collagen deposited in these wounds depended on the congenic donor interval. Animals with resistant C57BL/6 lmr haplotypes deposited more collagen, and the collagen was ordered in consistently oriented bundles, quite the converse of collagen deposited in the wounds of either BALB/c or B6.lmr1/2 congenics. The differences in the kinetics of in vivo wound healing were also observed in the healing of scratches in fibroblast monolayers, implying a common mechanism.
The observation that only fibroblasts isolated from mice with the resistance intervals respond to molecules produced by C57BL/6-infected macrophages strongly ties the healing of the scratches in fibroblast monolayers to the original observations in the microarray experiments, showing differential expression in macrophages of genes involved in tissue repair and wound healing. Macrophages from mice with the susceptible BALB/c lmr loci do not produce stimulatory factors, and their fibroblasts do not respond to those produced by C57BL/6-infected macrophages. The totally synergistic nature of this response implies a lock-and-key-style mechanism, for example, a ligand/receptor interaction where alleles of both ligand and receptor interact epistatically.
The genetic association of the wound-healing phenotype with the severity of disease phenotype strongly suggests the former as the mechanism through which Leishmania resistance is mediated by the lmr1-3 loci. Therefore, the postulated mechanism of resistance mediated through lmr1-3 is an enhanced wound-healing response that is stimulated by molecules produced by infected macrophages, acting on fibroblasts genetically primed for response, resulting in an accelerated healing of the L. major-induced lesion. It is likely that this effect is due to the differential expression of one or more of the genes described in Table 1. Genes that are not necessarily present in the congenic interval may become mobilized, such as connective tissue growth factor that shows an increased expression in strains with the C57BL/6 resistance interval. Connective tissue growth factor plays a role in the expression of type 1 collagen and in the mobilization and proliferation of fibroblasts (25). The breeding and use of congenic animals that demonstrate no differential T cell response (19, 20) has allowed the dissection of the “healing” part of the granulomatous response away from the much studied T cell responses that are so important in the development of granulomas. It is possible that similar mechanisms play a role in humans. An extensive remodeling process involving fibroblasts and macrophages has been described in the early leishmaniasis lesion (11). Moreover, abnormalities in wound healing have been noted in patients treated for cutaneous leishmaniasis (26).
It is also possible that this mechanism of resistance involving cells of the innate immune system may be common for other granulomatous diseases (27). Indeed, resistance to M. tuberculosis in the mouse has been mapped to loci overlying lmr1 and -2 (28, 29), suggesting a resistance mechanism common between these two granulomatous diseases.
Acknowledgments
We thank Anne Voss and Ian Darby for advice and Lynn Buckingham for technical assistance. We are particularly grateful to Fiona Mitchell and Tracey Baldwin for their assistance. This work was supported by grants from the National Institutes of Health, the Australian National Health and Medical Research Council, the Howard Hughes Medical Institute, and the World Health Organization Special Program for Research and Training in Tropical Diseases (TDR).
Author contributions: A.S., C.M.E., E.H., and S.J.F. designed research; A.S., C.M.E., J.M.C., and B.K. performed research; A.S., C.M.E., K.S., T.P.S., E.H., and S.J.F. analyzed data; and E.H. and S.J.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Olds, G. R., Griffin, A. & Kresina, T. F. (1985) Gastroenterology 89, 617–624. [DOI] [PubMed] [Google Scholar]
- 2.Andrade, Z. A. (1991) Mem. Inst. Oswaldo Cruz. 86, Suppl. 3, 61–73. [DOI] [PubMed] [Google Scholar]
- 3.Vaalamo, M., Kariniemi, A. L., Shapiro, S. D. & Saarialho-Kere, U. (1999) J. Invest. Dermatol. 112, 499–505. [DOI] [PubMed] [Google Scholar]
- 4.Majka, S. M., Kasimos, J., Izzo, L. & Izzo, A. A. (2002) Med. Mycol. 40, 323–328. [DOI] [PubMed] [Google Scholar]
- 5.Izzo, A. A., Izzo, L. S., Kasimos, J. & Majka, S. (2004) Tuberculosis (Edinburgh) 84, 387–396. [DOI] [PubMed] [Google Scholar]
- 6.Singh, K. P., Gerard, H. C., Hudson, A. P. & Boros, D. L. (2004) Ann. Trop. Med. Parasitol. 98, 581–593. [DOI] [PubMed] [Google Scholar]
- 7.Price, N. M., Gilman, R. H., Uddin, J., Recavarren, S. & Friedland, J. S. (2003) J. Immunol. 171, 5579–5586. [DOI] [PubMed] [Google Scholar]
- 8.Raja, A. (2004) Indian J. Med. Res. 120, 213–232. [PubMed] [Google Scholar]
- 9.Sacks, D. & Noben-Trauth, N. (2002) Nat. Rev. Immunol. 2, 845–858. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann, S. H. (2002) Ann. Rheum. Dis. 61, Suppl. 2, ii54–ii58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esterre, P., Dedet, J. P., Guerret, S., Chevallier, M., Frenay, C. & Grimaud, J. A. (1991) Pathol. Res. Pract. 187, 924–930. [DOI] [PubMed] [Google Scholar]
- 12.Alrajhi, A. A. (2003) Skin Therapy Lett. 8, 1–4. [PubMed] [Google Scholar]
- 13.Blackwell, J. M. (1996) Parasitology 112, S67–S74. [PubMed] [Google Scholar]
- 14.Blackwell, J. (2001) Trends Mol. Med. 7, 521–526. [DOI] [PubMed] [Google Scholar]
- 15.Roberts, L. J., Baldwin, T. M., Curtis, J. M., Handman, E. & Foote, S. J. (1997) J. Exp. Med. 185, 1705–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts, L. J., Baldwin, T. M., Speed, T. P., Handman, E. & Foote, S. J. (1999) Eur. J. Immunol. 29, 3047–3050. [DOI] [PubMed] [Google Scholar]
- 17.Mock, B. A., Fortier, A. H., Potter, M., Blackwell, J. & Nacy, C. A. (1985) Curr. Top. Microbiol. Immunol. 122, 115–121. [DOI] [PubMed] [Google Scholar]
- 18.Lipoldova, M., Svobodova, M., Krulova, M., Havelkova, H., Badalova, J., Nohynkova, E., Holan, V., Hart, A. A., Volf, P. & Demant, P. (2000) Genes Immun. 1, 200–206. [DOI] [PubMed] [Google Scholar]
- 19.Elso, C. M., Roberts, L. J., Smyth, G. K., Thomson, R. J., Baldwin, T. M., Foote, S. J. & Handman, E. (2004) Genes Immun. 5, 93–100. [DOI] [PubMed] [Google Scholar]
- 20.Elso, C., Kumar, B., Smyth, G., Foote, S. & Handman, E. (2004) Genes Immun. 5, 188–196. [DOI] [PubMed] [Google Scholar]
- 21.Scott, C. L., Roe, L., Curtis, J., Baldwin, T., Robb, L., Begley, C. G. & Handman, E. (2000) Microbes Infect. 2, 1131–1138. [DOI] [PubMed] [Google Scholar]
- 22.Irizarry, R. A., Bolstad, B. M., Collin, F., Cope, L. M., Hobbs, B. & Speed, T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed]
- 23.Efron, B., Tibshirani, R., Storey, J. D. & Tusher, V. G. (2001) J. Am. Stat. Assoc. 96, 1151–1160. [Google Scholar]
- 24.DeBiasio, R., Bright, G. R., Ernst, L. A., Waggoner, A. S. & Taylor, D. L. (1987) J. Cell Biol. 105, 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leask, A. & Abraham, D. J. (2004) FASEB J. 18, 816–827. [DOI] [PubMed] [Google Scholar]
- 26.Romero, G. A., Lessa, H. A., Macedo Vde. O., de Carvalho, E. M. & Marsden, P. D. (1996) Rev. Soc. Bras. Med. Trop. 29, 285. [DOI] [PubMed] [Google Scholar]
- 27.Shi, Z., Wakil, A. E. & Rockey, D. C. (1997) Proc. Natl. Acad. Sci. USA 94, 10663–10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavebratt, C., Apt, A. S., Nikonenko, B. V., Schalling, M. & Schurr, E. (1999) J. Infect. Dis. 180, 150–155. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez, F., Radaeva, T. V., Nikonenko, B. V., Persson, A. S., Sengul, S., Schalling, M., Schurr, E., Apt, A. S. & Lavebratt, C. (2003) Infect. Immun. 71, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]