Abstract
The structure of α-synuclein (α-syn) amyloid was studied by hydrogen-deuterium exchange by using a fragment separation–MS analysis. The conditions used made it possible to distinguish the exchange of unprotected and protected amide hydrogens and to define the order/disorder boundaries at close to amino acid resolution. The soluble α-syn monomer exchanges its amide hydrogens with water hydrogens at random coil rates, consistent with its natively unstructured condition. In assembled amyloid, long N-terminal and C-terminal segments remain unprotected (residues 1–≈38 and 102–140), although the N-terminal segment shows some heterogeneity. A continuous middle segment (residues ≈39–101) is strongly protected by systematically H-bonded cross-β structure. This segment is much too long to fit the amyloid ribbon width, but non-H-bonded amides expected for direction-changing loops are not apparent. These results and other known constraints specify that α-syn amyloid adopts a chain fold like that suggested before for amyloid-β [Petkova et al. (2002) Proc. Natl. Acad Sci. USA 99, 16742–16747] but with a short, H-bonded interlamina turn. More generally, we suggest that the prevalence of accidental amyloid formation derives mainly from the exceptional ability of the main chain in a structurally relaxed β-conformation to adapt to and energy-minimize side-chain mismatching. Seeding specificity, strain variability, and species barriers then arise because newly added parallel in-register chains must faithfully reproduce the same set of adaptations.
Many proteins and polypeptides are able to adopt the generic massively aggregated structure known as amyloid (1). The macroscopic fibrillar character of amyloid is obvious by direct electron microscopic observation, but its detailed structure and the structural basis of its unusual behavior remains a challenging problem (2–9).
Methods based on the hydrogen exchange (HX) behavior of polypeptides can provide useful information. The backbone amide hydrogens of proteins engage in continual exchange with the hydrogens of solvent water. These hydrogens, uniformly distributed at every amino acid (except proline) in every protein molecule, provide built-in, structure-sensitive, nonperturbing probes that can be used to study soluble or insoluble proteins under any desired conditions. Hydrogens that are freely exposed to solvent exchange at known rates that depend on pH, temperature, neighboring residues, and the hydrogen isotopes used (10, 11). Hydrogens that are protected by structure, almost always in H bonds, exchange far more slowly. Their exchange is modulated by dynamic structural events that reversibly separate protecting H bonds and transiently expose them to the normal chemical exchange process. Accordingly, HX measurements can distinguish the presence and absence of protecting structure, determine the thermodynamic stability and dynamic properties of local and surrounding structure, and probe the effects of mutations, manipulations, and conditions, in principle, at amino acid resolution (12).
The development of multidimensional solution NMR methods (13–15) has made high-resolution analysis of HX behavior routine for small soluble proteins (16). A fragment separation method that does not depend on solution NMR measurement extends HX studies to large proteins and insoluble protein systems (17–19). In this method, hydrogen isotope exchange can be performed under conditions that are most pertinent for the protein system being studied. Timed samples are then placed into slow HX conditions (low pH and temperature) and quickly fragmented, and the fragments are separated and analyzed for their H isotope content (20). Fragmentation and separation in a fast, online immobilized enzyme/HPLC/MS mode now make HX studies possible at high resolution (21–24).
The present work applied the fragment separation method to the study of α-synuclein (α-syn) amyloid. Monomeric α-syn is a highly soluble, 140-aa neuronal peptide that is thought to be important for vesicle trafficking. It can, however, assemble into macroscopic insoluble amyloid fibrils that are implicated in the death of substantia nigra neurons and the onset of Parkinson's disease. Amyloid formation by α-syn and other proteins can be duplicated in the laboratory, making possible a broad range of physical and biological studies (25).
Previous studies of amyloid structure have used HX in various modes (26–36). We used conditions (low pH and temperature) where the exchange of even unprotected amide hydrogens can be measured so that unstructured as well as structured regions can be directly determined. The fact that the initial monomeric α-syn is unstructured makes it straightforward to distinguish residues that become protected in amyloid. The many overlapping fragments that we obtain indicate the positions of H-bonded and non-H-bonded structure at near amino acid resolution, the absence of unprotected turns, and the presence of structural heterogeneity. These results limit the structural models that can be considered.
We also consider how adaptive energy-minimizing interactions at the amino acid level can determine the important structural details of amyloid and therefore its characteristic self-recognition behavior.
Materials and Methods
α-syn. Human α-syn was expressed in Escherichia coli BL21(DE3) and purified as described in ref. 37. Secondary structure content was inferred from CD spectra [5 μM protein in 5 mM phosphate buffer and an Aviv 202 spectrometer (Aviv Associates, Lakewood, NJ) were used]. Amyloid fibrils were viewed by transmission electron microscope (negative staining with 2% uranyl acetate and a JEOL 1010 microscope were used).
HX Labeling and Analysis. Amyloid was prepared by incubating for 4 days (at 37°C with shaking), pelleting twice (in an Eppendorf tube at 20,800 × g for 5 min) to remove nonmature species, and resuspending in D2O buffer [200 mM NaCl/0.1% formic acid, pDr (uncorrected glass electrode reading in D2O) of 4.0, 5°C, final amount of D2O 70%] to initiate H-to-D exchange. At various time points, aliquots were diluted 1:1 with 4 M guanidinium thiocyanate (GdmSCN) for 1 min for rapid dissociation into monomer and then diluted 4-fold into quench buffer (0.4% formic acid/H2O, pH 2.3, 5°C), centrifuged for 1 min to remove any remaining aggregate, frozen on dry ice, and stored (55 μl) at -80°C for later MS analysis (final protein concentration of 2 mg/ml in 0.5 M GdmSCN; 10% glycerol was added). Exchange experiments with monomeric α-syn were done similarly, starting with a small volume of the soluble protein.
For the determination of carried deuterium (D) label, frozen samples were quickly thawed, passed through two immobilized protease columns (porcine pepsin and Aspergillus saitoi fungal protease type XIII from Sigma, 0.05% TFA, 250 μl/min) and then a C18 column [Vydac C-18 300A (Hesperia, CA), 50 μl/min, 5–45% AcCN gradient, 0.05% TFA, 30 min), all at pH 2.3 and 0°C, and injected directly into a mass spectrometer (ESI Q-TOF, Micromass, Manchester, U.K., capillary temperature of 200°C) (38, 39). Proteolytic fragments were initially identified by analysis of parallel MS1–MS2 data acquired on a Finnigan Classic ion trap-type mass spectrometer using the sequest software program (ThermoFinnigan, San Jose, CA) and dxms data-reduction software (Sierra Analytics, Modesto, CA) (38, 39).
MS data analysis used the dxms program and software that identifies each fragment in the MS profiles and measures its centroid mass (38, 39). The measured D label for each fragment was corrected for the 70% D2O present during the exchange-in and for the loss of D label during the analysis, which was obtained by running fully deuterated samples through the same analysis. Recovery of D label was mainly in the 70–80% range, although some fragments were consistently lower.
Results
Amyloid Characterization. Assembled α-syn appears by electron microscopy as typical amyloid fibrils with ribbon width of ≈10 nm (Fig. 1A). CD spectra (Fig. 1B) of suspended slurries after cleansing by two centrifugal pelletings exhibit β-structure content in the range of 40–50%.
Fig. 1.
Characterization of α-syn. (A) Typical amyloid fibrils are seen by transmission electron microscopy (negative staining by uranyl acetate). (B)CD spectra for monomeric α-syn (random coil) and for suspensions of amyloid at pH 7 and a pDr of 4, the condition used for HX labeling experiments.
The present experiments were done at a pDr of 4. CD spectra for both the random coil monomer and assembled amyloid change slightly when the pH is lowered from 7 to 4 because of the protonation of acidic side chains. The CD changes occur immediately upon solvent pH change and therefore seem unlikely to signal a major alteration in amyloid or random coil structure. It can be noted that the uranyl acetate solutions typically used for negative staining are at a pH of ≈3.
dxms Analysis. To initiate H-to-D exchange, a centrifugal pellet of α-syn was resuspended in D2O (pDr = 4, 5°C). Timed samples were analyzed for carried D label by the fragmentation analysis illustrated in Fig. 2.
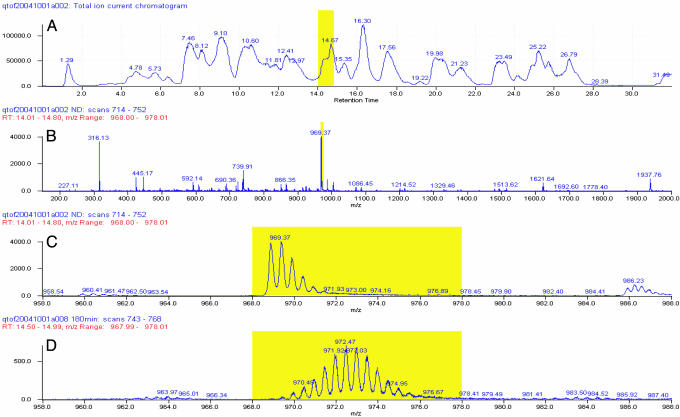
Fig. 2.
Illustration of the fragmentation–separation analysis (at pH 2.3 and 0°C). Samples of α-syn were fragmented by passage through two immobilized acid protease columns, and the fragments were roughly separated by reverse-phase HPLC and further resolved by continuous online scanning of the HPLC eluant by using electrospray Q-TOF MS. (A) MS-measured total ion current trace of the HPLC column eluant. (B) MS scan of the region marked in A. (C) Expansion of the mass peak marked in B. (D) The same peptide after deuterium labeling.
Fig. 2 A shows a total ion current (TIC) trace of an HPLC column eluant. Fig. 2B shows the online MS scan of the part of the TIC elution profile indicated in A. Fig. 2C expands the narrow region indicated in B, which represents the fragment 125–140. The several isotopic lobes are due to the Poisson (binomial) distribution of the natural abundance of 13C atoms in the 125–140 fragment population. In deuterium-labeled samples, the same fragment is shifted to higher mass (Fig. 2D) with the deuterium-distribution pattern convoluted with the 13C pattern. Subtraction of the computed centroid mass of the unlabeled fragment from the labeled fragment yields the number of D-labeled sites recovered per fragment. This value was corrected for loss of exchangeable D label during the analysis and other factors as indicated in Materials and Methods.
We reproducibly obtained 46 fragments with good signal-to-noise ratio, often in more than one charge state. For each fragment, Fig. 3 compares the time course measured for H-to-D exchange labeling of the monomeric (green) and amyloid (red) forms with the labeling expected (10, 11) in the absence of protection (black). The fragments cover the entire α-syn chain and have multiple overlaps, which allows details of the HX labeling to be quantified at near amino acid resolution. Fig. 4 summarizes the HX protection results.
Fig. 3.
HX of α-syn (pDr of 4 and temperature of 5°C). D-labeling kinetics measured by the fragment separation method is shown for soluble α-syn monomer (green) and insoluble amyloid (red). Black curves compare the labeling expected for an unprotected random coil (10, 11). (Spreadsheet is available from the authors upon request.) Proline residues that have no exchangeable amide hydrogen are at positions 108, 117,120, 128, and 138. A low background of labeling through the amyloid region, in the range 1 ± 0.6 D per fragment, represents some unexplained experimental artifact. It is present from the earliest time point rather than accumulating at the free peptide rate, and its level is not correlated with fragment length.
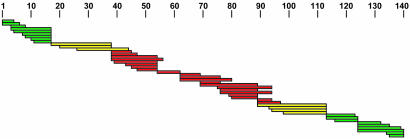
Fig. 4.
HX protection map. Fragments measured are placed along the length of the α-syn molecule. Colors denote unprotected (green) and well protected (red) regions and fragments that overlap the boundaries (yellow).
Soluble Monomer Structure. The experimental conditions used here made it possible to directly measure and recognize the HX of unprotected amides. At a pDr of 4 and 5°C, HX half-times for unprotected amides are ≈10 min, and they vary in detail with residue type and neighbors (10, 11). Amide hydrogens protected by H-bonded structure exchange more slowly by orders of magnitude. Therefore, one can unambiguously distinguish amyloid-dependent structure from unstructured regions including intervening loops. This distinction is not obtained when labeling is performed under more aggressive HX conditions (e.g., neutral pH, higher temperature, and long HX times) as has typically been done in prior studies.
The D-labeling kinetics measured for the soluble α-syn monomer (Fig. 3) compares well with expected HX curves for unprotected amides, although generally a little faster. The measured labeling of each fragment reaches the final expected level by the 3-hr time point. These results confirm the natively unstructured condition of the α-syn monomer (40). It has no significant main-chain H bonding.
Assembled Amyloid Structure. Within assembled amyloid, D labeling through the C-terminal length of α-syn matches the HX rates and amplitudes expected for unprotected amides, just as for the unstructured monomer. The final level of labeling measured for two fragments in the C-terminal length, 117–124 and 125–135, is lower than expected by one and two amides, respectively (compare red and black curves), but the expected level is found in four other fragments that encompass the same sequence.
A middle portion of the α-syn chain is sharply distinguished by strong protection against HX labeling, which presumably represents the systematically H-bonded cross-β structure. Within the cross-β region, no non-H-bonded loop segment is found, even at much longer (>1,000 times) effective HX time (see also ref. 33).
Fragments derived from the N-terminal length of the molecule (Fig. 3) consistently reach ≈75% labeling at the rate expected for freely exposed amides (τ ≈ 10 min). The remaining ≈25% of sites accumulate label only very slowly at much longer times, indicating strong protection. The fractional labeling is not due to partial protection dynamically shared by all of the molecules; rather, it is due to static heterogeneity (see also ref. 41). Some molecules are strongly protected, and some are wholly unprotected. In agreement, the MS data for D labeling of these fragments display a bimodal mass distribution (more apparent for longer fragments). These results also show that any dynamic interchange of the differently protected populations must occur very slowly (>24 hr).
Order–Disorder Boundaries. The HX data in Fig. 3 show a midchain structured region and unstructured lengths on both ends. The degree of HX protection for fragments that overlap structured and unstructured regions defines the boundary positions.
Fragments 39–45, 39–47, 39–54, and 39–56 have a maximum of one unprotected amide, indicating that the N-terminal unprotected region does not extend past residue 41. Note that the condition of residues 39 and 40 in these fragments is indeterminate because the D isotope label on the N-terminal and penultimate residues of all fragments (and on polar side chains) exchanges rapidly (10) and is lost during the analysis (proteolysis, HPLC and MS in H2O). An exception occurs when the penultimate residue is valine or isoleucine where approximately half of the amide label is retained.
The number of unprotected amides found for the fragments 18–38 and 21–38 (15 and 13, respectively) indicates that the N-terminal unprotected region reaches at least to residue 34 (residue 22 is Val). The 12 freely exposed amides measured for the 27–44 segment extend the unprotected region to residue 39 (residue 28 is Ile). If 25% of the chain population is protected and not labeled in these segments, then the estimate for the unprotected length in the unprotected fraction of the population extends to residue 38 and 43, respectively. However, exposure past residue 41 is ruled out, as just noted.
At the C terminus of the protected midregion, the segments 90–94 and 90–97 are fully protected, placing the boundary past residue 97. The fragments 90–113, 94–113, 95–113, and 99–113 overlap the boundary. The number of sites labeled on each of these fragments estimates the last protected amide at residue 100, 100, 101, and 103, respectively.
In summary, these results indicate a somewhat less well defined boundary from mostly unstructured to structured at about position 38/39 (±3) and a sharp discontinuity back to nonstructure at position 101/102 (±1).
Discussion
Structure in α-syn Amyloid. HX results indicate that the α-syn amyloid protects a continuous stretch of amides reaching approximately from residue 39 to 101. The supposition that this region represents the well documented cross-β structure of amyloid is supported by the fact that the amount of structure indicated (45%) agrees with the β-structure content found by CD (40–50%).
Long N- and C-terminal segments extend outside of the continuous cross-β amyloid region. The absence of HX protection indicates that the C-terminal length is disordered. This 40-residue chain segment is rich in residues with low β-structure propensity (five prolines, four glycines, and 15 ionic residues). Through the N-terminal length, ≈75% of the molecules show no HX protection, whereas 25% appear to be strongly protected. The chain heterogeneity may be due to structural or environmental heterogeneity in different regions of the fibril or perhaps to populations of fibril subtypes with somewhat different structure (42).
Comparison with Prior Results. Der-Sarkissian et al. (41) prepared 36 different α-syn constructs with individual cysteine residues distributed through the protein, bound nitroxide spin labels to each cysteine, and measured EPR spectra of the soluble monomer and the assembled amyloid at warm and subzero temperatures. These side-chain data agree with the present main-chain results in that an N-terminal length shows some kind of structural heterogeneity, a midmolecule segment adopts systematic cross-β structure, and a long C-terminal region appears unstructured. N-terminal heterogeneity to proteolysis was also noticed in the N-terminal segment of amyloid-β (43). The heterogeneity imposes some uncertainty on the boundary position, placed at ≈33/34 by the EPR results and at ≈38/39 by HX. Both experiments place the last protected residue at position 101.
Miake et al. (44) exposed α-syn amyloid assembled in vitro and in vivo to fragmentation by proteinase K, which has broad specificity. They identified a protease-resistant segment through the central region of the α-syn chain, reaching from residue 31 to 109, that was longer on both ends than the structured core identified here, as might be expected because of protease specificity and structural interference. Other studies are also consistent with the structure we find, including work done with α-syn fragments that can copolymerize and even independently fibrillize (37, 45, 46) and with mutations (47) and truncations that affect the seeding, nucleation, and assembly processes (48, 49).
Amyloid Models: Constraints and Freedom. Possible models for amyloid structure must satisfy certain constraints (2–4). The polypeptide chains are known to form an unlimited β-sheet, which appears as an elongated ribbon. The chains lie in the ribbon plane with average direction normal to the ribbon length and H bonds parallel to it, which is known as the cross-β motif. β-Sheets can stack to form lamina joined by their interacting side chains. The major uncertainties concern side-chain interactions and how they determine amyloid properties.
Amyloids designed in the laboratory (50) or by biological evolution for functional purposes (9) can have highly ordered side-chain interactions. Accidental amyloids are far more likely to fall into a format with poorly ordered side chains because they are distorted forms of proteins that have been designed for other purposes. Net stability of accidental amyloids must then emerge from a mix of favorable and unfavorable side-chain interactions, both side by side within a given sheet and head to head between lamina.
Proteins in general are able to tolerate and energy-minimize some degree of poor side-chain matching. Amyloid is likely to exhibit an exaggerated ability to do so because of the great freedom available to the main chain. Main-chain dihedrals are free to occupy both the β-basin and the neighboring β-compatible polyproline II basin, which together occupy approximately half of available Ramachandran space. [The polyproline II configuration is adopted by 23% of β-strand residues in well structured Protein Data Bank proteins (51).] Further, the poor packing of side chains will relax the usual side-chain-dependent constraints for specific angular twist within strands and a 30° angle between β-sheet lamina (52). The large main-chain freedom that results, together with variable side-chain packing, may make amyloid more akin to molten globular proteins (53) than to familiar tightly structured proteins.
Importantly, different side-chain defects and sterically different accommodations to energy-minimize them will occur at multiple positions through any given polypeptide sequence. In different amyloids, different sets of accommodations will occur. Thus, different amyloids, although visually similar in gross morphology, must differ greatly in local steric detail.
Remarkably, the long-chain accidental amyloids α-syn and amyloid-β form in-register, parallel β-sheets (5, 41, 54). We take this observation to mean that an in-register orientation increases the probability for finding stable combinations, apparently because it naturally produces favorable side-by-side interactions between identical side chains, especially hydrophobe-to-hydrophobe but also aromatic-to-aromatic (50) and amide-to-amide (9) interactions. Designed short-chain amyloids can overwhelm the in-register self-interaction benefit and drive β-strands to orient either parallel or antiparallel and in or out of register (9, 35, 50, 55–58). However, accidental long-chain amyloids must achieve net stability by some chance selection from their many mostly unfavorable options. Here, the in-register advantage can remain significant.
These observations lead to a key deduction. Because chains newly added to propagating amyloid line up reproducibly, in parallel and in register, they must be able to closely duplicate the preexisting set of varied steric adaptations. Accordingly, because amyloids formed from different polypeptides require sterically different adaptations, they are likely to be structurally incompatible.
These considerations reflect on the structure of α-syn amyloid and on amyloid properties more generally.
Structure of α-syn Amyloid. The present results specify the cross-β region of α-syn (see also ref. 41) and impose two additional constraints. If the protected β-strand segment found here were fully extended, it would reach >20 nm, much greater than the typically observed width of the amyloid ribbon (≤10 nm). Therefore, the main chains must somehow bend back on themselves, and this bending must occur in a way that does not interrupt the pattern of continuous amide H bonding. Given all of these constraints, the one remaining broad degree of freedom that α-syn can search through to find some stable amyloid form concerns the alternative sets of side-chain interactions that connect the stacked sheets. The choice is determined by the position of main-chain bends.
Amyloid models can be considered that incorporate one or more distinct main-chain bends. The bend(s) might run in the plane of the ribbon, as in a typical β-hairpin. Each β-strand folds to juxtapose its own main-chain amides so that the strand segments run antiparallel and form connecting H bonds in the ribbon plane. The requirement for parallel chain orientation (see above) might then be satisfied by alternating neighboring molecules. However, amides in the bend region are then removed from their nearest possible systematically H-bonding partners by the laminated side-chain thickness (≈10 Å), so that systematic H bonding between turn amides does not naturally occur for a sharp turn. For a more elongated turn in an accidental amyloid like α-syn, systematic H bonding seems very unlikely to occur simply by chance. Thus, this model does not match the present constraints.
Alternatively, the bend(s) might run perpendicular to the ribbon plane so that each chain folds to occupy adjacent lamina separated by their side-chain distance. Neighboring parallel, in-register chains (41) do the same, and they are connected by the usual β-sheet H bonds parallel to the ribbon length. To bridge the interlamina distance (≈10 Å), the connecting bend requires three or more residues. The conformation of long connecting loops is not limited by Ramachandran constraints and would have to form systematic H-bonding protection simply by chance, which seems unlikely. In a tight bend, amides are constrained to point parallel to amides in the β-sheets and so will naturally continue systematic main-chain-to-main-chain H bonding. This configuration matches the present constraints. The addition of identically bent, in-register parallel chains will then construct a long amyloid ribbon as previously inferred for amyloid-β (59).
Finally, one can relax the need for distinct turns. The polypeptide chains might wrap continuously [for example, around a cylinder in a wide β-helix as suggested by Perutz et al. (60) for certain peptides] (see also ref. 61). However, this model seems inconsistent with more recent high-resolution crystallographic results for the similarly amide-rich Sup35 amyloid (9).
Amyloid Properties More Generally. The considerations just reviewed suggest that the surprisingly high probability for accidental amyloid formation depends on the unusual ability of the structurally relaxed main chain to adapt to and energy-minimize side-chain mismatching (aided by the easy accessibility and inherent stability of the β-conformation). If the amino acid sequence of any polypeptide allows some net stable side-chain-plus-main-chain combination to be found, a thermodynamic search will ultimately find it. Additional polypeptides can be subsequently bound in-register only if they can faithfully reproduce many of the varied steric details of the previously established configuration.
Given these principles, the properties of amyloid follow. The time-consuming conformational search necessary to discover some rare set of steric configurations that achieves net stability naturally generates nucleation and seeding phenomena. Once a stable seed is found, it will propagate indefinitely because an open-ended β-sheet adds new chains without closing, forming an elongated sheet. The stringent requirement for slavishly repeating the identical set of sequence-dependent configurations initially found will produce seeding specificity, strain variability, and species barriers (42). The kinetic and equilibrium situation is very much like protein crystallization.
Finally, other work shows how native proteins can be recruited into growing amyloid. Proteins repeatedly unfold, in whole (62) and in part (63, 64), even under fully native conditions. Refolding is guided and stabilized by collisional interactions with compatible preexisting templating structure (65, 66). Accordingly, amyloid growth can, over time, compete with normal protein refolding because it is thermodynamically favored even though kinetically obstructed.
Acknowledgments
We thank M. Goedert (Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.) for the α-syn clone, C. Gessner and S. Li for technical help, and R. Wetzel and Y. Hamuro for comments on the manuscript. This work was supported by National Institutes of Health Grants GM031847 (to S.W.E.) and CA099835, CA118595, GM205001, and GM074150 (to V.L.W.) and the Mathers Charitable Foundation.
Author contributions: E.A.G. designed research; C.D.M., E.A.G., and L.M. performed research; V.L.W. contributed new reagents/analytic tools; C.D.M. and S.W.E. analyzed data; and S.W.E. wrote the paper.
Abbreviations: α-syn, α-synuclein; HX, hydrogen exchange; pDr, uncorrected glass electrode reading in D2O.
References
- 1.Dobson, C. M. (2001) Philos. Trans. R. Soc. London B 356, 133-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eanes, E. D. & Glenner, G. G. (1968) J. Histochem. Cytochem. 16, 673-677. [DOI] [PubMed] [Google Scholar]
- 3.Sunde, M. & Blake, C. C. (1997) Adv. Protein Chem. 50, 123-159. [DOI] [PubMed] [Google Scholar]
- 4.Sunde, M. & Blake, C. C. (1998) Q. Rev. Biophys. 31, 1-39. [DOI] [PubMed] [Google Scholar]
- 5.Benzinger, T. L., Gregory, D. M., Burkoth, T. S., Miller-Auer, H., Lynn, D. G., Botto, R. E. & Meredith, S. C. (2000) Biochemistry 39, 3491-3499. [DOI] [PubMed] [Google Scholar]
- 6.Torok, M., Milton, S. C., Kayed, R., Wu, P., McIntyre, T., Glabe, C. G. & Langen, R. (2002) J. Biol. Chem. 277, 40810-40815. [DOI] [PubMed] [Google Scholar]
- 7.Tycko, R. (2004) Curr. Opin. Struct. Biol. 14, 96-103. [DOI] [PubMed] [Google Scholar]
- 8.Sumner, M. O. & Serpell, L. C. (2004) J. Mol. Biol. 335, 1279-1288. [DOI] [PubMed] [Google Scholar]
- 9.Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. O., Riekel, C., Grothe, R. & Eisenberg, D. (2005) Nature 435, 773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai, Y., Milne, J. S., Mayne, L. & Englander, S. W. (1993) Proteins 17, 75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connelly, G. P., Bai, Y., Jeng, M. F. & Englander, S. W. (1993) Proteins 17, 87-92. [DOI] [PubMed] [Google Scholar]
- 12.Krishna, M. M. G., Hoang, L., Lin, Y. & Englander, S. W. (2004) Methods 34, 51-64. [DOI] [PubMed] [Google Scholar]
- 13.Ernst, R. R., Bodenhausen, G. & Wokaun, A. (1987) Principles of Nuclear Magnetic Resonance in One and Two Dimensions (Clarendon, Oxford).
- 14.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 15.Bax, A. (1994) Curr. Opin. Struct. Biol. 4, 738-744. [Google Scholar]
- 16.Wand, A. J. & Englander, S. W. (1996) Curr. Opin. Biotechnol. 7, 403-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosa, J. J. & Richards, F. M. (1979) J. Mol. Biol. 133, 399-416. [DOI] [PubMed] [Google Scholar]
- 18.Englander, S. W., Calhoun, D. B., Englander, J. J., Kallenbach, N. R., Liem, R. K. H., Malin, E. L., Mandal, C. & Rogero, J. R. (1980) Biophys. J. 32, 577-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Englander, J. J., Rogero, J. R. & Englander, S. W. (1985) Anal. Biochem. 147, 234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Englander, S. W. & Englander, J. J. (1994) in Methods in Enzymology, eds. Everse, J., Vandegriff, K. D. & Winslow, R. M. (Academic, New York), Vol. 232, pp. 26-42.8057864 [Google Scholar]
- 21.Zhang, Z. & Smith, D. L. (1993) Protein Sci. 2, 522-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engen, J. R. & Smith, D. L. (2001) Anal. Biochem. 73, 256A-265A. [DOI] [PubMed] [Google Scholar]
- 23.Hoofnagle, A. N., Resing, K. A. & Ahn, N. G. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 1-25. [DOI] [PubMed] [Google Scholar]
- 24.Eyles, S. J. K. & Igor, A. (2004) Methods 34, 88-99. [DOI] [PubMed] [Google Scholar]
- 25.Norris, E. H., Giasson, B. I. & Lee, V. M. (2004) Curr. Top. Dev. Biol. 60, 17-54. [DOI] [PubMed] [Google Scholar]
- 26.Kheterpal, I., Zhou, S., Cook, K. D. & Wetzel, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13597-13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kheterpal, I., Lashuel, H. A., Hartley, D. M., Walz, T., Lansbury, P. T., Jr., & Wetzel, R. (2003) Biochemistry 42, 14092-14098. [DOI] [PubMed] [Google Scholar]
- 28.Nazabal, A., Maddelein, M.-L., Bonneu, M., Saupe, S. J. & Schmitter, J.-M. (2005) J. Biol. Chem. 280, 13220-13228. [DOI] [PubMed] [Google Scholar]
- 29.Whittemore, N. A., Mishra, R., Kheterpal, I., Williams, A. D., Wetzel, R. & Serpersu, E. H. (2005) Biochemistry 44, 4434-4441. [DOI] [PubMed] [Google Scholar]
- 30.Kraus, M., Bienert, M. & Krause, E. (2003) Rapid Commun. Mass Spectrom. 17, 222-228. [DOI] [PubMed] [Google Scholar]
- 31.Wang, S. S., Tobler, S. A., Good, T. A. & Fernandez, E. J. (2003) Biochemistry 42, 9507-9514. [DOI] [PubMed] [Google Scholar]
- 32.Ippel, J. H., Olofsson, A., Schleucher, J., Lundgren, E. & Wijmenga, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 8648-8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol. 9, 332-336. [DOI] [PubMed] [Google Scholar]
- 34.Kuwata, K., Matumoto, T., Cheng, H., Nagayama, K., James, T. L. & Roder, H. (2003) Proc. Natl. Acad. Sci. USA 100, 14790-14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritter, C., Maddelein, M. L., Siemer, A. B., Luhrs, T., Ernst, M., Meier, B. H., Saupe, S. J. & Riek, R. (2005) Nature 435, 844-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi, K., Katou, H., Hoshino, M., Hasegawa, K., Naiki, H. & Goto, Y. (2004) J. Mol. Biol. 338, 559-571. [DOI] [PubMed] [Google Scholar]
- 37.Giasson, B. I., Murray, I. V., Trojanowski, J. Q. & Lee, V. M. (2001) J. Biol. Chem. 276, 2380-2386. [DOI] [PubMed] [Google Scholar]
- 38.Englander, J. J., DelMar, C., Li, W., Englander, S. W., Kim, J. S., Stranz, D. D., Hamuro, Y. & Woods, V. L., Jr. (2003) Proc. Natl. Acad. Sci. USA 100, 7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pantazatos, D., Kim, J. S., Klock, H. E., Stevens, R. C., Wilson, I. A., Lesley, S. A. & Woods, V. L., Jr. (2004) Proc. Natl. Acad. Sci. USA 101, 751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinreb, P. H., Zhen, W., Poon, A. W., Conway, K. A. & Lansbury, P. T., Jr. (1996) Biochemistry 35, 13709-13715. [DOI] [PubMed] [Google Scholar]
- 41.Der-Sarkissian, A., Jao, C. C., Chen, J. & Langen, R. (2003) J. Biol. Chem. 278, 37530-37535. [DOI] [PubMed] [Google Scholar]
- 42.Chien, P., Weissman, J. S. & DePace, A. H. (2004) Annu. Rev. Biochem. 73, 617-656. [DOI] [PubMed] [Google Scholar]
- 43.Kheterpal, I., Williams, A. D., Murphy, C., Bledsoe, B. & Wetzel, R. (2001) Biochemistry 40, 11757-11767. [DOI] [PubMed] [Google Scholar]
- 44.Miake, H., Mizusawa, H., Iwatsubo, T. & Hasegawa, M. (2002) J. Biol. Chem. 277, 19213-19219. [DOI] [PubMed] [Google Scholar]
- 45.Ueda, K., Fukushima, H., Masliah, E., Xia, Y., Iwai, A., Yoshimoto, M., Otero, D. A., Kondo, J., Ihara, Y. & Saito, T. (1993) Proc. Natl. Acad. Sci. USA 90, 11282-11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han, H., Weinreb, P. H. & Lansbury, P. T., Jr. (1995) Chem. Biol. 2, 163-169. [DOI] [PubMed] [Google Scholar]
- 47.Greenbaum, E. A., Graves, C. L., Mishizen-Eberz, A. J., Lupoli, M. A., Lynch, D. R., Englander, S. W., Axelsen, P. H. & Giasson, B. I. (2005) J. Biol. Chem. 280, 7800-7807. [DOI] [PubMed] [Google Scholar]
- 48.Murray, I. V., Giasson, B. I., Quinn, S. M., Koppaka, V., Axelsen, P. H., Ischiropoulos, H., Trojanowski, J. Q. & Lee, V. M. (2003) Biochemistry 42, 8530-8540. [DOI] [PubMed] [Google Scholar]
- 49.Kessler, J. C., Rochet, J. C. & Lansbury, P. T. J. (2003) Biochemistry 42, 672-678. [DOI] [PubMed] [Google Scholar]
- 50.Makin, O. S., Atkins, E., Sikorski, P., Johansson, J. & Serpell, L. C. (2005) Proc. Natl. Acad. Sci. USA 102, 315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jha, A. K., Colubri, A., Zaman, M. H., Koide, S., Sosnick, T. R. & Freed, K. F. (2005) Biochemistry 44, 9691-9702. [DOI] [PubMed] [Google Scholar]
- 52.Chothia, C. (1984) Annu. Rev. Biochem. 53, 537-572. [DOI] [PubMed] [Google Scholar]
- 53.Ptitsyn, O. B. (1995) Trends Biochem. Sci. 20, 376-379. [DOI] [PubMed] [Google Scholar]
- 54.Antzutkin, O. N., Balbach, J. J., Leapman, R. D., Rizzo, N. W., Reed, J. & Tycko, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13045-13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lansbury, P. T., Costa, P. R., Griffiths, J. M., Simon, E. J., Auger, M., Halverson, K. J., Kocisko, D. A., Hendsch, Z. S., Ashburn, T. T., Spencer, R. G. S. et al. (1995) Nat. Struct. Biol. 2, 990-998. [DOI] [PubMed] [Google Scholar]
- 56.Balbach, J. J., Ishii, Y., Antzutkin, O. N., Leapman, R. D., Rizzo, N. W., Dyda, F., Reed, J. & Tycko, R. (2000) Biochemistry 39, 13748-13759. [DOI] [PubMed] [Google Scholar]
- 57.Petkova, A. T., Buntkowsky, G., Dyda, F., Leapman, R. D., Yau, W. M. & Tycko, R. (2004) J. Mol. Biol. 335, 247-260. [DOI] [PubMed] [Google Scholar]
- 58.Gordon, D., Balbach, J., Tycko, R. & Meredith, S. C. (2004) Biophys. J. 86, 428-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742-16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perutz, M., Finch, J., Berriman, J. & Lesk, A. (2002) Proc. Natl. Acad. Sci. USA 99, 5591-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wetzel, R. (2002) Structure 10, 1031-1036. [DOI] [PubMed] [Google Scholar]
- 62.Bai, Y., Milne, J. S., Mayne, L. & Englander, S. W. (1994) Proteins 20, 4-14. [DOI] [PubMed] [Google Scholar]
- 63.Bai, Y., Sosnick, T. R., Mayne, L. & Englander, S. W. (1995) Science 269, 192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bai, Y. & Englander, S. W. (1996) Proteins 24, 145-151. [DOI] [PubMed] [Google Scholar]
- 65.Rumbley, J., Hoang, L., Mayne, L. & Englander, S. W. (2001) Proc. Natl. Acad. Sci. USA 98, 105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maity, H., Maity, M., Krishna, M. M. G., Mayne, L. & Englander, S. W. (2005) Proc. Natl. Acad. Sci. USA 102, 4741-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]