Abstract
The costimulatory ligands B7-1 and B7-2 are expressed on the surface of antigen-presenting cells and interact with the costimulatory receptors CD28 and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) expressed on T cells. Although B7-1 and B7-2 are homologous ligands having common receptors, they exhibit distinct biochemical features and roles in immune regulation. Several biochemical and structural studies have indicated differences in the oligomeric state of B7-1 and B7-2. However, the organization of B7 ligands on the cell surface has not been examined. By using photobleaching-based FRET (pbFRET), we demonstrate that B7-1 and B7-2 adopt different oligomeric states on the cell surface. Our study shows that B7-2 exists as a monomer on the cell surface whereas B7-1 exists predominantly as dimers on the cell surface. A series of mutations in B7-1 result in the expression of a predominantly monomeric species on the cell surface and validate the dimer interface proposed by prior crystallographic analysis. The difference in the oligomeric states of B7-1 and B7-2 provides insight into the geometric organization of the costimulatory receptor–ligand complexes in the immunological synapse and suggests constraints on signal transduction mechanisms involved in T cell activation.
Keywords: oligomerization, photobleaching FRET, T cell costimulation
The engagement of B7-1 and B7-2 with CD28 and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) represents the best characterized T cell costimulatory interactions. CTLA-4 and CD28 share a similar molecular architecture, existing as disulfide linked homodimers of Ig variable (IgV) domains (1, 2). CTLA-4 and CD28 bind the B7 molecules with different affinities (3) and direct different T cell functional outcomes. Interaction of CD28 with B7 ligands delivers a positive signal to T cells that promotes proliferation and cytokine secretion and prevents the induction of T cell tolerance (4, 5). In contrast, the interaction of CTLA-4 with B7 ligands attenuates T cell activation and induces T cell anergy (6–8). Furthermore, recent studies have shown that CD28 and CTLA-4 binding to B7 ligands delivers different signals to dendritic cells. Binding of CD28 to B7 ligands leads to up-regulation of IL-6 production by dendritic cells, resulting in immunostimulatory activity (9). On the other hand, binding of CTLA-4 to B7 ligands up-regulates IFN-γ, which, in turn, up-regulates the expression of the enzyme indolamine, 2,3-dioxygenase in dendritic cells, resulting in tryptophan catabolism and suppression of T cell proliferation (10, 11).
B7-1 and B7-2 are both type 1 transmembrane proteins with a membrane distal IgV and a membrane proximal Ig constant (IgC) domain and share ≈25% sequence identity. Although interacting with the same receptors, B7-1 and B7-2 show several distinct features (12, 13). B7-1 binds CTLA-4 and CD28 with equilibrium dissociation constants (Kd) of 0.2 μM and 4 μM, respectively, whereas B7-2 exhibits ≈5- to 10-fold lower affinities, with Kd values of 2.6 μM and 20 μM for CTLA-4 and CD28, respectively (3). Although B7-2 is constitutively present on the cell surface of the antigen-presenting cell (APC) and is rapidly up-regulated upon interaction with CD28, the expression of B7-1 is induced (14). Furthermore, previous studies have shown that B7-2 augments overall T helper 2 (Th2)-type T cell responses whereas B7-1 preferentially favors Th1 type T cell differentiation. In some in vivo studies, antibody blockade of B7-1 resulted in enhancement of immune responses whereas blockade of B7-2 resulted in attenuation (15, 16). It has also been suggested that cross-linking B7-2 up-regulates B cell proliferation, enhances the expression of antiapoptotic molecule Bcl-xL and stimulates the production of IgG1, IgG2a, and IgE (17, 18). In contrast, cross-linking of B7-1 on B cells and B lymphomas leads to reduced proliferation and up-regulation of proapoptotic molecules caspase-3, caspase-8, Fas, FasL, Bak, and Bax (18, 19).
The structural mechanisms underlying the different T cell and APC responses upon ligation of B7 ligands with CD28 and CTLA-4 are not completely understood. Analytical ultracentrifugation has demonstrated that soluble B7-1 exists as a weak dimer, with a Kd of 20–50 μM (20). Furthermore, the B7-1 crystal structure revealed a highly conserved dimer interface that involves V11, V22, G45, M47, I58, D60, I61, T62, and L70 contributed from the B, C″, D, and E β-strands on the back sheet of the IgV domain (20). Additional support for the B7-1 dimer is provided by the crystal structure of B7-1 complexed with CTLA-4, which exhibits the same B7-1 dimer interface present in the unbound B7-1 structure (21). On the other hand, the structure of B7-2 IgV domain alone shows that it is a monomer in the crystalline state (22), which is consistent with the analytical ultracentrifugation and gel filtration studies. Interestingly, the crystal structure of the complex of CTLA-4 with B7-2 reveals a network of alternating CTLA-4 and B7-2 dimers, similar to that observed for the B7-1:CTLA-4 complex, suggesting the possibility that B7-2 can undergo receptor-induced dimerization, at least in the crystalline state (23). These findings suggest that the structural and oligomeric properties of B7-1 and B7-2 may be important determinants for the regulation and organization of macromolecular signaling complexes at the immunological synapse (IS). However, the biological relevance of these structural observations must await confirmation of the oligomeric state of the costimulatory molecules in the restricted environment of the cell surface.
Here, we have assessed the cell surface organization of the B7 ligands using confocal microscopy-based FRET, a phenomenon that occurs due to nonradiative energy transfer from an excited donor to an acceptor molecule (24, 25). Because energy transfer depends on the inverse sixth power of the distance between the two fluorophores, this phenomenon typically reports the proximity of two molecules on a scale of 10–100 Å, a distance that correlates with the size of the molecules on the cell surface (26, 27). Our data support a model in which B7-2 is present as a monomer on the cell surface, whereas B7-1 exists as a mixed population of monomers and dimers, with dimers predominating. Point mutations made in the putative dimer interface of B7-1 disrupt the dimeric state of the B7-1 molecule and result in the generation of a population with enhanced monomer representation. These mutagenesis studies support the validity of the B7-1 dimer interface previously suggested by the B7-1 crystal structure. The different oligomeric states of B7-1 and B7-2 indicate that these molecules form costimulatory receptor–ligand complexes with distinct cell surface organizations, which may represent an important mechanistic determinant for the assembly and functional properties of signaling complexes at the T cell–APC interface.
Materials and Methods
cDNA, Mutagenesis, Cell Lines, and Transfections. cDNAs for murine B7-1 and B7-2 were obtained as gifts from G. J. Freeman (Dana–Farber Cancer Institute, Boston, MA). These cDNAs were cloned into ECFP-N1 or EYFP-N1 (Clontech) expression vectors that had been earlier modified by site-directed mutagenesis to contain the A206K mutation that prevents the self-dimerization of the fluorophores (R. Tsien, personal communication). The mutants of B7-1 were generated by site-directed mutagenesis by using specific 5′ and 3′ primers. The “cysteine trap” mutants (CYS) contained an 8-aa residue linker (GAGAGCGA) in the stalk region, proximal to the transmembrane domain. The control (CTRL) for the cysteine trap mutant had an 8-aa residue linker lacking the cysteine (GAGAGAGA). The dimer interface mutants of B7-1 had the control linker in their stalk region. For flow cytometry, see Supporting Methods, which is published as supporting information on the PNAS web site.
CHO cells were maintained in RPMI medium 1640 supplemented with 10% FCS, penicillin-streptomycin and GlutaMAX. CHO cells (5–6 × 104) were seeded on glass cover slips (Fisher), grown overnight, and then cotransfected with the vectors coding for cyan fluorescent protein (CFP)- or yellow fluorescent protein (YFP)-tagged proteins, by using FuGENE 6 (Roche Biochemicals) as per the manufacturer's protocol. Twenty-four hours posttransfection, cells were fixed with 4% paraformaldehyde and washed with PBS (containing Ca2+ and Mg2+), and the coverslips were mounted onto glass slides by using Anti-Fade reagent (Molecular Probes).
Western Blotting. CHO cells expressing either the WT mouse B7-1, B7-2, or their mutants were lysed on ice in 20 mM Tris·HCl (pH 7.6), 40 mM NaCl, 0.5% Triton X-100, 5 mM EDTA, 0.1% SDS and protease inhibitor mixture (Sigma). Lysates were clarified after centrifugation at 10,000 × g for 15 min. The amount of total protein in each sample was quantified with the Micro BCA assay (Pierce), and equal loads of protein were analyzed by SDS/PAGE and transferred onto poly(vinylidene difluoride) (PVDF) membrane (Bio-Rad). The membranes were probed with specific Abs against B7-1 or B7-2 (Santa Cruz Biotechnology and R & D Systems, respectively). Blots were developed by using horseradish peroxidase (HRP)-conjugated appropriate secondary antibodies (Santa Cruz Biotechnology) and the ECL Chemiluminescent Detection System (Amersham Pharmacia).
Photobleaching FRET by Confocal Microscopy. CHO cells transfected with CFP- and YFP-fused plasmid constructs were examined with ×63 objective lens on a Leica TCS SP II AOBS laser-scanning confocal microscope at the Analytical Imaging Facility, Albert Einstein College of Medicine. An excitation wavelength of 405 nm and an emission range of 416–492 nm, and an excitation wavelength of 514 nm and an emission range of 525–600 nm were used to acquire images of CFP and YFP, respectively. YFP was photobleached by using full power of the 514-nm line for 1–2 min. An image of CFP fluorescence and YFP fluorescence after photobleaching was obtained by using the respective filter sets. Such data were collected from 30–50 different cells in different fields from the same coverslip. Four to five regions of interest (located on the cell membrane) in the photobleached area were selected per cell, and the mean CFP fluorescence before and after photobleaching was obtained by using leica confocal software (lcs, Leica, Vienna). FRET efficiency was calculated by using the following relationship: FRET efficiency (E) % = [(Dpost – Dpre)/Dpost] × 100. Here, Dpost is the fluorescence intensity of the CFP (donor; D) after photobleaching and Dpre is the fluorescence intensity of the CFP before photobleaching. All of the data were acquired in 8-bit format.
Statistical Analysis. Student's t test (two tailed) was performed to demonstrate the level of significance between various groups. A P value of <0.01 was considered significant.
Results
Analysis and Experimental Validation of Photobleaching-Based FRET (pbFRET). pbFRET (28, 29) was used to examine the oligomeric state of the B7-1 and B7-2 costimulatory ligands on the cell surface. Using a combination of imaging and FRET allows energy transfer between the molecules of interest to be directly determined on a single cell basis, and, thus, the interaction of molecules of interest can be studied on intact cell membranes. Because the fluorescent FRET donor is quenched when in the proximity of an acceptor, FRET efficiency (E) can be measured by comparing donor fluorescence before and after photobleaching the acceptor. An increase in donor fluorescence after acceptor photobleaching indicates that donor and acceptor fluorophores were within FRET range (28, 29). Importantly, FRET results can also be analyzed to distinguish between a population of randomly distributed molecules (where there is a small but finite probability that some donors will be within FRET distance of acceptors) and specific clustering of acceptor molecules. Physiologically, the clusters of proteins can be envisaged to range from simple dimers to complex higher order oligomers. A mixed distribution is characterized by the presence of both clustered and randomly organized molecules. Based on theoretical models (30, 31), a clustered distribution results in an E that is independent of acceptor density whereas, in a random distribution, E increases with an increase in the acceptor density. Furthermore, in a clustered distribution, E increases with an increase in acceptor-to-donor ratio whereas, in a random distribution, E does not vary with the acceptor to donor ratio. A mixed distribution exhibits the features of both random and clustered distribution as E increases both with the acceptor density and the acceptor-to-donor ratio (30, 31).
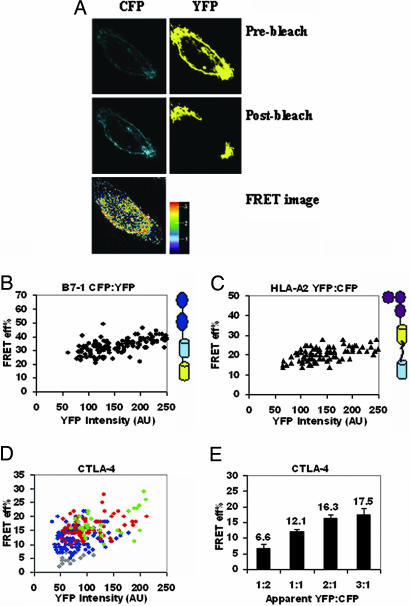
We validated our pbFRET system (schematically shown in Fig. 6, which is published as supporting information on the PNAS web site) by transiently expressing a construct in CHO cells in which CFP and YFP (separated by 7-aa residues) were tandemly fused at the C terminus of B7-1 (Fig. 1A). Because CFP and YFP are contained within the same polypeptide chain, the resulting intramolecular FRET represents the FRET expected between clustered molecules. Thus, for the tandem CFP-YFP construct, E was independent of YFP intensity over a broad range of intensities, a feature expected for clustered molecules (Fig. 1B). Only when the YFP intensity, which is a measure of cell surface density of the molecules, was >200 arbitrary units (AU), indicating high expression of the tandem molecule, did we observe a slight dependence of E on YFP intensity. We attribute this slight dependence of E on YFP intensity to intermolecular FRET between CFP and YFP on two different molecules, which are in proximity due solely to surface crowding effects and not specific interactions. In subsequent experiments, only YFP intensities <200 AU were considered for the analyses. Our findings with another tandem positive control, HLA-A2 tagged with YFP and CFP (separated by 26-aa residues) at its C terminus (31), were similar to those with the tandem B7-1 (Fig. 1C). Taken together, these results set the appropriate threshold for experimental interpretation and support our approach for image-based pbFRET to assess the oligomeric state of B7-1 and B7-2.
Fig. 1.
Validation of pbFRET for covalently linked CFP and YFP tandem proteins and characterization of CTLA-4. (A) Pre- and postbleaching of cells expressing CFP and YFP (Top and Middle row; note the enhancement in the intensity of CFP postbleaching). (Bottom) The pseudocolored FRET image with a color gradient from blue to red with values 1 (minimum) to 3 (maximum) for FRET. FRET efficiency, E, for tandem (B7-1)-CFP-YFP (B) and tandem (HLA-A2)-YFP-CFP (C) is independent of YFP intensity, over a wide range of YFP intensities, expressed as arbitrary units (AU). (D) E for CTLA-4 is independent of YFP intensity at each acceptor (YFP) to donor (CFP) ratio: gray (1:2), blue (1:1), red (2:1). and green (3:1) YFP-to-CFP ratio. (E) Average E for CTLA-4 increases with an increase in YFP-to-CFP ratio. The average E was calculated from the E corresponding to the range of 50–220 AUs of YFP to exclude the very low and very high intensity points that either give rise to noise or are outside the experimentally observed range. The error bars represent the 0.01% confidence limits. The P values between the adjacent ratios of YFP to CFP (i.e., 1:2 and 1:1, 1:1 and 2:1) were <0.01. The data are pooled from three independent experiments.
CTLA-4 Exists as a Dimer on the Cell Surface. Biochemical and structural analyses have demonstrated that CTLA-4 forms a disulfide-linked homodimer, and it is likely that this state represents the oligomeric form present on the cell surface (2, 32). The distribution pattern of CTLA-4 on the cell surface was examined by pbFRET with the rationale of establishing it as a positive control for the detection of a dimer on the cell surface. CFP- and YFP-tagged CTLA-4 were coexpressed in CHO cells. As described above, theoretical models were used to distinguish between random, mixed, or clustered distributions of molecules on the cell surface (30–32). For CTLA-4, the E was found to be independent of YFP intensity (Fig. 1D) at any given apparent YFP-to-CFP ratio and increased with an increase in YFP-to-CFP ratio (Fig. 1E). These results correlate with a clustered distribution of CTLA-4 molecules and are completely consistent with the theoretical prediction for a covalent dimer, thereby supporting a model in which CTLA-4 is present as a dimer on the cell surface.
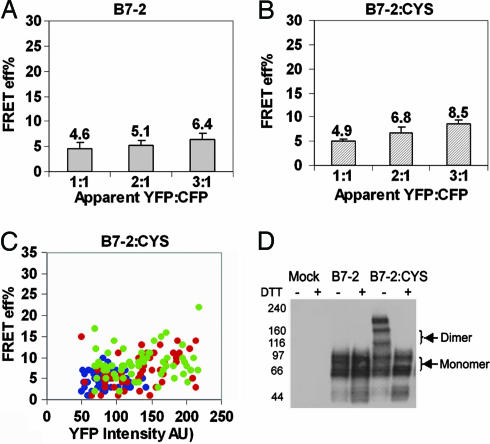
B7-2 Exists as a Monomer on the Cell Surface. To examine the membrane organization of B7-2, CFP- and YFP-tagged B7-2 were coexpressed in CHO cells. The average E for these B7-2 constructs was 5–7% (Fig. 7, which is published as supporting information on the PNAS web site), a value that we considered the instrument threshold, because our experiments with a previously reported negative control (33, 34) showed an average E of 5% (data not shown). Because of a very low E for B7-2, the dependence of E on the YFP intensity could not be measured (Fig. 7). The possibility that the absence of FRET might be due to unfavorable relative molecular orientations of the fluorophores was ruled out by a significant E (≈10%) that was observed at very high YFP intensities (YFP AU >220; data not shown). Our previously published results with programmed death-1 (PD-1) also ruled out the length of the cytoplasmic tail as one of the possible factors contributing to the lack of FRET (35). Furthermore, there was no statistically significant increase in the E as the ratio of YFP to CFP was increased (P > 0.1; Fig. 2A). Altogether, these observations suggest a random distribution for B7-2 implying that B7-2 exists as a monomer on the cell surface.
Fig. 2.
B7-2 as a monomer on the cell surface. (A) Average E (calculated as in Fig. 1E) for B7-2 does not change with an increase in YFP-to-CFP ratio. (B) Average E for B7-2:CYS does not show a consistently significant increase with increase in YFP-to-CFP ratio. (C) E for B7-2:CYS modestly increases with an increase in YFP intensity at different acceptor-to-donor ratios; 1:1 (blue), 2:1 (red), and 3:1 (green) YFP-to-CFP ratios. The data are pooled from two independent experiments. The P value between the adjacent ratios of YFP to CFP in A was >0.05. For B, the P value was <0.01 between 1:1 and 2:1 and >0.01 between 2:1 and 3:1 YFP-to-CFP ratio. The error bars in A and B represent the 0.01% confidence limits. (D) Whole cell lysates of CHO cells transiently expressing B7-2-YFP or B7-2:CYS-YFP were immunoblotted with anti-B7-2. Western blot analysis shows that B7-2:CYS exists as a mixture of monomers and dimers. The “artifactual” dimers of B7-2:CYS arise from cells expressing very high levels of B7-2:CYS.
Earlier studies with signaling molecules such as CD45 have shown that the introduction of a cysteine into the stalk region of the molecule allowed the formation of disulfide-linked dimers, which were detectable by conventional techniques such as Western blotting (36). By using a similar strategy, a cysteine trap mutant of B7-2 (B7-2:CYS) was generated with the rationale of trapping any population of dimers that might exist and that would be below the FRET detection limit. Western blot data revealed that B7-2:CYS could potentially form dimers (≈50%) although it still existed as a mixture of monomers and dimers (Fig. 2D). A higher order oligomer (band above the indicated dimers) was also detected, which presumably resulted due to overexpression usually observed in transient transfections (no such higher order oligomers were observed in a similar experiment using cells with stable expression of B7-2:CYS; data not shown). With our detailed pbFRET-based analysis, we found that the E for B7-2:CYS increased modestly with an increase in YFP intensity (Fig. 2C). The increase in E value observed at high concentration of B7-2:CYS can be attributed to high expression resulting in an increase in the random colocalization, allowing FRET to occur (Fig. 2C). Moreover, E did not show a consistent dependence on YFP-to-CFP ratio (Fig. 2B). This behavior suggests a random distribution of molecules on the cell surface, implying that the B7-2:CYS molecules are still present predominantly as monomers, although the possibility of concentration-dependent dimerization, which might be below the FRET detection limit, cannot be completely ruled out. The seemingly contradictory observations made by Western blot and FRET-based analysis can be readily reconciled by the fact that, whereas a Western blot characterizes the organization of a protein in a cell population with diverse expression levels, image-based FRET reports on the protein organization on the cell surface on a single cell basis. In the FRET-based results, the cells with high YFP expression (which correspond to the cells with high expression of B7-2:CYS) showed high E (data not shown). Thus, we believe that B7-2:CYS molecules on the cells with high expression levels contribute to the dimers observed by the Western blot. Thus, caution should be taken while analyzing data solely on the basis of Western blotting. Taken together, these results support a model in which B7-2 exists as a monomer on the cell surface.
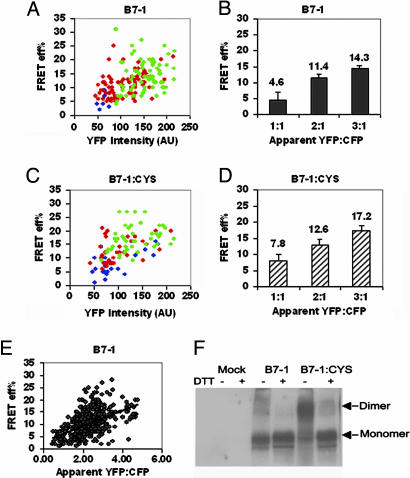
B7-1 Is Present Predominantly as a Dimer on the Cell Surface. To examine the cell surface organization of B7-1, the interaction between B7-1 molecules expressed as CFP or YFP fusion proteins was studied. We found that the E increased with an increase in YFP intensity (Fig. 3A). Moreover, a statistically significant increase in E was observed with an increase in ratio of YFP to CFP (P < 0.01; Fig. 3B). This behavior is consistent with a mixed population, suggesting that B7-1 molecules are present as a mixture of monomers and clusters on the cell surface. Furthermore, there was a linear dependence of E on the YFP-to-CFP ratio (slope of the plot in Fig. 3E). The linear dependence of FRET efficiency on acceptor-to-donor ratio has been reported to indicate a monomer/dimer equilibrium and absence of higher order oligomers (37, 38). These data show that B7-1 is present as a mixture of monomers and dimers.
Fig. 3.
B7-1 is predominantly a dimer on the cell surface. (A) E for B7-1 increases with an increase in YFP intensity at each YFP-to-CFP ratio. (B) Average E (calculated as in Fig. 1E) increases with an increase in YFP-to-CFP ratio. (C) E for B7-1:CYS increases with an increase in YFP intensity at each YFP-to-CFP ratio. (D) Average E for B7-1:CYS increases with an increase in YFP-to-CFP ratio. The symbols in A and C are 1:1 (blue), 2:1 (red), and 3:1 (green) YFP-to-CFP ratios. The error bars in B and D represent the 0.01% confidence limits. The data are representative of two independent experiments. The P value between the adjacent ratios of YFP to CFP in B and D was <0.01. (E) E for B7-1 shows a linear dependence on YFP-to-CFP ratio. (F) Western blot of whole cell lysates of cells transiently expressing B7-1 or B7-1:CYS shows the presence of dimers of B7-1:CYS.
To determine the relative fraction of monomer and dimer, the cysteine trap mutant of B7-1 (B7-1:CYS) was generated. Western blot analyses revealed that B7-1:CYS exists as a mixture of covalent dimers (≈80%) and monomers (Fig. 3F). No higher order oligomers were observed. In our FRET-based experiments with B7-1:CYS, we hypothesized that a change in either the absolute value of E or its dependence on YFP intensity would be indicative of the relative proportion of monomeric and dimeric forms of B7-1. The presence of cysteine in B7-1 did not change the trend of the dependence of E on YFP intensity (the slope of the plot in Fig. 3 A and C) and acceptor-to-donor ratio (Fig. 3 B and D). However, there was a modest increase in the absolute E at different ratios of YFP to CFP compared with WT B7-1 (Fig. 3 B and D). This small change could be explained by a small overall increase in the population of dimeric B7-1 molecules when a cysteine is introduced, suggesting that, under steady-state conditions, most of the WT B7-1 molecules are present as noncovalent dimers on the cell surface.
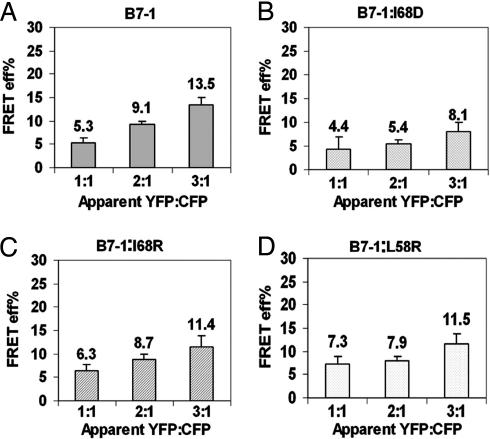
L58 and I68 Mutations in the Putative Dimer Interface Interfere with the Dimerization of B7-1. Based on existing structural data, mutations were made that would be predicted to disrupt the putative dimer interface (26). Substitution point mutations at Leu-58 (B7-1:L58R) and Ile-68 (B7-1:I68R, B7-1:I68D) were made. These mutations did not affect the cell surface expression nor binding to CTLA-4 and CD28 because the mutants and WT B7-1 showed equivalent equilibrium binding to CTLA-4:Ig and CD28:Ig (Fig. 8, which is published as supporting information on the PNAS web site). CHO cells were cotransfected with CFP- and YFP-tagged B7-1:L58R, -:I68D, or -:I68R, and FRET efficiency was analyzed as a function of the acceptor-to-donor ratio. For these mutants, there was no systematic dependence of E on acceptor-to-donor ratio (Fig. 4 B–D) compared with WT B7-1 (Fig. 4A). This behavior is characteristic of a system that contains randomly distributed molecules on the cell surface. However, of all of the mutants tested, the I68D mutation was observed to be the most effective in interfering with the dimeric nature of B7-1 (Fig. 4B). Thus, the data suggest that these residues are involved in the physiological dimerization of B7-1 on the cell surface.
Fig. 4.
Mutations at L58 and I68 residues interfere with the dimeric state of B7-1. (A) E for WT B7-1 increases with an increase in YFP-to-CFP ratio whereas E for I68D (B), I68R (C), and L58R (D) does not show any systematic dependence on YFP-to-CFP ratio. The error bars represent the 0.01% confidence limits. The data are pooled from three independent experiments. The P value between 1:1 and 2:1 YFP-to-CFP ratio in B, C, and D was ≈0.5 whereas in A it was <0.01.
Discussion
Numerous biochemical and structural studies have provided information on the potential for costimulatory molecules to form oligomers in vitro (20–23, 39). However, it is the oligomeric states adopted by these molecules when confined to the restricted environment of the plasma membrane that is biologically relevant because they provide important constraints on the mechanisms governing signal transduction in both T cells and APCs.
Solution and structural studies suggest that B7-2 exists as a monomer (22). Our FRET studies with B7-2 support a model in which B7-2 is a monomer on the plasma membrane. Consistent with the apparent lack of propensity to dimerize, sequence and structural analysis reveal that the putative dimer interface of B7-2 is comprised mainly of hydrophilic amino acid residues, in contrast to the hydrophobic interdimer surface present in B7-1 (22). Notably, both B7-1 and B7-2 exhibit roughly similar dimeric organizations in their crystalline complexes with CTLA-4, raising the possibility of receptor-induced dimerization although the B7-2 dimer deviates modestly from ideal 2-fold symmetry. The deviation from 2-fold symmetry and the hydrophilic nature of the putative B7-2 interface raise questions as to whether the observed interactions may be solely the consequence of crystal packing, and to date there are no data supporting the dimerization of B7-2 in vivo (23).
Although the FRET-based data suggest that B7-2 is a monomer, the sensitivity of this approach would likely preclude the detection of a small population of dimers. Accordingly, we used a strategy previously applied in the study of CD45 to further probe for any potentially weak tendency for B7-2 to dimerize (36). The fact that B7-2 gives a very low FRET efficiency even upon introducing the cysteine suggests that B7-2 may have a tendency to resist dimerization. Thus, B7-2 alone has little or no propensity to exist as a dimer when expressed on the cell surface.
In contrast to B7-2, several lines of evidence based on crystallographic and biochemical studies have indicated that B7-1 tends to homodimerize spontaneously (20, 21). Our FRET analysis clearly supports a model in which B7-1 exists as a mixture of monomers and dimers, predominantly dimers, on the cell surface. The L58R, I68R, and I68D mutations in the crystallographically observed dimer interface result in a significant decrease in the dependence of FRET efficiency on acceptor to donor ratio (Fig. 4A-4D). This behavior is characteristic of a random distribution of molecules and is consistent with the disruption of the B7-1 dimer on the cell surface. These data are also consistent with the absence of any additional higher order oligomerization states of B7-1 on the cell surface. It is important to stress that the observed behavior of the FRET efficiency is a qualitative effect and these data do not allow the relative levels of dimers and monomers to be quantified. This limitation is likely to be an important consideration for the interpretation of functional responses, which may be far more sensitive than global biophysical characterization.
B7-1 and B7-2 costimulatory ligands have been directly implicated as signaling molecules in B cells and more recently in dendritic cells (9, 10, 17, 18). For example, antibody-mediated cross-linking of B7-1 caused suppression of B cell proliferation and up-regulation of proapoptotic genes. On the other hand, cross-linking of B7-2 promoted proliferation of B cells and up-regulation of antiapoptotic genes (17, 18). These differences in the biological function of B7-1 and B7-2 are likely related to the nature of their cytoplasmic tails and the signaling molecules that are recruited to them. It is also likely that the different cell surface oligomeric states of B7-1 and B7-2 molecules play a role, because these features contribute to the local concentration and geometric organization of signaling complexes.
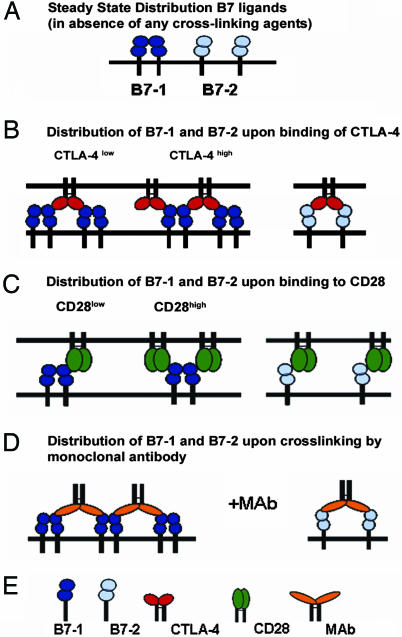
The adoption of different oligomeric states of B7-1 and B7-2 suggest possible interpretations for the observed effects of antibody-mediated triggering. Because B7-1 exists predominantly as a dimer, cross-linking it with monoclonal antibodies could result in the formation of an extended, ordered array as is observed in the crystal structure of the B7-1:CTLA-4 complex (Fig. 5D Left). On the other hand, for B7-2, which exists as a monomer, the antibodies may simply induce clustering in which two B7-2 molecules are bridged by a single bivalent monoclonal antibody (Fig. 5D Right). This proposed behavior would result in an increased local concentration of dimeric B7-1 but a uniform, random distribution of cross-linked B7-2 monomers. The organization of B7 ligands upon binding bivalent antibodies is likely to have relevance to the physiological state when the ligands bind to their preferred receptors, which have been suggested by recent studies to be CTLA-4 for B7-1 and CD28 for B7-2 (ref. 40; Fig. 5 B and C). For instance, the dimeric nature of B7-1 would allow it to participate in an organized assembly with CTLA-4 in the central zone of the IS. In contrast, the monomeric form of B7-2 would result in a uniform distribution of B7-2 and its associated receptor at the IS. Altogether, these differences between the organization states of B7-2 and B7-1 may play an important role in modulating the function of APCs.
Fig. 5.
Models for the interaction of CTLA-4 and CD28 with B7-1 and B7-2. (A) Steady-state distribution of B7-1 (dimer) and B7-2 (monomer). (B) Assemblies of B7-1 and B7-2 upon binding to CTLA-4. At low concentration of CTLA-4 (relative to B7-1), a bivalent homodimeric CTLA-4 molecule can bind two B7-1 molecules, but, at high concentration of CTLA-4, it may form an extended array with B7-1. However, with monomeric B7-2, CTLA-4 can engage only two B7-2 monomers. (C) Assemblies of B7-1 and B7-2 upon binding to CD28. At low concentration of monovalent CD28, a dimeric B7-1 could engage a single CD28 molecule; however, at high concentration of CD28, two molecules of CD28 may be bridged by one molecule of B7-1 without the possibility of forming higher order assemblies. With monomeric B7-2, monovalent CD28 may form single solitary complexes. (D) Assemblies of B7-1 and B7-2 upon cross-linking by bivalent monoclonal antibodies. Anti-B7-1 antibodies may interact with dimeric B7-1 like CTLA-4 and form an ordered network whereas anti-B7-2 antibodies may simply induce bridging of two B7-2 monomers. (E) Key for the symbols.
The different oligomeric states of B7-1 and B7-2 are also likely to have functional consequences for T cells. It is well documented that both the B7 ligands and their receptors, CD28 and CTLA-4, are concentrated at the central zone of the IS (39, 41). However, the nature of the complexes formed by B7-1 or B7-2 with their receptors at the IS may differ qualitatively (Fig. 5 B and C). Consistent with the observed B7-1:CTLA-4 complex crystal structure, dimeric B7-1 may be able to form ordered arrays with CTLA-4 as well as lead to increased concentration of CTLA-4 at the IS (Fig. 5B Left). On the other hand, binding of CTLA-4 to monomeric B7-2 may result in localization or enrichment of isolated complexes at the IS (Fig. 5B Right). It is also possible that, at high local concentration, as may be achieved in the IS, B7-2 may undergo dimerization upon binding to CTLA-4 as has been suggested from crystallographic studies (23).
In contrast to CTLA-4, recent studies have suggested that CD28 is monovalent (42), which would result in either the sequestration of “solitary” complexes or the bridging of two CD28 molecules by a single B7-1 molecule (Fig. 5C Left). The relative abundance of these two types of complexes (as well as all of the other complexes discussed) will depend on the concentration of the two species at the immunological synapse. Only single isolated complexes would be expected between monomeric B7-2 and monovalent CD28; no evidence currently exists that supports dimerization of B7-2 upon binding to CD28 (Fig. 5C Right). Hence, because of the unique organization of B7 ligands and their complexes with CD28 or CTLA-4, it becomes important to quantitate the number of molecules of both the ligands and the receptors at the immunological synapse.
In summary, T cell activation is a complex process, characterized by a differential expression pattern, as well as different affinities between competing receptors and ligands. In addition, our data suggest that the unique oligomeric states of the costimulatory ligands may also represent a key mechanistic feature of T cell activation.
Supplementary Material
Acknowledgments
We thank Drs. T. Dilorenzo, S. Porcelli, M. Scharff, E. Lázár-Molnár, D. Brims, K. Chattopadhyay, and M. Roden and Mr. E. Cao for their insightful discussions; M. Cammer and the staff at the Analytical Imaging Facility for assistance; and H. Qian for statistical analysis. We also acknowledge the help of M. Everett and G. Sexton at the Johns Hopkins University. This work was supported by National Institutes of Health Grants AI07289, and T32CA09173 (to S.G.N.) and AI14584 (to M.E.).
Author contributions: S.B., S.C.A., and S.G.N. designed research; S.B. performed research; S.B. contributed new reagents/analytic tools; S.B., M.E., and S.C.A. analyzed data; S.B. wrote the paper; M.E. and S.C.A. provided a critical reading of the paper; and S.G.N. provided critical discussion of the results and the manuscript.
Abbreviations: CTLA, cytotoxic T lymphocyte-associated antigen; IgV, Ig variable; APC, antigen-presenting cell; IS, immunological synapse; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; pbFRET, photobleaching-based FRET; E, FRET efficiency; AU, arbitrary unit.
References
- 1.Peach, R. J., Bajorath, J., Brady, W., Leytze, G., Greene, J., Naemura, J. & Linsley, P. S. (1994) J. Exp. Med. 180, 2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greene, J. L., Leytze, G. M., Emswiler, J., Peach, R., Bajorath, J., Cosand, W. & Linsley, P. S. (1996) J. Biol. Chem. 271, 26762–26771. [DOI] [PubMed] [Google Scholar]
- 3.van der Merwe, P. A. & Davis, S. J. (2003) Annu. Rev. Immunol. 21, 659–684. [DOI] [PubMed] [Google Scholar]
- 4.Thompson, C. B., Lindsten, T., Ledbetter, J. A., Kunkel, S. L., Young, H. A., Emerson, S. G., Leiden, J. M. & June, C. H. (1989) Proc. Natl. Acad. Sci. USA 86, 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linsley, P. S., Brady, W., Grosmaire, L., Aruffo, A., Damle, N. K. & Ledbetter, J. A. (1991) J. Exp. Med. 173, 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walunas, T. L., Lenschow, D. J., Bakker, C. Y., Linsley, P. S., Freeman, G. J., Green, J. M., Thompson, C. B. & Bluestone, J. A. (1994) Immunity 1, 405–413. [DOI] [PubMed] [Google Scholar]
- 7.Waterhouse, P., Penninger, J. M., Timms, E., Wakeham, A., Shahinian, A., Lee, K. P., Thompson, C. B., Griesser, H. & Mak, T. W. (1995) Science 270, 985–988. [DOI] [PubMed] [Google Scholar]
- 8.Tivol, E. A., Borriello, F., Schweitzer, A. N., Lynch, W. P., Bluestone, J. A. & Sharpe, A. H. (1995) Immunity 3, 541–547. [DOI] [PubMed] [Google Scholar]
- 9.Orabona, C., Grohmann, U., Belladonna, M. L., Fallarino, F., Vacca, C., Bianchi, R., Bozza, S., Volpi, C., Salomon, B. L., Fioretti, M. C., et al. (2004) Nat. Immunol. 5, 1134–1142. [DOI] [PubMed] [Google Scholar]
- 10.Grohmann, U., Orabona, C., Fallarino, F., Vacca, C., Calcinaro, F., Falorni, A., Candeloro, P., Belladonna, M. L., Bianchi, R., Fioretti, M. C. & Puccetti, P. (2002) Nat. Immunol. 3, 1097–1101. [DOI] [PubMed] [Google Scholar]
- 11.Munn, D. H., Sharma, M. D. & Mellor, A. L. (2004) J. Immunol. 172, 4100–4110. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, G. J., Freedman, A. S., Segil, J. M., Lee, G., Whitman, J. F. & Nadler, L. M. (1989) J. Immunol. 143, 2714–2722. [PubMed] [Google Scholar]
- 13.Freeman, G. J., Gribben, J. G., Boussiotis, V. A., Ng, J. W., Restivo, V. A., Jr., Lombard, L. A., Gray, G. S. & Nadler, L. M. (1993) Science 262, 909–911. [DOI] [PubMed] [Google Scholar]
- 14.Greenwald, R. J., Freeman, G. J. & Sharpe, A. H. (2005) Annu. Rev. Immunol. 23, 515–548. [DOI] [PubMed] [Google Scholar]
- 15.Lenschow, D. J., Walunas, T. L. & Bluestone, J. A. (1996) Annu. Rev. Immunol. 14, 233–258. [DOI] [PubMed] [Google Scholar]
- 16.Kuchroo, V. K., Das, M. P., Brown, J. A., Ranger, A. M., Zamvil, S. S., Sobel, R. A., Weiner, H. L., Nabavi, N. & Glimcher, L. H. (1995) Cell 80, 707–718. [DOI] [PubMed] [Google Scholar]
- 17.Jeannin, P., Delneste, Y., Lecoanet-Henchoz, S., Gauchat, J. F., Ellis, J. & Bonnefoy, J. Y. (1997) J. Biol. Chem. 272, 15613–15619. [DOI] [PubMed] [Google Scholar]
- 18.Suvas, S., Singh, V., Sahdev, S., Vohra, H. & Agrewala, J. N. (2002) J. Biol. Chem. 277, 7766–7775. [DOI] [PubMed] [Google Scholar]
- 19.Hirokawa, M., Kuroki, J., Kitabayashi, A. & Miura, A. B. (1996) Immunol. Lett. 50, 95–98. [DOI] [PubMed] [Google Scholar]
- 20.Ikemizu, S., Gilbert, R. J., Fennelly, J. A., Collins, A. V., Harlos, K., Jones, E. Y., Stuart, D. I. & Davis, S. J. (2000) Immunity 12, 51–60. [DOI] [PubMed] [Google Scholar]
- 21.Stamper, C. C., Zhang, Y., Tobin, J. F., Erbe, D. V., Ikemizu, S., Davis, S. J., Stahl, M. L., Seehra, J., Somers, W. S. & Mosyak, L. (2001) Nature 410, 608–611. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, X., Schwartz, J. C., Almo, S. C. & Nathenson, S. G. (2003) Proc. Natl. Acad. Sci. USA 100, 2586–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz, J. C., Zhang, X., Fedorov, A. A., Nathenson, S. G. & Almo, S. C. (2001) Nature 410, 604–608. [DOI] [PubMed] [Google Scholar]
- 24.Edidin, M. (2002) Curr. Protoc. Immunol. 18.10.1–18.10.18, 10.1002/0471142735.im1810s52. [DOI]
- 25.Szollosi, J., Damjanovich, S. & Matyus, L. (1998) Cytometry 34, 159–179. [PubMed] [Google Scholar]
- 26.Kenworthy, A. K. (2001) Methods 24, 289–296. [DOI] [PubMed] [Google Scholar]
- 27.Kenworthy, A. K. & Edidin, M. (1999) Methods Mol. Biol. 116, 37–49. [DOI] [PubMed] [Google Scholar]
- 28.Jovin, T. M. & Arndt-Jovin, D. J. (1989) Annu. Rev. Biophys. Biophys. Chem. 18, 271–308. [DOI] [PubMed] [Google Scholar]
- 29.Jares-Erijman, E. A. & Jovin, T. M. (2003) Nat. Biotechnol. 21, 1387–1395. [DOI] [PubMed] [Google Scholar]
- 30.Kenworthy, A. K. & Edidin, M. (1998) J. Cell Biol. 142, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pentcheva, T. & Edidin, M. (2001) J. Immunol. 166, 6625–6632. [DOI] [PubMed] [Google Scholar]
- 32.Linsley, P. S., Nadler, S. G., Bajorath, J., Peach, R., Leung, H. T., Rogers, J., Bradshaw, J., Stebbins, M., Leytze, G., Brady, W., et al. (1995) J. Biol. Chem. 270, 15417–15424. [DOI] [PubMed] [Google Scholar]
- 33.Chan, F. K., Chun, H. J., Zheng, L., Siegel, R. M., Bui, K. L. & Lenardo, M. J. (2000) Science 288, 2351–2354. [DOI] [PubMed] [Google Scholar]
- 34.Siegel, R. M., Frederiksen, J. K., Zacharias, D. A., Chan, F. K., Johnson, M., Lynch, D., Tsien, R. Y. & Lenardo, M. J. (2000) Science 288, 2354–2357. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, X., Schwartz, J. C., Guo, X., Bhatia, S., Cao, E., Lorenz, M., Cammer, M., Chen, L., Zhang, Z. Y., Edidin, M. A., et al. (2004) Immunity 20, 337–347. [DOI] [PubMed] [Google Scholar]
- 36.Majeti, R., Xu, Z., Parslow, T. G., Olson, J. L., Daikh, D. I., Killeen, N. & Weiss, A. (2000) Cell 103, 1059–1070. [DOI] [PubMed] [Google Scholar]
- 37.Adair, B. D. & Engelman, D. M. (1994) Biochemistry 33, 5539–5544. [DOI] [PubMed] [Google Scholar]
- 38.Li, E., You, M. & Hristova, K. (2005) Biochemistry 44, 352–360. [DOI] [PubMed] [Google Scholar]
- 39.Egen, J. G. & Allison, J. P. (2002) Immunity 16, 23–35. [DOI] [PubMed] [Google Scholar]
- 40.Pentcheva-Hoang, T., Egen, J. G., Wojnoonski, K. & Allison, J. P. (2004) Immunity 21, 401–413. [DOI] [PubMed] [Google Scholar]
- 41.Bromley, S. K., Iaboni, A., Davis, S. J., Whitty, A., Green, J. M., Shaw, A. S., Weiss, A. & Dustin, M. L. (2001) Nat. Immunol. 2, 1159–1166. [DOI] [PubMed] [Google Scholar]
- 42.Evans, E. J., Esnouf, R. M., Manso-Sancho, R., Gilbert, R. J., James, J. R., Yu, C., Fennelly, J. A., Vowles, C., Hanke, T., Walse, B., et al. (2005) Nat. Immunol. 6, 271–279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.