Abstract
Members of the three kingdoms of life contain tRNA genes with introns. The introns in pre-tRNAs of Bacteria are self-splicing, whereas introns in archaeal and eukaryal pre-tRNAs are removed by splicing endonucleases. We have studied the structures of the endonucleases of Archaea and the architecture of the sites recognized in their pre-tRNA substrates. Three endonuclease structures are known in the Archaea: a homotetramer in some Euryarchaea, a homodimer in other Euryarchaea, and a heterotetramer in the Crenarchaeota. The homotetramer cleaves only the canonical bulge–helix–bulge structure in its substrates. Variants of the substrate structure, termed bulge–helix–loops, appear in the pre-tRNAs of the Crenarcheota and Nanoarcheota. These variant structures can be cleaved only by the homodimer or heterotetramer forms of the endonucleases. Thus, the structures of the endonucleases and their substrates appear to have evolved together.
Keywords: molecular evolution, RNA–protein interactions, splicing
In Archaea, the tRNA splicing endonuclease is responsible for the correct removal of introns from pre-tRNAs and is also involved in the processing of pre-rRNA and presumably certain pre-mRNA (1–4). An RNA motif consisting of a bulge–helix–bulge (BHB) is the universal substrate of the endonucleases from all archaeal lineages and eukaryotes (5). This motif has been shown by biochemical and NMR studies to be comprised of two bulges of three nucleotides symmetrically disposed on opposite strands and separated by a helix of four base pairs (6, 7). Although a consensus sequence has been derived (8), the conformation of this structure appears to be more relevant than its sequence (9).
The development of the genomics of Archaea made possible a characterization of the genes coding for pre-tRNA substrates (10) and the genes coding for the tRNA splicing endonucleases (11). Most introns of archaeal pre-tRNA genes are located in the anticodon loop, between nucleotides 37 and 38, the unique location of their eukaryotic counterparts. However, in several Archaea, mostly in Crenarchaeota, introns have been found at other positions: the anticodon stem and loop, the D- and T-loops, the V-arm, or the amino acid arm. Marck and Grosjean (10) renamed the BHB as hBHBh′, indicating with the new name that the canonical BHB motif should be enlarged to include two outer helices having at least two Watson–Crick base pairs. For introns located at 37/38 and elsewhere in the pre-tRNA, canonical hBHBh′ motifs were not always found. Instead, a relaxed hBH or HBh′ motif, including the constant central 4-bp helix H flanked by one helix (h or h′) with at least two Watson–Crick base pairs on either side, could be discerned (10).
We recently detected two paralogs of the tRNA endonuclease gene of Methanocaldococcus jannaschii (METJA) in the genome of the crenarchaeote Sulfolobus solfataricus (SULSO) (11). This finding led to the discovery of a previously unrecognized heterotetrameric form of the enzyme. The two genes code for two different subunits, both of which are required for cleavage of the pre-tRNA substrate. Thus, three different forms of tRNA endonuclease can now be recognized in the Archaea: a homotetramer in some Euryarchaea (such as METJA), a homodimer in other Euryarchaea (such as Archeoglobus fulgidus, ARCFU), and a heterotetramer in the Crenarchaeota (such as SULSO) and Nanoarchaeota. The heterotetrameric form of the enzyme, arising most likely by gene duplication and subsequent “sub-functionalization,” requires the products of both genes to be active (12, 13).
Marck and Grosjean (10) were correct to recognize the several forms of the substrates but, missing the second subunit of the endonuclease from the Crenarcheota, they incorrectly assigned particular forms of the substrate to particular enzyme structures.
In the present article, we analyze the relationship of the intron-containing motif of the pre-tRNAs to the tRNA endonuclease architecture in the Archaea and show that the relaxed form of the substrate requires either the dimeric or the heterotetrameric endonuclease to be cleaved properly.
Materials and Methods
Expression and Purification of the Protein Constructs. The genes encoding the endonucleases from ARCFU, SULSO, and METJA were PCR-amplified from the respective genomic DNA using two primers designed to obtain an amplified fragment presenting an NdeI site upstream of the gene and a BamHI site downstream. The digested PCR fragments of the genes coding for ARCFU, METJA, and the α-subunit of SULSO were cloned in pET28b (Novagen), and the gene coding for the β-subunit of SULSO was cloned in pCYCA11b (14). All of the clones obtained were verified by sequencing. The proteins were overexpressed as hexa-histidine-tagged forms (pET28b) with the exception of β-SULSO (pCYAC-11b) that was untagged to be coexpressed together with the tagged form of α-SULSO (pET28b) in Escherichia coli BL21DE3 (Novagen). Cells were grown in 1-liter cultures of Luria-Bertani broth at 37°C in the presence of 30 μg/ml kanamycin (pET28) with the addition of 30 μg/ml chloramphenicol (pCYCA-11b) in the case of coexpression. The proteins were purified on a metal affinity column as described (11). Homogeneity of the enzymes was assessed by Coomassie blue staining of SDS polyacrylamide gels. The tRNA endonuclease from the toad Xenopus laevis (XENLA) was purified according to ref. 15.
In Vitro Transcription and Splicing. DNA templates prepared as described (11) were transcribed by T7 RNA polymerase by using the Ambion (Austin, TX) T7-Megashortscript kit in the presence of [α-32P]UTP (800 Ci/mmol; Amersham Pharmacia). Products of the correct size were purified on a 10% denaturing polyacrylamide gel after phenol extraction and ethanol precipitation. Labeled tRNA precursors (20 fmol) were incubated with purified splicing endonucleases in reaction mixtures containing 25 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl, and 10% glycerol at 65°C for 1 h with the exception of the reaction containing the XENLA enzyme that was incubated at 22°C. Cleavage products were analyzed, after phenol extraction and ethanol precipitation, by electrophoresis on 10% denaturing polyacrylamide gels. Image analysis was done by using a Molecular Dynamics model Storm 860 PhosphorImager with image quant software, version 4.
Results
A Common Fold for All of the Archaeal Enzyme Subunits Is Stabilized by Two Conserved Residue Signatures. The genes coding for tRNA splicing endonucleases encoded in 19 different archaeal genomes have been described and characterized. Three different forms of the endonuclease have been distinguished; the ancestor of the archaeal enzyme was probably a homotetramer, which, after two independent gene duplication events (or horizontal gene transfer), gave rise to a homodimeric and a heterotetrameric form. One event took place in the ancestor of Crenarcheota, resulting in two genes coding for two subunits, whereas the other occurred in the common ancestor of Archaeoglobales, Halobacteriales, and Methanosarcinales, resulting in an in-frame duplication, giving rise to a single gene coding for two fused subunits (Fig. 1). We have shown how the homodimeric subunits can work as a heterotetramer, by cutting the gene into two independent segments each expressing a polypeptide (11). In all of the natural tetramers and the artificially generated one, each set of two subunits plays a specific role. One set contains the catalytic sites, and the other has the structural role of positioning the subunits with the active sites. These two different roles have resulted in the acquisition of mutually exclusive features that allow one to distinguish two functional classes of subunits (11).
Fig. 1.
tRNA intron motifs and enzyme architectures. Vertical bars indicate the species sharing the same endonuclease architecture (11). α4 refers to the homotetramer, α2 refers to the homodimer, and α2β2 refers to the heterotetramer (11). The numbers indicate the number of hBHBh (BHB) and HBh or hBH (BHL) motifs present in the genome, according to ref. 10.
Despite the existence of the two classes of subunits, a modular organization is conserved among them. The conserved residues in (β4)-(α2)-(β5) in the N-terminal domain and in (α4)-loop-(β6) in the C-terminal domain delineate motifs that provide a specific signature for the endonuclease family (Fig. 2). Using this universally conserved motif we can retrieve the subunits of Archaeal endonucleases selectively in the SwissProt database. The signature residues, represented in a cartoon model (Fig. 2 A) of the METJA structure (16), stress the functional importance of helix α2 (blue) and helix α4 (purple) for the stabilization of each of the two domains and their positioning with respect to one another. When the residues are plotted every 100° consecutively around a spiral, the conserved residues in α2 are clustered on two opposite sides of the helix (Fig. 2B), presenting hydrophobic side chains. One face (residues 47, 50, 53, and 54) interacts with conserved residues in the β-sheet of the N-terminal domain. The other face (residues 48, 49, 52, and 56) interacts with the C-terminal domain and also directly with helix α4.
Fig. 2.
Archaeal endonucleases common fold. (A) Cartoon representation of the monomeric subunit of METJA (16). All of the side chains represented are conserved among all of the subunits of archaeal endonucleases. The residues constituting the two signatures, as specified in the text, are colored blue and purple; the other conserved residues are yellow. (B) Helical wheel representation of helix α2. Hydrophobic residues are shaded in gray; only the conserved amino acids are numbered. (C) Helical wheel representation of helix α4. Representation as above.
Helix α4 also presents conserved residues on two different faces (Fig. 2C). One face (residues 88, 91, 94, and 95) is packed against the β-sheet of the C-terminal domain, where it forms specific interactions with conserved residues. The other face (residues 85, 89, 92, and 96) interacts with helix α2 and the N-terminal domain. These observations support the existence of a canonical structure shared by all of the subunits, which implies that they all share a common origin (17, 18).
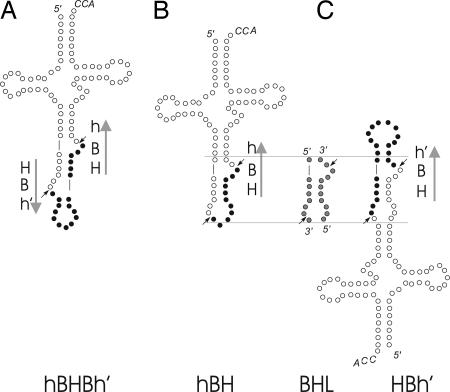
Canonical and Noncanonical Motifs in Intron-Containing Archaeal Pre-tRNAs. Following Marck and Grosjean (10), we examined the sequences spanning intron–exon junctions in intron-containing pre-tRNAs of 19 Archaea. Particularly interesting are those introns whose length is too short to form a second 3-nt bulge followed by a helix consisting of at least two Watson–Crick pairs. Fig. 3 shows that both hBH and HBh′ motifs are characterized by a bulge and an internal loop and can be represented by a structure that resembles the bulge–helix–loop (BHL), as described in some eukaryotic pre-tRNAs (19). Because, presumably, the archaeal endonucleases do not contact the mature domain, hBH and HBh′ do not appear as different to their enzymes. Hereafter, we shall refer to both hBH and HBh′ motifs as BHL-like motifs. Of 139 intron-containing pre-tRNAs, 82 contain a BHB and 57 contain a BHL–like motif. Fig. 1 shows that genes coding for intron-containing pre-tRNAs characterized by the BHL-like motif are absent from species that carry a homotetrameric (α4) tRNA endonuclease. Pre-tRNAs containing BHL-like motifs are found only in those species characterized by the heterodimeric (α2) or the heterotetrameric (α2β2) forms of the tRNA endonuclease.
Fig. 3.
Canonical and noncanonical motifs. (A) hBHBh′, BHB splicing motif flanked by two helices (h, h′) presenting at least two Watson–Crick base pairs. Arrows indicate the 5′ 32 → 3′ sense. (B and C) hBH and HBh′ relaxed BHB motifs. The two superimposed motifs are shaded in gray to show the similarity to a BHL motif.
In Vitro Cleavage of Pre-tRNAs Presenting Either a BHB or a BHL Motif. These observations lead to the prediction that α4 endonucleases require a BHB substrate, whereas α2 or α2β2 endonucleases can cleave BHL substrates. These predictions were tested as follows: two different uniformly labeled pre-tRNA substrates were used for the cleavage assay (Figs. 4 and 5). These comprise a pre-tRNA presenting a motif with the intron and the boundary region of the 5′ exon and 3′ exon of the molecule folded into either one of a 2- or 3-nt bulge separated by a 4-bp helix (BHB motif) (Fig. 4A), and a pre-tRNA presenting a 3-nt bulge and an internal loop separated by a 4-bp helix (BHL-like) (Fig. 5A). We used as a control the partially purified endonuclease from XENLA, because it can process both substrates correctly, based on previous observations (19). Each substrate was incubated with three different tRNA endonucleases from Archaea, representing each one of the three different architectures: the homodimer of ARCFU, the homotetramer of METJA, and the heterotetramer of SULSO.
Fig. 4.
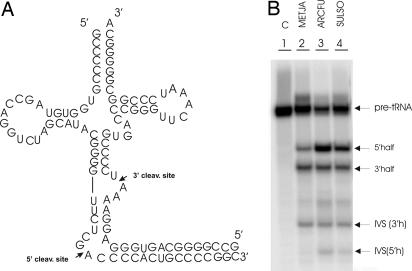
In vitro cleavage of a BHB-containing substrate. (A) The pre-tRNA Archeuka was constructed by using two regions derived from yeast pre-tRNAPhe (nucleotides 1–31 and 38–76) joined by a 25-nt insert that corresponds to the BHB motif of archaeal tRNA Trp. (B) The pre-tRNA Archeuka was incubated with four different enzymes. The conditions of the reactions are reported in Materials and Methods. The cleavage products were analyzed by electrophoresis on 10% polyacrylamide gel containing 29:1 monomer to bis and 8 M Urea, followed by autoradiography. The identification of the reaction products is indicated. Lane 1 is control (C, no enzyme added); lanes 2–5 show the products after incubation with the endonucleases from METJA, SULSO, ARCFU, and XENLA, respectively. The 2/3 molecules are produced by single cleavage. The slowly migrating class corresponds to intron–3′ exon molecules and the fast migrating class to 5′ exon–intron molecules. The heterogeneity of the 3′ halves results from the run-off transcription by T7 RNA polymerase, used to prepare the substrate.
Fig. 5.
In vitro cleavage of a BHL-containing substrate. (A) The pre-tRNATyr from Caenorhabditis elegans. The synthetic substrate presents a residue change at the 5′ terminus (C to G) required for T7 transcription and a corresponding change (G to C) in the complementary strand. (B) The pre-tRNATyr was incubated with four different enzymes. The conditions of the reactions and product analysis were the same as described in the legend to Fig. 3.
Fig. 4B shows the expected result that all of the enzymes cleave the BHB substrate correctly. On the contrary, there are differences in the processing of the BHL-like intron. Fig. 5B, lane 2 clearly shows that the homotetrameric enzyme from METJA cannot cleave the BHL-like structure correctly. The enzyme cleaves inefficiently, only at the 3′ site, resulting in accumulation of the 5′ half-intron product, whereas the heterotetrameric enzyme from SULSO and the homodimeric enzyme from ARCFU cleave the BHL-like structure correctly (Fig. 5B, lanes 3 and 4), as does the (eukaryotic) XENLA endonuclease (Fig. 5B, lane 5).
Cis and Trans Splicing Motifs. All of the substrates described above were characterized by BHB or BHL targets produced by annealing strands belonging to the same RNA molecule. We turn now to describe a substrate characterized by a BHL-like intron–exon junction produced by the assembly of two different RNA molecules. Nanoarchaeum equitans is a small hyperthermophilic archaeal parasite (20). The N. equitans genome contains nine genes that encode tRNA halves; together they account for the missing genes encoding the glutamate, histidine, tryptophan, and initiator methionine transfer RNA species (21). The tRNA sequences are split after the anticodon-adjacent position 37, the normal location of tRNA introns (18). Terminal segments of these half-tRNAs constitute an intervening sequence that includes a 12- to 14-nt, GC-rich RNA duplex formed between the 3′ end of the 5′ tRNA half and the 5′ beginning of the 3′ tRNA half (Fig. 6A). The missing N. equitans tRNAs reveal the necessity for assembly of two tRNA half-molecules. Randau et al. (22) proposed a model based on their discovery of extended reverse complementarity in the intervening sequences: an extended GC-rich duplex in the split intron would facilitate base-pairing of the two halves. This stable duplex in the intron could facilitate folding of the whole tRNA body and promote the cloverleaf structure of the tRNA.
Fig. 6.
In vitro cleavage of a substrate presenting a trans BHL. (A) Proposed structure of the N. equitans pre-tRNAGlu formed by annealing the products of two half-genes. The left side of the structure contains the 5′ half-gene, including the 5′ cleavage site in the loop formed by CGAC. The right side of the structure contains the 3′ cleavage site comprising the AAU bulge. The long GC-rich stem at the bottom of the structure is not covalently closed but that does not matter, because it is removed by the splicing endonuclease. (B) Cleavage of the pre-tRNAGlu by purified archaeal endonucleases. The substrate was incubated with three different recombinant proteins. The cleavage products were analyzed as described in the legend to Fig. 3. The identification of the reaction products is indicated. Lane 1 is control (C, no enzyme added); lanes 2–4 show the products after incubation with the endonucleases from METJA, ARCFU, and SULSO, respectively. Because the substrate molecule is open at the bottom, the enzymes produce, in addition to 5′ and 3′ halves, two intron fragments. IVS (3′h) is the fragment resulting from cleavage at the 3′ cleavage site, and IVS (5′h) is the fragment resulting from cleavage at the 5′ cleavage site.
Fig. 6A shows a schematic representation of a 5′ tRNA half-gene product (tRNAGlu) and the corresponding 3′ tRNA half-gene product formed in N. equitans. The region between the folded tRNA and the intervening RNA duplex at the bottom resembles the BHB. Fig. 6A shows that the substrate derived by annealing the RNAs coded by the two tRNAGlu split genes (pre-tRNAGlu) has a 3′ site bulge, a 4-bp helix (with one UG pair), and a 5′ site in a four-base loop. Therefore, we are dealing with a BHL-like motif. Because this structure is located at the position where most archaeal tRNA introns occur, the fusion of the two halves might involve cleavage by the tRNA endonuclease followed by ligation. Here, we demonstrate that tRNA splicing endonucleases can cleave at the expected positions in pre-tRNAGlu (Fig. 6A).
We used a representative for each of the three forms of archaeal tRNA endonuclease: the homotetramer from METJA, the homodimer from ARCFU, and the heterotetramer from SULSO. Fig. 6B, lanes 3 and 4 shows that pre-tRNAGlu is correctly cleaved by the ARCFU and SULSO enzymes. The products are 5′ and 3′ halves plus the two intron fragments. The identity of the bands was verified by sequencing. The METJA enzyme, on the contrary, requires a strict BHB structure, and it has problems with the recognition and cleavage of the 5′ site (Fig. 6B, lane 2). We produced a minisubstrate, lacking the mature domain (5). Consistent with the fact that the archaeal enzymes operate independently of the mature domain, the minisubstrate behaves exactly like the complete pre-tRNAGlu molecule (data not shown).
Discussion
The BHB, renamed hBHBh′ by Marck and Grosjean (10), is a universal substrate (5). It is cleaved twice by all of the characterized tRNA splicing endonucleases, both archaeal and eukaryal. This observation constitutes the main argument to postulate the existence of conserved features among the sites cleaved by the archaeal and the eukaryal enzymes. Certainly conserved is the distance between the active sites (16).
The only available BHB structure, determined by NMR spectroscopy, is that of a 38-nt RNA, derived from Haloferax volcanii pre-tRNA Trp (7). The conformation of the two 3-nt bulges is stabilized by stacking interactions between bulge nucleotides and bases in helices H, h, and h′. Both bulges appear on the same minor groove face of the 4-bp helix H. Not all of the archaeal intron–exon junctions can fold into a canonical BHB (hBHBh′) structure. A relaxed hBH or HBh′ motif, including the constant central 4-bp helix H flanked by one helix (h or h′) with at least 2 bp on each side, is often found (10). We can, therefore, conclude that only helix H and one of the two helices h or h′ are strictly necessary for cleavage in certain Archaea. Because, presumably, the archaeal endonucleases do not contact the mature domain, hBH and HBh′ do not appear different to the enzymes. For this reason we refer collectively to both of them as BHL-like motifs.
Three different forms of tRNA endonuclease can be recognized in Archaea: a homotetramer in some Euryarchaea, a homodimer in other Euryarchaea, and a heteroteramer in the Crenoarchaea and Nanoarchaea. On the basis of the combination of data derived from the study of the phylogenetic distribution of the motifs at the exon–intron junctions (10) and the endonuclease architectures (11) we were led to hypothesize that all three forms of the enzyme can cleave the canonical BHB and that the relaxed BHL-like motif can be cleaved only by the homodimeric (α2) and the heterotetrameric (α2β2) forms. Our biochemical experiments were designed to explore this hypothetical evolutionary relationship.
Only homodimers or heterotetramers can cleave the BHL-like structures (Figs. 4 and 5). The intron–exon junction motifs and the structures of the enzymes are, therefore, evolutionarily related. Although major questions regarding the origin of tRNA introns are still unanswered, we can speculate that if BHL-like motifs appeared as a consequence of events that modified the BHB motif, it would be necessary to have on hand forms of the enzyme capable of removing the intron correctly. Only those archaeal species that, after gene duplication, present an endonuclease that is either a homodimer or a heterotetramer could process the new substrates. This idea is supported by the fact that some Euryarcheota present a homodimeric endonuclease, but pre-tRNA genes with a BHL-like motif are not encoded in their genomes. It appears that the enzyme specificity for the BHB and BHL-like substrates is the result of adaptation of similar active sites, because the enzymes capable of processing the BHL–like structure are also capable of processing the BHB. This substrate ambiguity is a conspicuous feature that will be evolutionarily exploited in eukaryotic organisms (12, 19, 23).
The intron excision reaction in Eukaryotes is characterized by exquisite dependence on the mature domain (24, 25). The hBH type motif resembles the motif found in most yeast pre-tRNAs presenting introns at 37/38 (18). In this case the bulge is often >3 nt and a conserved base pair between a pyrimidine of the 5′ exon (position 32 in tRNA) and a base in the single-stranded loop of the intron (position 3) is required for correct cleavage of the 3′ splice site (26). The conserved base pair has been called the A-I pair, where A stands for anticodon and I for intron (27). We propose that the relaxed motifs and the consequent ambiguity are a prelude, in the archaeal world, to the loss of autonomy of the BHB-type motifs and the advent of the domination by the mature domain. A common fold, stabilized by the two-conserved-residue signature, characterizes all archaeal endonucleases, despite the existence of the different enzyme forms. The archaeal common fold is not found in the eukaryal enzyme. Again, changes in substrate structure correspond to changes in enzyme structure, according to the paradigms of coevolution (12, 23).
The BHB or the relaxed BHL structures can be formed both in cis and trans. We show that annealing the two tRNA split genes from N. equitants produces a substrate for certain archaeal tRNA endonucleases. Because the archaeal tRNA endonuclease does not contact the mature domain of the pre-tRNA, but simply and directly binds to and cleaves the BHB or BHB-like structures, we can expect that transsplicing mediated by the tRNA endonuclease is not restricted to tRNA. In fact, we recently reported that an archaeal endonuclease (from METJA) can catalyze nonspliceosomal mRNA splicing in mouse cells (28).
Note. Kate Calvin, Michelle D. Hall, Fangmin Xu, Song Xue, and Hong Li (29), in agreement with our results, found that the splicing endonuclease from SULSO contains two different subunits and accepts a broad range of substrates.
Acknowledgments
We thank A. Ferrara for secretarial assistance and G. Di Franco for technical assistance. This work was supported by the Programma Biomolecole per la Salute Umana Ministero dell'Universitá e della Ricerca Scientifica e Tecnologica–Consiglio Nazionale delle Ricerche L95/955%; Fondo Investimenti Ricerca di Base Ministero Instruzione Universitá Ricerca; Progetto Genomica Funzionale L449/97, Ministero Instruzione Universitá Ricerca–Consiglio Nazionale delle Ricerche; Progetto Strategico Tecnologie di Base della Postgenomica, Consiglio Nazionale delle Ricerche; Progetto Strategico Biotecnologie, Ministero dell'Universitá e della Ricerca Scientifica e Tecnologica–Consiglio Nazionale delle Ricerche; Progetto Strategico Genetica Molecolare L449/97 Ministero dell'Universitá e della Ricerca Scientifica; and European Networks of Excellence (EUMORPHIA and MUGEN).
Author contributions: G.D.T.-V., P.F., and G.P.T.-V. designed research, performed research, analyzed data, and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BHB, bulge–helix–bulge; BHL, bulge–helix–loop; METJA, Methanocaldococcus jannaschii; SULSO, Sulfolobus solfataricus; ARCFU, Archeoglobus fulgidus; XENLA, Xenopus laevis.
References
- 1.Kjems, J., Jensen, J., Olesen, T. & Garrett, R. A. (1989) Can J. Microbiol. 35, 210-214. [DOI] [PubMed] [Google Scholar]
- 2.Kjems, J. & Garrett, R. A. (1991) Proc. Natl. Acad. Sci. USA 88, 439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kjems, J. & Garrett, R. A. (1988) Cell 54, 693-703. [DOI] [PubMed] [Google Scholar]
- 4.Tang, T. H., Rozhdestvensky, T. S., d'Orval, B. C., Bortolin, M. L., Huber, H., Charpentier, B., Branlant, C., Bachellerie, J. P., Brosius, J. & Huttenhofer, A. (2002) Nucleic Acids Res. 30, 921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabbri, S., Fruscoloni, P., Bufardeci, E., Di Nicola Negri, E., Baldi, M. I., Attardi, D. G., Mattoccia, E. & Tocchini-Valentini, G. P. (1998) Science 280, 284-286. [DOI] [PubMed] [Google Scholar]
- 6.Thompson, L. D. & Daniels, C. J. (1990) J. Biol. Chem. 265, 18104-18111. [PubMed] [Google Scholar]
- 7.Diener, J. L. & Moore, P. B. (1998) Mol. Cell 1, 883-894. [PubMed] [Google Scholar]
- 8.Kleman-Leyer, K., Armbruster, D. W. & Daniels, C. J. (1997) Cell 89, 839-847. [DOI] [PubMed] [Google Scholar]
- 9.Lykke-Andersen, J. & Garrett, R. A. (1997) EMBO J. 16, 6290-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marck, C. & Grosjean, H. (2003) RNA 9, 1516-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tocchini-Valentini, G. D., Fruscoloni, P. & Tocchini-Valentini, G. P. (2005) Proc. Natl. Acad. Sci. USA 102, 8933-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, R. A. (1976) Annu. Rev. Microbiol. 30, 409-425. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, A. L. & Friedman, R. (2005) Evol. Dev. 7, 196-200. [DOI] [PubMed] [Google Scholar]
- 14.Fribourg, S., Romier, C., Werten, S., Gangloff, Y. G., Poterszman, A. & Moras, D. (2001) J. Mol. Biol. 306, 363-373. [DOI] [PubMed] [Google Scholar]
- 15.Di Nicola Negri, E., Fabbri, S., Bufardeci, E., Baldi, M. I., Gandini Attardi, D., Mattoccia, E. & Tocchini-Valentini, G. P. (1997) Cell 89, 859-866. [DOI] [PubMed] [Google Scholar]
- 16.Li, H., Trotta, C. R. & Abelson, J. (1998) Science 280, 279-284. [DOI] [PubMed] [Google Scholar]
- 17.Li, H. & Abelson, J. (2000) J. Mol. Biol. 302, 639-648. [DOI] [PubMed] [Google Scholar]
- 18.Abelson, J., Trotta, C. R. & Li, H. (1998) J. Biol. Chem. 273, 12685-12688. [DOI] [PubMed] [Google Scholar]
- 19.Fruscoloni, P., Baldi, M. I. & Tocchini-Valentini, G. P. (2001) EMBO Rep. 2, 217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber, H., Hohn, M. J., Rachel, R., Fuchs, T., Wimmer, V. C. & Stetter, K. O. (2002) Nature 417, 63-67. [DOI] [PubMed] [Google Scholar]
- 21.Waters, E., Hohn, M. J., Ahel, I., Graham, D. E., Adams, M. D., Barnstead, M., Beeson, K. Y., Bibbs, L., Bolanos, R., Keller, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Randau, L., Munch, R., Hohn, M. J., Jahn, D. & Soll, D. (2005) Nature 433, 537-541. [DOI] [PubMed] [Google Scholar]
- 23.Hughes, A. L. (2005) Proc. Natl. Acad. Sci. USA 102, 8791-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes, V. M. & Abelson, J. (1988) Cell 55, 719-730. [DOI] [PubMed] [Google Scholar]
- 25.Mattoccia, E., Baldi, I. M., Gandini-Attardi, D., Ciafre, S. & Tocchini-Valentini, G. P. (1988) Cell 55, 731-738. [DOI] [PubMed] [Google Scholar]
- 26.Baldi, M. I., Mattoccia, E., Bufardeci, E., Fabbri, S. & Tocchini-Valentini, G. P. (1992) Science 255, 1404-1408. [DOI] [PubMed] [Google Scholar]
- 27.Abelson, J. (1992) Science 255, 1390. [DOI] [PubMed] [Google Scholar]
- 28.Deidda, G., Rossi, N. & Tocchini-Valentini, G. P. (2003) Nat. Biotechnol. 21, 1499-1504. [DOI] [PubMed] [Google Scholar]
- 29.Calvin, K., Hall, M. D., Xu, F., Xue, S. & Li, H. (2005) J. Mol. Biol., in press. [DOI] [PubMed]