Abstract
Most proteins that are to be imported into the mitochondrial matrix are synthesized as precursors, each composed of an N-terminal targeting sequence followed by a mature domain. Precursors are recognized through their targeting sequences by receptors at the mitochondrial surface and are then threaded through import channels into the matrix. Both the targeting sequence and the mature domain contribute to the efficiency with which proteins are imported into mitochondria. Precursors must be in an unfolded conformation during translocation. Mitochondria can unfold some proteins by changing their unfolding pathways. The effectiveness of this unfolding mechanism depends on the local structure of the mature domain adjacent to the targeting sequence. This local structure determines the extent to which the unfolding pathway can be changed and, therefore, the unfolding rate increased. Atomic force microscopy studies find that the local structures of proteins near their N and C termini also influence their resistance to mechanical unfolding. Thus, protein unfolding during import resembles mechanical unfolding, and the specificity of import is determined by the resistance of the mature domain to unfolding as well as by the properties of the targeting sequence.
Keywords: protein unfolding, mitochondria, protein import, protein sorting, atomic force microscopy
Protein unfolding is intimately associated with protein import into the mitochondrial matrix (1). Many mitochondrial proteins are synthesized in the cytosol as precursors containing an N-terminal targeting sequence or presequence followed by a folded mature domain. These proteins must be translocated across the two mitochondrial membranes to reach the matrix (2). During translocation, precursors are fully unfolded and threaded through the import channel amino acid by amino acid (1, 3). Mitochondria can import some precursor proteins many hundreds of times faster than these proteins can unfold spontaneously (4, 5), indicating that mitochondria can catalyze unfolding. Mitochondria accelerate the unfolding of synthetic precursors derived from the Bacillus amyloliquefaciens ribonuclease barnase by altering their spontaneous unfolding pathway to a mechanical one in which the mature domain is unraveled from its N terminus (4). Import and unfolding require both an electrical potential across the inner membrane and ATP hydrolysis by mitochondrial heat shock protein 70 in the matrix (2).
Fusing a mitochondrial targeting sequence to the N terminus of a cytosolic protein is usually sufficient to target it to mitochondria (6), and the efficiency of import is influenced by the amino acid sequence of the targeting signal. However, several observations suggest that targeting information is not limited to the presequences themselves. First, the targeting signal of the mitochondrial matrix protein superoxide dismutase induced efficient import of mouse dihydrofolate reductase but not yeast invertase when fused to these proteins (7). Second, the folding state of the mature domain can influence import efficiency. Precursors whose mature domains are destabilized by urea are imported better than native proteins in vitro (5, 8). Third, substituting the mitochondrial targeting signal of cytochrome oxidase subunit IV with randomly generated sequences resulted in one-quarter of the chimeric proteins still being targeted to mitochondria (9, 10).
Here, we investigate the effect of a mature domain's structure on precursor import. We find that the susceptibilities of proteins to unfolding by mitochondria, and thus their import efficiencies, are determined by their local structures adjacent to the targeting sequences. Specifically, mitochondria can unravel precursors much more effectively when the targeting sequence leads into a surface α-helix than when it leads into a buried β-strand. The explanation of this observation appears to be that the extent to which the import machinery can modify the unfolding pathway of the targeted protein, lowering its activation energy, varies with the local structure first to encounter the entrance to the import channel.
Materials and Methods
Precursor Proteins. Mitochondrial precursor proteins were constructed by fusing varying lengths of the N-terminal portion of precytochrome b2 to the N termini of B. amyloliquefaciens ribonuclease barnase (5), the E. coli chemotaxis response regulator CheY (11), or circular permutants (CPs) of E. coli dihydrofolate reductase (DHFR) (12) in pGEM-3Zf(+) vectors. The CheY mutational analysis was conducted in F14N/V54T double mutant as pseudo wild type into which the mutations V11T, V33T, A42G, V83T, A98G, and A103G were introduced (11). All barnase precursors contained the inactivating mutation H102A. The effects of mutations on barnase import were taken from Huang et al. (4). The targeting sequences were mutated to prevent disulfide-bridge formation between targeting sequences (C14V), to prevent processing by the mitochondrial matrix processing protease (R30G) (13), and to inactivate the stop–transfer signal (L62P) (14), ensuring that this protein was targeted to the mitochondrial matrix. Radioactive precursors were expressed from a T7 promoter by in vitro transcription and translation in a rabbit reticulocyte lysate supplemented with [35S]methionine (Promega). Ribosomes and associated incompletely translated polypeptide chains were removed by centrifugation at 150,000 × g for 15 min. Precursor proteins were then partially purified by precipitation in 50% saturated ammonium sulfate for at least 1 h on ice, pelleted by centrifugation at 20,800 × g for 15 min, and resuspended in import buffer (0.6 M sorbitol/50 mM Hepes-KOH, pH 7.4/50 mM KCl/10 mM MgCl2/2 mM KH2PO4/5 mM unlabeled methionine/1 mg/ml fatty acid-free BSA). In the DHFR experiments, the buffer also contained the appropriate concentrations of methotrexate.
Mitochondria were isolated from Saccharomyces cerevisiae strain D273-10B (MATα, American Type Culture Collection 2567) (15) and purified by centrifugation through a Nycodenz gradient (Nycomed, Oslo) (16).
Import Experiments. Import kinetics were determined as described in ref. 5. Thirty microliters of radiolabeled precursors were added to 570 μl of mitochondrial suspension at 0.5 mg of mitochondrial protein per ml in import buffer (described above) containing 4 mM ATP, 10.2 mM creatine phosphate, and 0.15 mg/ml creatine kinase. At defined time intervals, 50-μl samples were transferred to 400 μl of ice-cold mitoplasting buffer (20 mM Hepes-KOH, pH 7.4/1 mg/ml fatty acid-free BSA/0.1 mg/ml proteinase K/25 mM carbonylcyanide p-trifluoromethoxyphenylhydrazone). Proteinase K was inhibited with 1 mM PMSF after a 25-min incubation on ice. Mitochondria were reisolated by centrifugation at 20,800 × g and resuspended in SDS/PAGE sample buffer containing 2 mM PMSF. Samples were analyzed by SDS/PAGE, and the amount of imported protein was quantified by electronic autoradiography (Instant Imager, Packard). The extent of import was plotted as a percentage of the total amount of modified precursor in the import reaction. The import kinetics were analyzed with kaleidagraph (Abelbeck Software, Reading, PA) assuming a first or second order process.
Single-Molecule Force Spectroscopy. Force vs. extension curves were obtained as previously described (17) by stretching single protein polymers between a silicon nitride cantilever and a gold surface in a custom-built atomic force microscopy (AFM) apparatus. The genes for the multidomain proteins were constructed by standard molecular biology techniques so that their expression in E. coli yielded polypeptides in which the C terminus of each DHFR or CP P25 DHFR moiety was connected to the N terminus of the next by a 15-aa linker. The N terminus of the full-length polyprotein contained the 95 C14V amino acid mitochondrial targeting sequence, and the C terminus contained a 10-aa linker followed by two cysteines. The AFM was constructed by using a Digital Instruments (Santa Barbara, CA) Nanoscope II detector head mounted over a closed-loop single axis piezoelectric positioner (NPS-Z-15B, Queensgate Instruments, Torquay, U.K.). The detector head was retrofitted with a fiber-coupled laser source (Schäfter+Kirchhoff, Hamburg). Data acquisition and instrument control were performed via a data-acquisition board (PCI-6052E, National Instruments) and custom labview software (National Instruments, Austin, TX). The spring constants of each AFM cantilever (cantilever A MLCT-AUNM, Digital Instruments) were measured in solution before each experiment by using the equipartition method (18). Unfolding forces were extrapolated to a loading rate of 135,000 pN/s.
Results
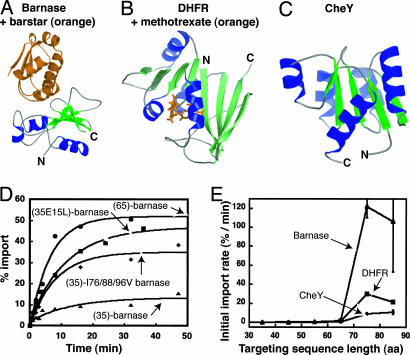
Properties of the Targeting Sequence and Mature Domain Affect the Efficiency of Mitochondrial Protein Import. To investigate the substrate specificity of the mitochondrial matrix import machinery, we constructed a series of precursor proteins whose mature regions were derived from barnase, DHFR, and CheY (Fig. 1 A–C) (see Materials and Methods). The proteins were targeted to the mitochondrial matrix by N-terminal targeting sequences of varying lengths derived from cytochrome b2.
Fig. 1.
Dependence of import efficiency on targeting sequence and mature domain. The color diagrams represent barnase (A), DHFR (B), and CheY (C), showing the N and C termini and the ligand-binding sites. (D) The dependence of mitochondrial import of (35)-barnase on the targeting sequence charge density, (35 E15L)-barnase, targeting sequence length, (65)-barnase, and mature domain's thermodynamic stability, (35)-I76/88/96V barnase. The percent of precursor imported is mapped over time. (E) The dependence of the initial import rate on the targeting signal length of precursors with barnase, DHFR, and CheY as the mature domains.
Import of barnase precursor proteins with a 35-aa-long targeting sequence [(35)-barnase] into mitochondria was inefficient, with only ≈12% of the protein having been transported after ≈50 min (Fig. 1D). However, import of barnase could be improved through three separate modifications of the precursors. First, increasing the positive charge density of the targeting sequence with the mutation E15L increased the extent of import approximately 4-fold [Fig. 1D, (35 E15L)-barnase]. The higher positive charge density in the targeting sequence presumably enhanced its interaction with the membrane potential, facilitating its threading through the import channel (19). Second, lengthening the targeting sequence from 35 to 65 amino acid residues without changing its linear charge density increased the extent of import 5-fold [Fig. 1D, (65)-barnase]. The longer targeting sequence probably allowed precursors to engage the mitochondrial unfolding machinery more effectively (4, 5, 19). Finally, removing three methyl groups in the hydrophobic core of the protein (I76/88/96V barnase), which destabilizes the protein against unfolding by 2.6 kcal/mol (A.M. and Alan R. Fersht, unpublished data); increased the extent of import 3-fold [Fig. 1D, (35)-I76/88/96V barnase]. Destabilization probably enhanced spontaneous unfolding of the precursor, which reduced the requirement for the mitochondrial unfolding machinery when dealing with a short (35 aa) targeting sequence and, hence, accelerated import. These results suggest that properties of both the targeting sequence and the mature domain affect import efficiency and, therefore, contribute to the specificity of protein sorting to the mitochondrial matrix.
The Structure of the Mature Domain Affects Mitochondrial Import. The susceptibility of a precursor to mitochondrial unfolding depends on the mature domain's structure. To investigate the effect of protein structure on translocation, we compared the import of barnase with that of CheY and DHFR when attached to a series of targeting sequences ranging in length from 35 to 95 aa. Precursor proteins with shorter targeting sequences cannot fully engage the mitochondrial unfolding machinery, and, as shown above, their import is largely dependent on the precursor's spontaneous unfolding at the surface of the mitochondria (5). In contrast, precursors harboring targeting sequences of 75 to 95 aa can fully connect to the mitochondrial unfolding machinery and are imported significantly faster than their shorter counterparts (5) (Fig. 1E). In this case, import rates of the precursors were not determined by the equilibrium or kinetic stabilities of the mature domains against global spontaneous unfolding.
CheY has approximately the same thermodynamic stability as barnase [unfolding free energy = 8.5 kcal/mol for CheY (11) and 8.8 kcal/mol for barnase (20) at 25°C] and spontaneously unfolds 10 times faster than barnase [unfolding rate constants = 1.4 × 10-3 s-1 for CheY (11) and 1.5 × 10-4 s-1 for barnase (21) at 25°C]. However, CheY was imported 12 times more slowly than barnase in precursors with identical 75-aa-long targeting sequences (Fig. 1E). Similarly, DHFR is thermodynamically less stable than barnase [unfolding free energy = 6.1 kcal/mol (12) at 15°C] and unfolds spontaneously more than 250 times faster than barnase [unfolding rate constant = 4 × 10–2 s-1 at 15°C (22)] but is imported 6-fold slower with an identical 75-aa-long targeting sequence (Fig. 1E). Previous studies have shown that fusing these targeting sequences to the barnase or DHFR domains does not destabilize them against unfolding (5).
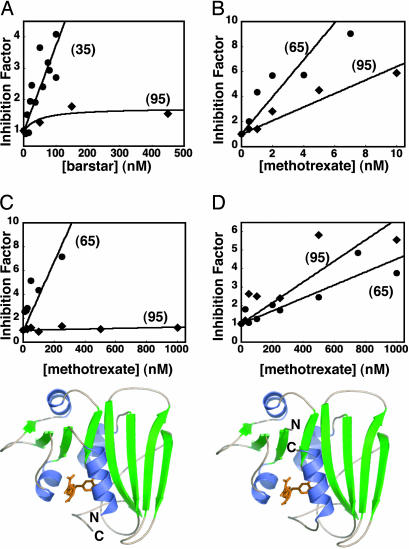
The differences in import efficiency between different precursors become even more pronounced when the proteins are stabilized against spontaneous unfolding by ligand binding. Barstar binds barnase tightly [KD ≈ 30 nM (4)] and stabilizes the protein against spontaneous unfolding (23). Barstar inhibited the import of barnase precursors with shorter (35) targeting sequences, which is governed by spontaneous precursor unfolding (4) (Fig. 2A). However, import of barnase precursors with 95-aa-long targeting sequences, which can fully engage the mitochondrial unfolding machinery, remained largely unaffected by barstar (Fig. 2 A). Similarly, DHFR can be stabilized against spontaneous unfolding by addition of its ligand methotrexate (24) (KD ≈ 0.7 nM, determined as described in ref. 25). In this case, however, ligand-stabilized DHFR precursors were not imported even when attached to long targeting sequences that engaged the import machinery fully (Fig. 2B). Together, these results suggest that a precursor's mitochondrial import efficiency depends on a property of the mature domain's structure that is not its thermodynamic or kinetic stability against spontaneous global unfolding.
Fig. 2.
Effect of ligand binding on protein import. The graphs show the inverse of the measured initial import rate vs. barstar concentration for (35)- and (95)-barnase precursors (A), or the inverse of the measured initial import rate vs. methotrexate concentration for (65)- and (95)-wild-type DHFR (B), (65)- and (95)-CP P25 DHFR (C), and (65)- and (95)-CP K38 DHFR precursors (D), standardized to the initial import rates of the unliganded precursors.
The Local Structure Adjacent to the Targeting Sequence Affects Mitochondrial Import. What aspect of the structures of barnase and DHFR precursors determines their import efficiency? Mitochondria can unfold barnase precursors by unraveling them from their N-terminal targeting sequences and, thus, changing their unfolding pathway from that of spontaneous global unfolding (4). Therefore, it is possible that the local structure of a precursor's mature domain immediately adjacent to the targeting signal has a strong influence on the protein's susceptibility to unfolding by mitochondria. We investigated the effect of local structure of proteins on their susceptibility to unfolding by studying the import of CPs of DHFR. In these experiments, the original N and C termini of DHFR were connected by a short linker, new N termini were created at Pro-25 (CP P25) or Lys-38 (CP K38), and mitochondrial targeting sequences were fused to the proteins at the new N termini (26). The DHFR mutants have similar structures and stabilities and differ primarily in their topology (12). The amino acids immediately adjacent to the N terminus of CP P25 DHFR form an α-helix at the surface of the protein, as does the N terminus of barnase. In contrast, the amino acids immediately following the N terminus of CP K38 DHFR form a β-strand buried in the core of the protein, as is the case for wild-type DHFR (Fig. 2 A–D).
Ligand binding inhibited the import of both CP P25 DHFR and CP K38 DHFR precursors with 65-aa targeting sequences, indicating that import of the two proteins remained governed by spontaneous global unfolding of the mature domains (5). This result was expected because the length of these targeting sequences is just below the threshold required for fully engaging the mitochondrial unfolding machinery (Fig. 1E). Increasing the length of the targeting sequences to 95 aa led to methotrexate binding losing its effect on import of CP P25 DHFR, whereas methotrexate binding retained its effect on import of CP K38 DHFR (Fig. 2 C and D). These results were surprising because CP P25 DHFR is more stable against global unfolding than CP K38 DHFR [unfolding-free energies 4.4 kcal/mol and 3.5 kcal/mol at 15°C, respectively (12)], and CP P25 DHFR precursors bind methotrexate more tightly than CP K38 DHFR precursors (Fig. 2 C and D) (KD ≈ 35 and KD ≈ 290 nM, respectively). Together, these results suggest that the susceptibility to unfolding by mitochondria is determined by a precursor protein's local structure adjacent to the targeting signal. In the examples analyzed here, structures in which the N termini formed α-helices at the surface of the folded domains (barnase and CP P25 DHFR) (Fig. 2 A and C) were more susceptible to unfolding by mitochondria than structures in which the N termini formed β-strands buried inside the folded domains (wild-type DHFR and CP K38 DHFR) (Fig. 2 B and D).
The Mechanism of Mitochondrial Unfolding Is Determined by the Mature Domain's Local Structure Adjacent to the Targeting Sequence. Wild-type and CP K38 DHFR may be more difficult to unfold by mitochondria than barnase because the import machinery is somehow unable to induce similarly extensive changes to their unfolding pathways. The unfolding kinetics of DHFR in vitro are complex (27), and it is not feasible to determine the unfolding pathway of DHFR precursors during import. Therefore, we compared the unfolding pathways of barnase and CheY precursors by analyzing the effects of point mutations throughout their mature domains on import. CheY resembles DHFR in that both proteins have α/β folds and N termini that lead into buried β-strands.
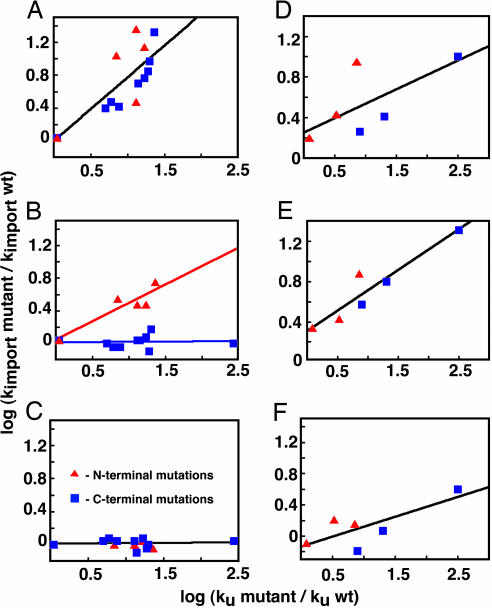
Mutations throughout both barnase (4) and CheY precursors with short (35 aa) targeting sequences accelerated import in proportion to the mutations' known effects on both the thermodynamics and kinetics of spontaneous global unfolding in vitro (Fig. 3 A and D). When the lengths of the targeting sequences were increased to 65 aa in length, mutations throughout barnase lost their effect on import and only mutations that destabilized the N terminus of the protein continued to accelerate translocation (4) (Fig. 3B). This observation indicates that mitochondria are able to change the unfolding pathway of barnase as the targeting sequence begins to engage the unfolding machinery (4). When the targeting sequence length was further increased to 95 aa to allow the unfolding machinery to engage the precursors fully, all mutations throughout barnase lost their effect on import, and unfolding no longer determined import rates (4, 5) (Fig. 3C). In CheY precursors, no such change was observed, and all mutations retained their full effect on import as the targeting sequence was increased to 65 aa (Fig. 3E). Moreover, all mutations retained a significant if reduced effect on CheY precursor import when the targeting sequence length was increased to 95 aa. Thus, the mitochondrial unfolding machinery is unable to induce the large changes in the unfolding pathway of CheY that it induces in barnase even when the mitochondrial import machinery fully engages the precursors.
Fig. 3.
Effect of mutations on mitochondrial import of barnase and CheY precursors. For each mutant analyzed, the initial import rate is plotted against the spontaneous unfolding rate (ku), both standardized to the wild-type import or spontaneous unfolding rates. The graphs show dependence of the mutational effects on import on their effect on spontaneous unfolding for (35)-barnase (A), (65)-barnase (B), (95)-barnase (C), (35)-CheY (D), (65)-CheY (E), and (95)-CheY (F).
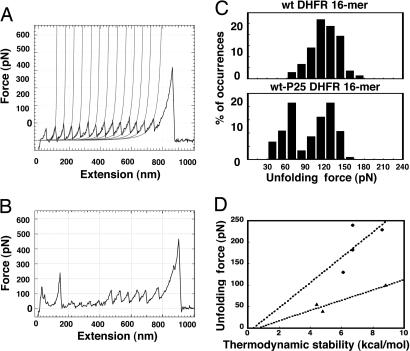
Mitochondrial Import Efficiency Correlates with the Mature Domain's Resistance to Mechanical Unfolding. The vectorial nature of protein translocation and the directed unraveling of precursors from their N termini by mitochondria suggest a scenario in which mitochondria unfold precursors by pulling at their N termini. Accordingly, the resistance of proteins against unfolding by direct mechanical pulling from their N termini as determined by AFM may be a better predictor of their susceptibility to mitochondrial unfolding than their global stability against unfolding measured by solvent or heat denaturation experiments. To test this hypothesis, we compared the amounts of force required to mechanically unfold several mature domains of the precursor proteins by AFM. We created recombinant DHFR polyproteins by fusing 16 wild-type DHFR domains to each other in frame. Plots of the unfolding force exerted on the proteins against the extension of the protein measured in AFM pulling experiments produced the expected sawtooth pattern (28). The collected force peaks for wild-type DHFR denaturation showed that individual domains unfolded at an average force of 131 ± 20 pN (n = 50, loading rate = 90,000 pN/s) (Fig. 4 A and C). To analyze the mechanical unfolding of CP P25 DHFR, we created a chimeric eight-domain protein composed of alternating wild-type and CP P25 DHFR entities. Both the N and C termini of wild-type DHFR lead into β-strands buried within a β-sheet, whereas the N and C termini of CP P25 DHFR lead into α-helices and unstructured loops, respectively. The force traces collected for wild-type CP P25 DHFR fusion proteins displayed two different types of force peaks (n = 84, loading rate = 135,000 pN/s) (Fig. 4 B and C), one at 130 ± 10 pN, which corresponded to the unfolding of wild-type DHFR, and another at 56 ± 12 pN, which we identified as corresponding to the unfolding of single CP P25 DHFR domains. Wild-type DHFR, therefore, resisted mechanical unfolding at least 2.3-fold more strongly than CP P25 DHFR, whereas its free energy of spontaneous global unfolding is only 1.7 kcal/mol or 40% larger. Previous mechanical unfolding experiments on barnase showed that wild-type DHFR resisted mechanical unfolding 1.7-fold more strongly than barnase despite its free energy of unfolding being ≈1.5- to 2-fold smaller (12) (Table 1).
Fig. 4.
Changing the location of the N terminus of DHFR alters its mechanical stability. (A) Force extension trace of wild-type DHFR 16-mer. Wild-type DHFR unfolds at a pulling force of 131 ± 20 pN. Data were recorded at a loading rate of 90,000 pN/s. (B) Force extension trace of wild-type-CP P25 DHFR 16-mer. Wild-type DHFR unfolds at a pulling force of 130 ± 10 pN. CP P25 DHFR unfolds at a pulling force of 56 ± 12 pN. Data were recorded at a loading rate of 135,000 pN/s. (C) Graph showing the fraction of recorded unfolding events at each force for experiments as those described in A and B. (D) The correlation between thermodynamic stability and unfolding forces of unrelated proteins is R2 = 0.58 (▴ and •). The correlation is stronger when similar structures are compared, i.e., for structures with at least one α-helical terminus R2 = 0.94 (▴) and for structures with β-strand termini R2 = 0.63 (•). Unfolding forces were extrapolated to a loading rate of 135,000 pN/s.
Table 1. Comparison of protein domain mechanical unfolding force, terminal structures, and thermodynamic stability.
| Protein | Unfolding force, pN (ref.)* | Terminal structures† | ΔGH2O, kcal/mol (ref.)‡ |
|---|---|---|---|
| Spectrin | 39 (39) | α | 4.8 (40) |
| DHFR CP P25 | 56 ± 12 | α, loop | 4.4 (12) |
| Barnase | 100 (41) | α, β | 8.8 (20) |
| DHFR wt | 130 ± 10 | β | 6.1 (12) |
| Tenascin | 180 (42) | β | 6.7 (43) |
| Ubiquitin | 240 (29) | β | 6.7 (44) |
| Titin | 229 (41) | β | 8.6 (41) |
Unfolding forces were extrapolated to a loading rate of 135,000 pN/s from cited papers or were taken as measured
α, α-helix; β, β-strand; loop, irregular loop
Thermodynamic stabilities were taken as published from cited papers
These results reflect a general trend according to which mechanical unfolding stability correlates poorly with overall thermodynamic stability (29–31) of proteins such as barnase, spectrin, ubiquitin, titin, and the fibronectin type III (FNIII) domain of tenascin (correlation coefficient R2 = 0.58) (Fig. 4D and Table 1). Mechanical stability does not appear to correlate with the rates of global unfolding either, although fewer data points can be analyzed (data not shown). Instead, stability against mechanical unfolding appears to depend on a combination of the local N- and C-terminal structures of the protein and its stability against spontaneous global unfolding. Proteins with β-strands at their N and C termini resist mechanical unfolding more strongly than proteins with α-helices or surface loops at those positions. The same trend is observed for the unfolding and transport of precursor proteins by mitochondria. If the subset of proteins we examine is restricted such that only proteins with similar terminal structures are compared (e.g., proteins with α-helices adjacent to their ends), the resistance of these proteins against mechanical unfolding then correlates better with their thermodynamic stabilities against global unfolding (Fig. 4D, ▴) (R2 = 0.94). Although the correlation is not as strong for proteins with buried β-strands at both ends (Fig. 4D, •) (R2 = 0.63), it appears that thermodynamic stability also relates to unfolding force amongst this subset of proteins.
Discussion
We propose that the efficiency of protein import into mitochondria and, hence, the specificity of protein sorting to mitochondria does not only depend on properties of the precursor's targeting sequence but also on properties of the precursor's mature domain. Import efficiency is influenced most strongly by the mature domain's local structure directly adjacent to the mitochondrial targeting sequence. Only when precursors have similar structures does import efficiency correlate with the mature domain's overall thermodynamic or kinetic stability against global unfolding. Thus, effective targeting to the mitochondrial matrix of a folded precursor requires two properties: an N-terminal targeting signal that is able to engage the unfolding machinery and a mature domain whose susceptibility to unfolding is such that the protein can be unraveled by the import machinery. Mitochondria import precursors whose targeting sequences lead directly into α-helices more easily than they import precursors whose targeting sequences lead directly into buried β-strands. Typically, α-helices are found on the surface of folded structures, whereas β-strands are generally buried in the hydrophobic core of the protein (32). Therefore, the location of the N-terminal structure within the domain may affect the efficiency of import. However, the identity of the secondary structure itself also contributes to import efficiency. In a CP of DHFR in which an N-terminal β-strand lies at the surface of the protein, methotrexate still inhibited unfolding and import significantly (data not shown).
Differences in proteins' susceptibilities to unfolding by mitochondria can contribute to protein sorting within the cell. One such example is provided in the medical condition known as primary hyperoxaluria type 1. About 40% of the cases of primary hyperoxaluria type 1 are caused by mistargeting of the enzyme alanine-glyoxylate aminotransferase from peroxisomes to mitochondria as a result of at least two mutations (33). Wild-type alanine-glyoxylate aminotransferase forms a stable homodimer in the cell (33) with a cryptic N-terminal mitochondrial targeting sequence and a functional C-terminal nonconsensus type 1 peroxisomal targeting sequence (34). One of the mutations associated with the disease appears to reactivate the mitochondrial-targeting signal, but this mutation by itself does not cause missorting (33). Mitochondrial targeting requires a second mutation that inhibits alanine-glyoxylate aminotransferase dimer formation (33) and, therefore, destabilizes the protein against unfolding. The second mutation accelerates mitochondrial over peroxisomal import because the latter does not require protein unfolding (35). Thus, a combination of two mutations, one that improves the functionality of the targeting sequence and another that increases the mature domain's susceptibility to unfolding, are required for alanine-glyoxylate aminotransferase mistargeting to mitochondria and subsequent disease development.
Why would mitochondria unfold mature domains with one terminal structure more efficiently than domains with another? The explanation appears to be that mitochondria are only able to significantly change the unfolding pathways of precursors in which the targeting sequences lead into surface α-helices but not that of precursors where they lead into buried β-strands. The results of AFM experiments suggest that a precursor's resistance to mitochondrial import may correlate with its resistance to mechanical unfolding by pulling. Unlike the tension on a string, the force applied by the mitochondrial machinery does not transmit uniformly throughout the polypeptide chain. Instead, the force concentrates at the first point of mechanical resistance in the structure, which is likely to be that of the local structure adjacent to the N-terminal targeting sequence. It is possible that the import machinery behaves like a motor with a power stroke that applies a mechanical force to lower the activation energy for dislodging the local structure from the rest of the protein. As the motor unfolds the local structure, the remainder of the protein collapses because protein folding is highly cooperative. Alternatively, the import machinery could trap local spontaneous unfolding fluctuations in the region of the protein directly adjacent to the targeting signal and opportunistically thread the unfolded region into the import channel, in effect pulling at the end of the protein. Mitochondria would catalyze precursor unfolding if they were able to trap local unfolding fluctuations that are faster than spontaneous global unfolding.
Mitochondria are unable to induce gross changes to the unfolding pathway of proteins in which the targeting sequence leads into β-strands, either because the power stroke of the motor is not strong enough to dislodge and unfold these structures or because the structures lack the appropriate trappable spontaneous fluctuations. The force required to unfold proteins in AFM experiments depends on the rate with which the force is applied; this relationship has been interpreted to mean that unfolding by AFM relies on existing unfolding fluctuations (28, 36). By this reasoning, we propose that mitochondrial unfolding similarly depends on local unfolding fluctuations in the substrate protein, although an applied force could accelerate these fluctuations.
There are also some clear differences between protein unfolding by pulling in AFM and mitochondrial import. In AFM, force is applied to both ends of the protein, whereas in mitochondria, force is applied from one end and the substrate is pulled against the narrow opening of the import channel. In the first case, the tension is concentrated on the first mechanical resistance points at the N- and C-terminal structures in the protein, whereas in the second, it is only applied to the point of resistance in the N-terminal structure adjacent to the target sequence. Nonetheless, the correlation between the susceptibility to mechanical unfolding of proteins by AFM and by the mitochondrial import machinery points to a basic similarity between these processes.
Protein unfolding during mitochondrial import is remarkably similar to protein unfolding during AAA+-protease degradation. In both processes, substrates are recognized by N- or C-terminal targeting sequences. Substrate unfolding requires ATP hydrolysis, and unfolding is coupled with the movement of the extended polypeptide through a channel. Thus, not surprisingly, the efficiency with which a precursor is imported into mitochondria correlates with the efficiency with which a similar substrate is degraded by AAA+-proteases (37, 38).
In summary, we show that the efficiency of protein targeting to the mitochondrial matrix is determined by the ability of the mitochondrial targeting sequence to engage the import machinery and the susceptibility of the mature domain to unfolding by mitochondria. This susceptibility to mitochondrial unfolding depends on the protein's local structure adjacent to the targeting sequence, just as is the case for mechanical unfolding. Accordingly, differences in local stability between protein substrates determine the extent to which mitochondria can change these substrates' unfolding pathway and therefore accelerate unfolding.
Acknowledgments
We thank R. Morimoto, A. Rosenzweig, E. Sontheimer, and members of the Matouschek laboratory for valuable discussion and advice. This work was supported by National Institutes of Health Grant GM063004 (to A.M.) and the Howard Hughes Medical Institute (C.B.). In addition, we gratefully acknowledge support from the Leukemia and Lymphoma Society (Scholar Award 1252-03 to A.M.) and the National Institutes of Health (Cellular and Molecular Biology of Disease Training Grant to A.J.W.), as well as use of the Keck Biophysics Facility at Northwestern University.
Author contributions: A.J.W., J.C., C.J.B., and A.M. designed research; A.J.W. and J.C. performed research; A.J.W., J.C., C.J.B., and A.M. analyzed data; and A.J.W., J.C., C.J.B., and A.M. wrote the paper.
Abbreviations: AFM, atomic force microscopy; CP, circular permutant; DHFR, dihydrofolate reductase.
References
- 1.Eilers, M. & Schatz, G. (1986) Nature 322, 228-232. [DOI] [PubMed] [Google Scholar]
- 2.Wiedemann, N., Frazier, A. E. & Pfanner, N. (2004) J. Biol. Chem. 279, 14473-14476. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz, M. P., Huang, S. & Matouschek, A. (1999) J. Biol. Chem. 274, 12759-12764. [DOI] [PubMed] [Google Scholar]
- 4.Huang, S., Ratliff, K. S., Schwartz, M. P., Spenner, J. M. & Matouschek, A. (1999) Nat. Struct. Biol. 6, 1132-1138. [DOI] [PubMed] [Google Scholar]
- 5.Matouschek, A., Azem, A., Ratliff, K., Glick, B. S., Schmid, K. & Schatz, G. (1997) EMBO J. 16, 6727-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurt, E. C. & van Loon, A. P. G. M. (1986) Trends Biochem. Sci. 11, 204-207. [Google Scholar]
- 7.Van Steeg, H., Oudshoorn, P., Van Hell, B., Polman, J. E. & Grivell, L. A. (1986) EMBO J. 5, 3643-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verner, K. & Lemire, B. D. (1989) EMBO J. 8, 1491-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allison, D. S. & Schatz, G. (1986) Proc. Natl. Acad. Sci. USA 83, 9011-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemire, B. D., Fankhauser, C., Baker, A. & Schatz, G. (1989) J. Biol. Chem. 264, 20206-20215. [PubMed] [Google Scholar]
- 11.Lopez-Hernandez, E. & Serrano, L. (1996) Fold. Des. 1, 43-55. [PubMed] [Google Scholar]
- 12.Iwakura, M., Nakamura, T., Yamane, C. & Maki, K. (2000) Nat. Struct. Biol. 7, 580-585. [DOI] [PubMed] [Google Scholar]
- 13.Arretz, M., Schneider, H., Guiard, B., Brunner, M. & Neupert, W. (1994) J. Biol. Chem. 269, 4959-4967. [PubMed] [Google Scholar]
- 14.Beasley, E. M., Müller, S. & Schatz, G. (1993) EMBO J. 12, 2303-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hase, T., Muller, U., Riezman, H. & Schatz, G. (1984) EMBO J. 3, 3157-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glick, B. S. & Pon, L. A. (1995) Methods Enzymol. 260, 213-223. [DOI] [PubMed] [Google Scholar]
- 17.Yang, G., Cecconi, C., Baase, W. A., Vetter, I. R., Breyer, W. A., Haack, J. A., Matthews, B. W., Dahlquist, F. W. & Bustamante, C. (2000) Proc. Natl. Acad. Sci. USA 97, 139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Florin, E. L., Rief, M., Lehmann, H., Ludwig, M., Dornmair, C., Moy, V. T. & Gaub, H. E. (1995) Biosens. Bioelectron. 10, 895-901. [Google Scholar]
- 19.Huang, S., Ratliff, K. S. & Matouschek, A. (2002) Nat. Struct. Biol. 9, 301-307. [DOI] [PubMed] [Google Scholar]
- 20.Serrano, L., Kellis, J. T., Jr., Cann, P., Matouschek, A. & Fersht, A. R. (1992) J. Mol. Biol. 224, 783-804. [DOI] [PubMed] [Google Scholar]
- 21.Serrano, L., Matouschek, A. & Fersht, A. R. (1992) J. Mol. Biol. 224, 805-818. [DOI] [PubMed] [Google Scholar]
- 22.Touchette, N. A., Perry, K. M. & Matthews, C. R. (1986) Biochemistry 25, 5445-5452. [DOI] [PubMed] [Google Scholar]
- 23.Martinez, J. C., Filimonov, V. V., Mateo, P. L., Schreiber, G. & Fersht, A. R. (1995) Biochemistry 34, 5224-5233. [DOI] [PubMed] [Google Scholar]
- 24.Kraut, J. & Matthews, D. A. (1987) in Biological Macromolecules and Assemblies, eds. Jurnak, F. A. & McPherson, A. (Wiley, New York), Vol. 3, pp. 1-72. [Google Scholar]
- 25.Huang, S., Murphy, S. & Matouschek, A. (2000) Proc. Natl. Acad. Sci. USA 97, 12991-12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura, T. & Iwakura, M. (1999) J. Biol. Chem. 274, 19041-19047. [DOI] [PubMed] [Google Scholar]
- 27.Wallace, L. A. & Robert Matthews, C. (2002) J. Mol. Biol. 315, 193-211. [DOI] [PubMed] [Google Scholar]
- 28.Rief, M., Gautel, M., Oesterhelt, F., Fernandez, J. M. & Gaub, H. E. (1997) Science 276, 1109-1112. [DOI] [PubMed] [Google Scholar]
- 29.Carrion-Vazquez, M., Li, H., Lu, H., Marszalek, P. E., Oberhauser, A. F. & Fernandez, J. M. (2003) Nat. Struct. Biol. 10, 738-743. [DOI] [PubMed] [Google Scholar]
- 30.Li, H., Carrion-Vazquez, M., Oberhauser, A. F., Marszalek, P. E. & Fernandez, J. M. (2000) Nat. Struct. Biol. 7, 1117-1120. [DOI] [PubMed] [Google Scholar]
- 31.Brockwell, D. J., Paci, E., Zinober, R. C., Beddard, G. S., Olmsted, P. D., Smith, D. A., Perham, R. N. & Radford, S. E. (2003) Nat. Struct. Biol. 10, 731-737. [DOI] [PubMed] [Google Scholar]
- 32.Branden, C. & Tooze, J. (1998) Introduction to Protein Structure (Garland, New York).
- 33.Purdue, P. E., Takada, Y. & Danpure, C. J. (1990) J. Cell Biol. 111, 2341-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motley, A., Lumb, M. J., Oatey, P. B., Jennings, P. R., De Zoysa, P. A., Wanders, R. J., Tabak, H. F. & Danpure, C. J. (1995) J. Cell Biol. 131, 95-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purdue, P. E. & Lazarow, P. B. (2001) Annu. Rev. Cell Dev. Biol. 17, 701-752. [DOI] [PubMed] [Google Scholar]
- 36.Carrion-Vazquez, M., Oberhauser, A. F., Fowler, S. B., Marszalek, P. E., Broedel, S. E., Clarke, J. & Fernandez, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3694-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, C., Schwartz, M. P., Prakash, S., Iwakura, M. & Matouschek, A. (2001) Mol. Cell 7, 627-637. [DOI] [PubMed] [Google Scholar]
- 38.Kenniston, J. A., Baker, T. A., Fernandez, J. M. & Sauer, R. T. (2003) Cell 114, 511-520. [DOI] [PubMed] [Google Scholar]
- 39.Rief, M., Pascual, J., Saraste, M. & Gaub, H. E. (1999) J. Mol. Biol. 286, 553-561. [DOI] [PubMed] [Google Scholar]
- 40.Kusunoki, H., Minasov, G., Macdonald, R. I. & Mondragon, A. (2004) J. Mol. Biol. 344, 495-511. [DOI] [PubMed] [Google Scholar]
- 41.Best, R. B., Li, B., Steward, A., Daggett, V. & Clarke, J. (2001) Biophys. J. 81, 2344-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oberhauser, A. F., Marszalek, P. E., Erickson, H. P. & Fernandez, J. M. (1998) Nature 393, 181-185. [DOI] [PubMed] [Google Scholar]
- 43.Hamill, S. J., Meekhof, A. E. & Clarke, J. (1998) Biochemistry 37, 8071-8079. [DOI] [PubMed] [Google Scholar]
- 44.Khorasanizadeh, S., Peters, I. D., Butt, T. R. & Roder, H. (1993) Biochemistry 32, 7054-7063. [DOI] [PubMed] [Google Scholar]