Abstract
Plasmodium vivax causes the most geographically widespread human malaria, accounting annually for 70-80 million clinical cases throughout the tropical and subtropical regions of the world's continents. We have analyzed the DNA sequences of the Csp (circumsporozoite protein) gene in 24 geographically representative strains of P. vivax and 2 of P. simium, which parasitizes several species of New World monkeys. The Csp sequences are of two types, VK210 and VK247, which differ by three diagnostic amino acid replacements, one in each of the 5′ and 3′ terminal regions [5′ nonrepeat (NR) and 3′ NR] of the gene and in an insertion sequence that precedes the 3′ NR region. The central region of the gene consists of ≈38 repetitive “motifs,” which are alternatively four and five amino acids long, which also are diagnostically different between the VK210 and VK247 types. There are very few synonymous substitutions within and between the two types of strains, which we hypothesize reflects that the worldwide spread of P. vivax is very recent. The two P. simium Csp sequences belong one to each of the two VK types and are genetically indistinguishable from the corresponding P. vivax strains, suggesting that at least two host transfers have occurred between humans and New World monkeys. We exclude as unlikely the possibility that the two types of sequences could have independently arisen in humans and platyrrhines by natural selection. There are reasons favoring each of the two possible directions of host transfer between humans and monkeys.
Keywords: circumsporozoite protein, clonal theory, Plasmodium population structure, host-parasite interactions, malaria
Malaria's human toll is appalling: 300-500 million clinical cases and 1-3 million deaths per year. Plasmodium falciparum accounts for 80% of human malaria's morbidity and mortality, mostly in sub-Saharan Africa. Most geographically widespread and prevalent in some regions is Plasmodium vivax, which accounts annually for 70-80 million clinical cases across much of the tropics and subtropics of the world.
The evolutionary origin of P. vivax has been placed by some authors in Southeast Asia (1-3). However, the high prevalence in sub-Saharan Africa of Duffy negativity (absence of the Duffy blood group antigen) that protects against P. vivax infection has been interpreted as evidence of the African origin of P. vivax (4-6). Most recently, phylogenetic and biogeographical evidence has been advanced supporting a Southeast Asia origin (7-9).
Recent investigations have shown a scarcity of selectively neutral genetic polymorphisms in P. vivax (8, 10, 11), which is consistent with a recent world expansion of P. vivax as a human parasite. The discovery that the platyrrhine parasite P. simium is genetically indistinguishable from P. vivax (2, 7, 8) manifests that a host transfer between humans and New World monkeys has happened in very recent evolutionary times.
The circumsporozoite protein (CSP) has been extensively studied in P. falciparum and other Plasmodium species because of its immune significance as the major surface protein of the sporozoites. We have investigated the nucleotide sequence of the Csp gene in 24 strains of P. vivax and 2 of P. simium. The Csp sequences of P. vivax are of two types, VK210 and VK247, which differ in the amino acid composition of the central repetitive region of the gene (12-14), but also by three diagnostic amino acid replacements, one in each of the 5′ and 3′ terminal regions of the gene and the third in an insertion region (IR) between the central repeat (CR) and the 3′ terminal region. Both variants are present in P. vivax form in the New and Old World. Nucleotide synonymous variation within and between the two VK types is meager, consistent with a recent world expansion of P. vivax malaria. Both Csp types, VK210 and VK247, are present in P. simium, suggesting that at least two host transfers have occurred between humans and monkeys.
Materials and Methods
The 26 strains and their geographic origin are listed in Table 1. The strains are representative of the P. vivax malaria regions. The sequences were obtained from available sources, except for strains 15 and 20, sequenced by us.
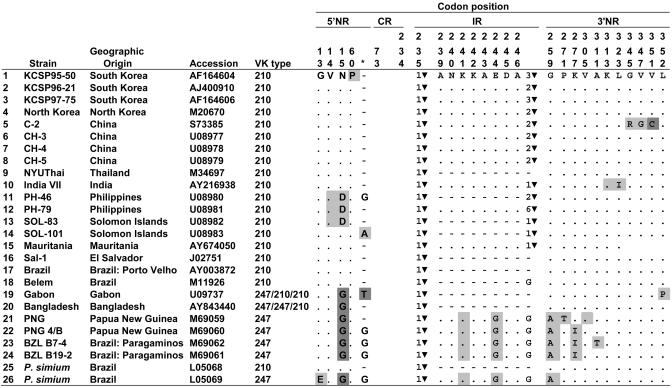
Table 1. Polymorphism in the 5′ NR, IR, and 3′ NR regions of Csp in P. vivax and P. simium.
Amino acids and their position are according to the reference strain AF164604. Dots indicate identity with reference strain; short dashes indicate deletions. Amino acid replacements are indicated; light shading indicates one, dark shading two nucleotide differences. ▾ is GGNA, preceded by the number of copies. The asterisk refers to an amino acid site between sites 72 and 73 of the reference strain that is absent from most VK210 strains.
The primers, sequencing, and phylogenetic and statistical methods are listed in Supporting Text, which is published as supporting information on the PNAS web site.
Results
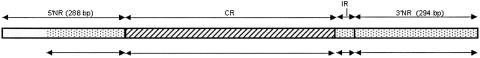
Fig. 1 displays the structure of the Csp gene in P. vivax and P. simium, identical in both species. Similarly as in P. falciparum and other Plasmodium species (3, 15), the Csp gene consists of two flanking, highly conserved and nonrepetitive (NR) regions, embracing a CR region consisting of numerous repetitions of short oligopeptides (amino acid motifs). The CR region varies in the number of amino acid motifs, and there is also sequence variation among the oligopeptides. Between the CR region and the 3′ NR terminal region, there is an IR of relatively simple structure.
Fig. 1.
Structure of the Csp gene in P. vivax, with two terminal NR regions, a CR with a variable number of tandem repeats, and a short IR. The analyzed sequences are indicated by the arrows below the diagram.
Table 1 shows the amino acid polymorphisms found in the 26 strains of P. vivax and P. simium in the 5′ NR, IR, and 3′ NR regions. The two types of strains, VK210 and VK247, differ by three diagnostic replacements, one in each of the three regions, at sites 15, 244, and 259. There is a fourth, nearly diagnostic, replacement at the end of the IR region: VK247 has G instead of zero, one, or several copies of the tetrapeptide GGNA; strain 18 from Belem also has G at this site. One P. simium strain (LO5068) has the VK210 type; the other (LO5069) has the VK247 type. Strains 19 and 20, from Gabon and Bangladesh, are recombinants with the 5′ NR of the VK247 type but the IR and 3′ NR of the VK210 type; the CR is VK210 in Gabon but VK247 in Bangladesh.
The phylogenetic relationships of the 26 Csp sequences for the 5′ NR and the 3′ NR regions are shown in Figs. 7 and 8, which are published as supporting information on the PNAS web site. The VK210 and VK247 types cluster separately on both trees, each cluster including one P. simium strain.
The presence of both VK types in P. simium, apparent in Table 1 and Figs. 7 and 8, suggests that the host transfer between humans and platyrrhine monkeys must have happened at least twice, once for each VK type. An unlikely alternative explanation is that one VK type evolved from the other VK type independently twice, once in P. vivax and again in P. simium.
Table 2 summarizes the polymorphism statistics. The values of π or θ, which measure polymorphism per site, are low within each type and species and in both types and both species combined. The number of synonymous segregating sites is low (only four among the 26 strains). Because synonymous substitutions are adaptively neutral, or nearly so, synonymous polymorphisms reflect the time elapsed since the sequences derived from a common ancestral sequence. The scarcity of Csp synonymous polymorphisms can be interpreted, as it has been for P. falciparum (15-18), as an indication of a recent bottleneck in the evolutionary history of P. vivax,sothat the current worldwide distribution of this species would be recent.
Table 2. Polymorphism in the 5′ NR and 3′ NR regions of the Csp gene.
| Sequences | n | A | S | πA | πS | πall | θA | θS | θall |
|---|---|---|---|---|---|---|---|---|---|
| P. vivax | |||||||||
| VK210 | 18* | 9 | 3 | 0.002 | 0.005 | 0.003 | 0.007 | 0.008 | 0.007 |
| VK247 | 6* | 5 | 1 | 0.007 | 0.003 | 0.006 | 0.006 | 0.004 | 0.005 |
| All | 24 | 16 | 4 | 0.006 | 0.004 | 0.006 | 0.011 | 0.010 | 0.011 |
| P. vivax and P. simium | |||||||||
| VK210 | 19 | 9 | 3 | 0.002 | 0.005 | 0.003 | 0.007 | 0.008 | 0.007 |
| VK247 | 7 | 7 | 1 | 0.007 | 0.002 | 0.006 | 0.008 | 0.004 | 0.007 |
| All | 26 | 17 | 4 | 0.006 | 0.004 | 0.006 | 0.012 | 0.009 | 0.011 |
n, sample size (*, the numbers of sequences shown are for 5′ NR; in 3′ NR, n = 20 and 4 for VK210 and VK247, respectively); A, segregating amino acid sites; S, segregating synonymous sites.
Table 3 compares replacement and amino acid polymorphisms between the two VK types and between them and P. cynomolgi. The Ka/Ks ratio is 1.57 between the two VK types, indicating that positive selection accounts for the differentiation between them. The high Ka/Ks ratios between P. vivax (plus P. simium) and P. cynomolgi suggest that Csp amino acid differentiation over time has been largely driven by natural selection. The Ka/Ks ratios are fairly similar between each VK type and P. cynomolgi, with no clear indication that one of the two VK types is ancestral to the other.
Table 3. Genetic differentiation in the Csp gene between the two VK types of P. vivax (including P. simium) and P. cynomolgi.
| Comparison | Ka | Ks | Ka/Ks |
|---|---|---|---|
| VK210 to VK247 | 0.011 | 0.007 | 1.57 |
| VK210 to P. cynomolgi | 0.096 | 0.112 | 0.86 |
| VK247 to P. cynomolgi | 0.095 | 0.106 | 0.90 |
| (VK210 & VK247) to P. cynomolgi | 0.096 | 0.111 | 0.87 |
The central part of the IR region is an 8-aa peptide present in 11 VK210 strains and all VK247 strains but absent in 10 VK210 strains (including one P. simium strain) (Table 1). The VK210 strains differ among themselves also in the number of copies of a GGNA tetrapeptide on the 3′ end of the IR region, which is absent in five VK210 (including one P. simium) strains, and is replaced by glycine (G) in one VK210 and all VK247 strains. The VK210 and VK247 strains also differ, as noted, by the diagnostic presence at site 244 of G in VK247 but E in VK210. In all strains of both species, the IR and CR regions are invariably separated by GGNA (positions 235-238).
The CR region of P. vivax has been described as consisting of repetitions of a 9-aa oligopeptide, with numerous amino acid replacements among the oligopeptides. We have decomposed the CR region into two kinds of repeating amino acid “motifs”: a tetrapeptide (GDRA in VK210 and ANGA in VK247) followed by a pentapeptide (AGQPA or DGQPA in VK210 and GNQPG or GDQPG in VK247) (Table 4 and Fig. 2). These motifs account for 82% to 98% of all motifs. The average number of motifs per strain is 37.9 ± 1.5 (Table 5).
Table 4. Amino acid motifs and their frequency in the CR region of the Csp gene.
| Number
|
Frequency within type, %
|
|||||
|---|---|---|---|---|---|---|
| Symbol | Motif | VK type | P. vivax | P. simium | P. vivax | P. simium |
| Four-aa motifs | ||||||
| VK210 | ||||||
| 1 | GDRA | 210 | 282 | 20 | 82.0 | 95.2 |
| 1a | GNGA | 210 | 31 | 1 | 9.0 | 4.8 |
| 1b | GDGA | 210 | 28 | 0 | 8.1 | 0 |
| 1c | GDRP | 210 | 1 | 0 | 0.3 | 0 |
| 1d | VDRA | 210 | 1 | 0 | 0.3 | 0 |
| 2 | ANGA* | 210 | 1 | 0 | 0.3 | |
| Total | 344 | 21 | 100 | 100 | ||
| VK247 | ||||||
| 2 | ANGA* | 247 | 105 | 20 | 98.1 | 95.2 |
| 2a | EDGA | 247 | 2 | 0 | 1.9 | 0 |
| 2b | ENGA | 247 | 0 | 1 | 0 | 4.8 |
| Total | 107 | 21 | 100 | 100 | ||
| Total | 451 | 42 | ||||
| Five-aa motifs | ||||||
| VK210 | ||||||
| 1 | AGQPA | 210 | 168 | 10 | 48.8 | 47.6 |
| 1a | DGQPA | 210 | 123 | 10 | 35.8 | 47.6 |
| 1b | GGQAA* | 210 | 24 | 1 | 7.0 | 4.8 |
| 1c | —GQPA | 210 | 9 | 0 | 2.6 | 0 |
| 1d | GGQPA | 210 | 8 | 0 | 2.3 | 0 |
| 1e | AGQAA | 210 | 7 | 0 | 2.0 | 0 |
| 1f | DGQAA | 210 | 4 | 0 | 1.2 | 0 |
| 1g | DGQP— | 210 | 1 | 0 | 0.3 | 0 |
| Total | 344 | 21 | 100 | 100 | ||
| VK247 | ||||||
| 2 | GNQPG | 247 | 72 | 15 | 67.3 | 71.4 |
| 2a | GDQPG | 247 | 20 | 4 | 18.7 | 19.0 |
| 1b | GGQAA* | 247 | 5 | 1 | 4.7 | 4.8 |
| 2b | DDQPG | 247 | 10 | 1 | 9.3 | 4.8 |
| Total | 107 | 21 | 100 | 100 | ||
| Total | 451 | 42 | ||||
These two motifs are present in both VK types; GGQAA as the terminal motif
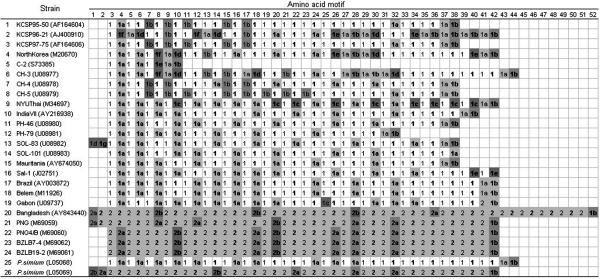
Fig. 2.
Alignment of the amino acid motifs in P. vivax and P. simium. The composition of the motifs is given in Table 4. The 4-aa (Roman) and 5-aa (boldface) motifs alternate in the sequences; they are different for the VK210 (number 1) and VK247 (number 2) types. The P. simium strains (at bottom) are one of each type.
Table 5. Number of strains with a given number of amino acid motifs in the CR region of the Csp gene.
| No. of motifs
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 8 | 30 | 36 | 38 | 40 | 42 | 52 | Mean ± SE | n | |
| P. vivax | 1 | 1 | 7 | 2 | 10 | 2 | 1 | 37.6 ± 1.4 | 24 |
| P. simium | 2 | 42.0 ± 0.0 | 2 | ||||||
| Total | 1 | 1 | 7 | 2 | 10 | 4 | 1 | 37.9 ± 1.5 | 26 |
n, number of strains. Mean ± SE is for the number of motifs per strain.
Fig. 2 displays the motif composition of each strain. The two VK types are sharply different (in P. vivax and P. simium), similarly as in the 5′ NR and 3′ NR regions. Both VK types, however, have the same terminal motif, GGQAA (1b) except for two strains, 14 and 16, with terminal motifs DGQPA (1a) and AGQAA (1e), respectively. As shown in Fig. 2 (see also Table 4), one P. simium strain is VK210 and the other VK247 in the CR region, similarly as in the 5′ NR and 3′ NR regions. The recombinant strain 19 (Gabon) is VK210 in the CR and 3′ NR regions but VK247 in the 5′ NR region; strain 20 (Bangladesh) is VK210 in the 3′ NR region but VK247 in the 5′ NR and CR regions (see Figs. 7 and 8).
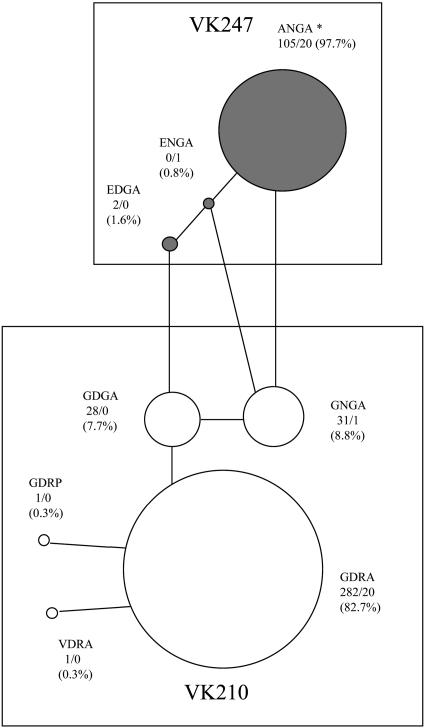
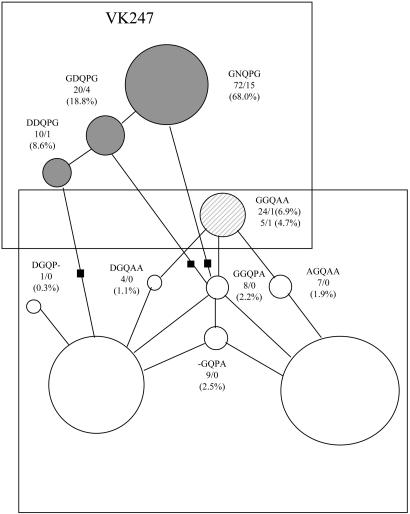
Figs. 3 and 4 display the relative abundance of the various motifs and the networks connecting them by one-mutation steps. The 4-aa network (Fig. 3) shows the distinct separation between the VK210 and VK247 types and the prevalence of ANGA and GDRA, which differ by 2 amino acid replacements. The ANGA motif is present as a singleton in the Gabon strain (19). The 5-aa network (Fig. 4) shows also the distinct motifs of each VK type. As pointed out earlier, GGQAA (1b) is the CR terminal motif for both types, VK210 and VK247. There are three possible transitions between the two networks in Fig. 4, each requiring two mutational steps, but the intermediate motif (black squares) is absent in all three cases. The prevalent motif in VK247 (GNQPG) is separated by a minimum of three amino acid replacements from the two prevalent motifs in VK210 (AGQPA and DGQPA).
Fig. 3.
Network connecting the 4-aa motifs of the two VK types by one-replacement steps. The numbers before and after the slash refer to the abundance of each motif in P. vivax and P. simium, respectively. The percents refer to the relative abundance of the motifs within each VK type. *, ANGA is present once in strain 19 from Gabon (see Fig. 2 and Table 1).
Fig. 4.
Network connecting the 5-aa motifs. Descriptions are as in Fig. 3. GGQAA (1b) is the terminal motif in both VK types.
The four-gamete test yields no evidence of recombination among the 26 Csp sequences. Nevertheless, recombination has occurred at least in strains 19 (Gabon) and 20 (Bangladesh). It seems likely that the recombinant event took place in the Gabon strain at the boundary between the 5′ NR region and the CR region but at the boundary between the CR region and the IR region in the Bangladesh strain (see Table 1).
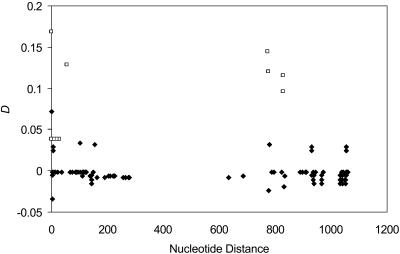
There are 17 segregating amino acid sites, 5 in the 5′ NR region and 12 in the 3′ NR region. There are 15 statistically significant (P < 0.05) disequilibrium values of 136 pairwise comparisons, four of them between the two NR regions (Fig. 5). Linkage disequilibrium does not decrease with increasing nucleotide distance, which is consistent with a predominantly clonal mode of reproduction and a low rate of intragenic recombination.
Fig. 5.
Linkage disequilibrium (D) as a function of nucleotide distance. Comparisons are made within and between 5′ NR and 3′ NR sites. Significant D values (P < 0.05) are represented by □.
Discussion
The Csp gene has been extensively studied in P. falciparum and other Plasmodium species because of its immune significance as the major surface protein of the parasite's sporozoite. Two CSP types have been distinguished in P. vivax, VK210 and VK247, with distinctive composition of the repeating units in the central region of the protein sequence (12-14). The two CSP types, VK210 and VK247, are diagnostically different in each of the four regions that make up the gene: the 5′ NR and 3′ NR terminal regions, the CR region, and the IR between CR and 3′ NR.
There is very little synonymous variation among the strains: only four segregating sites in the 5′ and 3′ NR regions (480 bp total) among the 26 strains of both species (Table 2). Amino acid replacements are considerably more numerous: 17 segregating sites among the 26 strains. Rich et al. (ref. 16; see also ref. 19) found a similar situation in the Csp gene of P. falciparum: no silent polymorphisms but 24 amino acid polymorphisms among 25 strains. These authors (16, 19) found, moreover, no silent variation in nine other gene loci of P. falciparum and favored as an explanation a recent bottleneck, so that the current distribution of P. falciparum throughout the world tropics would have come about by a recent expansion from one or very few propagules, probably within the last few thousand years and probably from sub-Saharan Africa. This explanation has been considerably reinforced in additional studies showing scarce nucleotide variation at synonymous sites in 10 other nuclear gene loci considered candidates for antimalarial vaccines (17), in 20 additional protein coding genes (18), in 25 introns (20), and in the mitochondrial genome (21, 22).
Consistent with the scarce synonymous variation in the Csp of P. vivax (Table 2), Leclerc et al. (8) found no variation at all in 12 microsatellite loci (and limited variation at a 13th locus) in 108 samples of P. vivax from eight localities in Asia, Africa, South America, and New Guinea; even less microsatellite variation than in P. falciparum (23), although microsatellite polymorphisms arise at high rates by replication spillage. There is also little polymorphism in 8 tandem repeats (TR) derived from 96 samples of P. vivax from eight localities, even though the TRs are located on a 100-kb DNA segment, previously described as highly polymorphic, from chromosome 3, where the Csp locus is located (8). Considerable polymorphism has been observed in P. vivax genes involved in drug resistance or coding for surface proteins (10, 12, 24-28). However, these polymorphisms are likely due to natural selection.
The scarcity in P. vivax of synonymous polymorphism at the Csp locus and in the microsatellite loci (8), as well as other neutral or nearly neutral sites, can be accounted most likely (16) by (i) a selective sweep or (ii) a recent demographic expansion. Directional selection favoring a particular allele will reduce or eliminate genetic polymorphism around the selective site (8, 29-31). If the selective sweep is recent and strong, a large region around the site will become genetically impoverished. The virtual absence of polymorphism at 12 microsatellite loci (8) could possibly be due to each locus being closely linked to a gene (site) recently subject to a selective sweep. There is no convincing direct evidence supporting such multiple recent selection events.
A recent demographic expansion (ii above) seems more likely. This recent demographic expansion could occur if a widely distributed species became at some point drastically reduced by a severe bottleneck and then expanded again; or because of the worldwide expansion of a new species or of a species that was previously restricted to a small population in some isolated locality (8, 16, 19). It seems unlikely that a severe population bottleneck would occur in a species such as P. vivax that consists of billions of parasites in each of millions of hosts. The worldwide expansion could be recent, however, if the species would have recently originated, which would be the case if P. vivax would have recently become a human parasite derived from P. simium, by host transfer from a platyrrhine monkey. Alternatively, P. vivax could have been geographically very restricted and expanded recently as a consequence of climatic or ecological changes and/or changes in the vectors or human hosts. This alternative is favored in the case of P. falciparum (16, 32, 33).
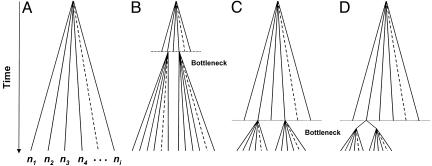
Is the divergence between the two VK types of Csp sequences ancient? Fig. 6 outlines four possible hypotheses. We will first consider the 5′ and 3′ NR regions. Fig. 6A is the general model of coalescent evolution, according to which neutral mutations accumulate over time at a rate determined by the rate of mutation and the time elapsed since the most recent common ancestor of the current sequences (34). In Fig. 6B, an ancient bottleneck is survived by only two strains, from which all current strains derive. But considerable neutral (synonymous) polymorphism would have accumulated since the bottleneck in each strain type, which is not the case. In Fig. 6C, the bottleneck is recent, and only two anciently diverged strains survive. Fig. 6C is consistent with substantial amino acid differentiation between the two VK types (as occurs particularly in the CR region) and can account for the scarcity of synonymous polymorphisms within each VK type. But Fig. 6C implies that there also be considerable synonymous differentiation between the two strain types, which is contradicted by our observations. In Fig. 6D, the divergence of the two strain types is recent. Fig. 6D is consistent with the scarcity of synonymous polymorphisms within and between the two strain types; their amino acid differences would have arisen by selection in response to the host's immune system.
Fig. 6.
Diagrams of neutral sequence evolution. (A) The general coalescence model. (B-D) Three possible hypotheses to account for the evolution of the two VK types. Only D is consistent with the data.
The data for the IR and CR regions are also consistent with Fig. 6D. In the IR region, there are not synonymous substitutions among the sequences of a given strain type and only one (at site 241) between the two types. There are 37.9 motifs per strain on average in the CR region (Table 5) and 986 motifs in total for the 26 strains. The total number of different amino acid motifs (counting 4-aa and 5-aa motifs in both VK210 and VK247) with frequency >0.01 is 10. Thus, one single ancestral strain with 38 motifs (the average number) could have carried all of the motifs with frequency >0.01; motifs with lower frequencies can arise by mutation. Rich et al. (15) called “repeat allotypes” (RATs) to the distinct nucleotide sequences that encode the amino acid motifs. Tables 6 and 7, which are published as supporting information on the PNAS web site, show that in our full sample, including 4-aa and 5-aa motifs in VK210 and VK247, there are 17 RATs with frequency >0.01. All these different RATs could also have been carried by one single ancestral strain of P. vivax with 38 motifs. After the bottleneck, natural selection could have segregated the motifs into the two VK types. Intragenic recombination, in addition to mutation, could have generated some of the motifs and RATs. In sequences with repetitive components, the frequency of intragenic recombination (unequal crossing over) occurs at frequencies several orders of magnitude greater than mutation.
It is possible that one of the two VK types may have been ancestral to the other and that the 4-aa motifs and the 5-aa motifs may have arisen one from the other. The ancestral motifs would be expected to have accumulated more synonymous mutations; that is, the older motifs would be encoded by a larger number of RATs. Within our sample of 26 sequences, there are not obvious differences in the number of RATs per motif between the two VK types (after correcting for the larger number of VK210 sequences). However, three of the 4-aa motifs, GDGA, ANGA, and GDRA, are each encoded, respectively, by three, five, and nine different RATs, whereas none of the 5-aa motifs is encoded by more than two RATs (Tables 6 and 7). Whether this difference is because the 4-aa motifs are ancestral might be assessed by investigating additional Csp sequences.
The VK210 and VK247 types do not have discrete geographic distributions (Table 1). At least at a macroscale, the two VK types appear to have worldwide distribution. This distribution is not consistent with the conclusion of Li et al. (35) that P. vivax parasites from Central and South America form a group distinct from those of Asia. The worldwide distribution of both VK types has been noted earlier (13). Moreover, Gonzalez et al. (14) found both types in localities from Colombia, South America. Cui et al. (36) also found both types in samples from a single locality, Mae Sod in Thailand.
Escalante et al. (3) noted the genetic identity between P. vivax and P. simium at the Csp locus and concluded that a recent host transfer had occurred between humans and New World monkeys. The genetic indistinguishability of the two species has been confirmed at 13 microsatellite loci and eight tandem repeats (8). The genetic identity of P. vivax and P. simium at rapidly evolving microsatellite loci confirms that the host transfer event must have occurred very recently in the evolutionary scale. The data reported in the present paper indicate that, moreover, the host transfer between humans and monkeys must have occurred twice, because both VK types are present in both species. Alternatively, one VK type might have evolved from the other independently in the two species. This explanation requires that the three diagnostic differences in the nonrepetitive regions of the Csp gene (Table 1) would be distinctly and jointly favored by natural selection. This improbable conjecture becomes even more implausible when examining the variant RATs of the amino acid motifs. P. vivax and P. simium are indistinguishable not only in the amino acid composition of motifs but also with respect to the silent nucleotide positions in the codons (Tables 6 and 7).
These observations favor a host switch between humans and monkeys, and also that the host switch has occurred recently and more than once. Determining the direction of the host switch, whether human to monkey or monkey to human, is relevant not only for unraveling the evolution of Plasmodium species but also for understanding the origins and expansion of P. vivax malaria in human populations. Humans and platyrrhine monkeys are distantly related (their most recent common ancestor lived some 35-40 million years ago) and have been geographically associated only after the first human colonization of the Americas, which occurred within the last 15,000 years. The host switch might, in fact, have occurred after the second influx of humans after the European colonizations of America in the 16th century. Whether 500 or 15,000 years have passed since the host switch, it would be a mere moment in evolutionary time, and so it is not surprising that P. vivax and P. simium are genetically so little diverged. Four considerations, described in the Supporting Information, favor a host transfer from humans to monkeys, whereas three considerations favor a transfer from monkey to human hosts (see also refs. 37 and 38). The matter can be resolved by comparing the genetic diversity of the human and primate parasites. If the transfer has been recently from human to monkeys, the amount of genetic diversity in silent nucleotide sites (and other neutral polymorphisms) will be greater in P. vivax than in P. simium. A transfer from monkey to humans would be evinced by greater polymorphism in P. simium than in P. vivax.
Supplementary Material
Acknowledgments
We thank A. Escalante for the DNA of strain 15 and A. Escalante, C. Machado, F. Renaud, and S. Rich for helpful comments to improve this paper.
Author contributions: F.J.A. designed research; C.S.L. and L.T. performed research; C.S.L. and F.J.A. analyzed data; and C.S.L. and F.J.A. wrote the paper.
Abbreviations: CR, central repeat; CSP, circumsporozoite protein; IR, insertion region; NR, nonrepeat; RAT, repeat allotypes.
References
- 1.Carter, R. (2003) Trends Parasitol. 19, 214-219. [DOI] [PubMed] [Google Scholar]
- 2.Escalante, A. A. & Ayala, F. J. (1994) Proc. Natl. Acad. Sci. USA 91, 11373-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escalante, A. A., Barrio, E. & Ayala, F. J. (1995) Mol. Biol. Evol. 12, 616-626. [DOI] [PubMed] [Google Scholar]
- 4.Carlton, J. (2003) Trends Parasitol. 19, 227-231. [DOI] [PubMed] [Google Scholar]
- 5.Carter, R. & Mendis, K. N. (2002) Clin. Microbiol. Rev. 15, 564-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livingstone, F. B. (1984) Hum. Biol. 56, 413-425. [PubMed] [Google Scholar]
- 7.Escalante, A. A., Cornejo, O. E., Freeland, D. E., Poe, A. C., Durrego, E., Collins, W. E. & Lal, L. L. (2005) Proc. Natl. Acad. Sci. USA 102, 1980-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leclerc, M. C., Durand, P., Gauthier, C., Patot, S., Billotte, N., Menegon, M., Severini, C., Ayala, F. J. & Renaud, F. (2004) Proc. Natl. Acad. Sci. USA 101, 14455-14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu, J., Joy, D. A., Duan, J., Huang, Y., Carlton, J., Walker, J., Barnwell, J., Beerli, P., Charleston, M. A., Pybus, O. G., et al. (2005) Mol. Biol. Evol. 22, 1686-1693. [DOI] [PubMed] [Google Scholar]
- 10.Feng, X., Carlton, J. M., Joy, D. A., Mu, J., Furuya, T., Suh, B. B., Wang, Y., Barnwell, J. W. & Su, X.-Z. (2003) Proc. Natl. Acad. Sci. USA 100, 8502-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, L., Escalante, A. A., Imwong, M. & Snounou, G. (2003) Trends Parasitol. 19, 220-226. [DOI] [PubMed] [Google Scholar]
- 12.Rosenberg, R., Wirtz, R. A., Lanar, D. E., Sattabongkot, J., Waters, A. P. & Prasittisuk, C. (1989) Science 245, 973-976. [DOI] [PubMed] [Google Scholar]
- 13.Kain, K. C., Brown, A. E., Webster, H. K., Wirtz, R. A., Keystone, J. S., Rodriguez, M. H., Kinahan, J., Rowland, M. & Lanar, D. E. (1992) J. Clin. Microbiol. 30, 1863-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González, J. M., Hurtado, S., Arévalo-Herrera, M. & Herrera, S. (2001) Mem. Inst. Oswaldo Cruz 96, 709-712. [DOI] [PubMed] [Google Scholar]
- 15.Rich, S. M., Hudson, R. R. & Ayala, F. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13040-13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rich, S. M., Licht, M. C., Hudson, R. R. & Ayala, F. J. (1998) Proc. Natl. Acad. Sci. USA 95, 4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escalante, A. A., Lal, A. A. & Ayala, F. J. (1998) Genetics 149, 189-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl, D. L. (2004) Nat. Rev. Microbiol. 2, 15-22. [DOI] [PubMed] [Google Scholar]
- 19.Rich, S. M. & Ayala, F. J. (2000) Proc. Natl. Acad. Sci. USA 97, 6994-7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volkman, S. K., Barry, A. E., Lyons, E. J., Nielsen, K. M., Thomas, S. M., Choi, M., Thakore, S. S., Day, K. P., Wirth, D. J. & Hartl, D. L. (2001) Science 293, 482-484. [DOI] [PubMed] [Google Scholar]
- 21.Conway, D. J., Fanello, C., Lloyd, J. M., Al-Joubori, B. M., Baloch, A. H., Somanath, S. D., Roper, C., Oduola, A. M. J., Mulder, B., Povoa, M. M., et al. (2000) Mol. Biochem. Parasitol. 111, 163-171. [DOI] [PubMed] [Google Scholar]
- 22.Joy, D. A., Feng, X., Mu, J., Furuya, T., Chotivanich, K., Krettli, A. U., Ho, M., Wang, A., White, N. J., Suh, E., et al. (2003) Science 300, 318-321. [DOI] [PubMed] [Google Scholar]
- 23.Anderson, T. J. C., Haubold, B., Williams, J. T., Estrada-Franco, J. G., Richardson, L., Mollindeo, R., Bockarie, M., Mokili, J., Mharakurwa, S., French, N., et al. (2000) Mol. Biol. Evol. 17, 1467-1482. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, M. K. & Hughes, A. L. (1995) Mol. Biochem. Parasitol. 71, 99-113. [DOI] [PubMed] [Google Scholar]
- 25.Putaporntip, C., Jongtuwiwes, S., Tanabe, K. & Thaithong, S. (1997) Mol. Biochem. Parasitol. 84, 49-56. [DOI] [PubMed] [Google Scholar]
- 26.Putaporntip, C., Jongwutiwes, S., Sakihama, N., Ferreira, M. U., Kho, W-G., Kaneko, A., Kanbara, H., Hattori, T. & Tanabe, K. (2002) Proc. Natl. Acad. Sci. USA 99, 16348-16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman, R. E., Sithiprasasna, R., Kankaew, P., Kiattibut, C., Ratanawong, S., Khuntirat, B. & Sattabongkot, J. (2002) J. Med. Entomol. 39, 556-559. [DOI] [PubMed] [Google Scholar]
- 28.Figtree, M., Pasay, C. J., Slade, R., Cheng, Q., Cloonan, N., Walker, J. & Saul, A. (2000) Mol. Biochem. Parasitol. 108, 53-66. [DOI] [PubMed] [Google Scholar]
- 29.Hudson, R. R., Bailey, K., Skarecky, D., Kwiatowski, J. & Ayala, F. J. (1994) Genetics 136, 1329-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson, R. R., Sáez, A. G. & Ayala, F. J. (1997) Proc. Natl. Acad. Sci. USA 94, 7725-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sáez, A. G., Tatarenkov, A., Barrio, E., Becerra, N. H. & Ayala, F. J. (2003) Proc. Natl. Acad. Sci. USA 100, 1793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayala, F. J., Escalante, A., Lal, A. & Rich, S. (1998) in Malaria: Parasite Biology, Pathogenesis, and Protection, ed. Sherman, I. W. (Am. Soc. Microbiol., Washington, DC), pp. 285-300.
- 33.Ayala, F. J., Escalante, A. A. & Rich, S. M. (1999) Parassitologia (Rome) 41, 55-68. [PubMed] [Google Scholar]
- 34.Slatkin, M. & Hudson, R. R. (1991) Genetics 129, 555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, J., Collins, W. E., Wirtz, R. A., Rathore, D., Lal, A. & McCutchan, T. F. (2001) Emerging Infect. Dis. 7, 35-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui, L., Mascorro, C. N., Fan, Q., Rzomp, K. A., Khuntirat, B., Zhou, G., Chen, H., Yan, G. & Sattabongkot, J. (2003) Am J. Trop. Med. Hyg. 68, 613-619. [DOI] [PubMed] [Google Scholar]
- 37.Escalante, A. A. & Ayala, F. J. (1995) Proc. Natl. Acad. Sci. USA 92, 5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rich, S. M. & Ayala, F. J. (2004) in Infectious Disease and Host-Pathogen Evolution, ed. Dronamraju, K. R. (Cambridge Univ. Press, Cambridge, U.K.), pp. 39-74.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.