Abstract
Several protein kinases from diverse eukaryotes known to perform important roles in DNA repair have also been shown to play critical roles in telomere maintenance. Here, we report that the human telomere-associated protein TRF2 is rapidly phosphorylated in response to DNA damage. We find that the phosphorylated form of TRF2 is not bound to telomeric DNA, as is the ground form of TRF2, and is rapidly localized to damage sites. Our results suggest that the ataxia-telangiectasia-mutated (ATM) protein kinase signal-transduction pathway is primarily responsible for the DNA damage-induced phosphorylation of TRF2. Unlike DNA damage-induced phosphorylation of other ATM targets, the phosphorylated form of TRF2 is transient, being detected rapidly at DNA damage sites postirradiation, but largely dissipated by 2 hours. In addition, we report that the phosphorylated form of TRF2 is present at telomeres in cell types undergoing telomere-based crisis and a recombination-driven, telomerase-independent, alternative lengthening of telomeres (ALT) pathway, likely as a consequence of a telomere-based DNA damage response. Our results link the induction of TRF2 phosphorylation to the DNA damage-response system, providing an example of direct cross-talk via a signaling pathway between these two major cellular processes essential for genomic stability, telomere maintenance, and DNA repair.
Telomeres are specialized DNA/protein structural protective caps that prevent chromosome ends from being treated as double-stand breaks (DSBs), thereby functioning to prevent end fusions (1–3). Painstaking maintenance of the telomere is essential, because dysfunction at the telomere is likely a key driving force behind the genomic instability observed in early cancer lesions and age-related disorders (4–6). An increasing number of proteins, specifically protein kinases of the phosphatidylinositol-3 kinase related kinase (PIKK) family, originally known to act in maintaining genomic stability via DNA repair pathways, have been shown to function in telomere maintenance (7). Additionally, DNA damage-response proteins localize in cellular senescence-associated DNA damage foci (8, 9) and telomere-dysfunction-induced foci created by inhibition of TRF2 (10). Investigations into mechanisms of direct communication between these two major cellular processes essential to safeguard genomic integrity, DNA repair, and telomere maintenance are actively being pursued.
A critical component of mammalian telomere maintenance involves the correct tissue-specific length of telomeric DNA (1). However, the proper regulation of the proteinaceous telomere cap must be maintained with its own set of unique tissue and developmental complexity (2, 3, 11). Telomere-associated proteins, such as TRF1 and TRF2, can bind telomeric DNA directly or can localize to the telomere through protein–protein interactions with telomere-repeat-binding proteins (2, 3, 12–14). Adding an additional layer of complexity, human telomeres end in a 3′ G-rich single-strand overhang, consisting of several hundred nucleotides that apparently displaces one strand of the telomeric repeat and hybridizes to its complementary sequence (15). The resulting structure of a large duplex loop, called the t-loop, contains the folded DNA and associated proteins, particularly TRF2, which is thought to bind and stabilize the t-loop junction (15). Implicating TRF2 in a new functional role as an early component of the DNA repair response system, it was recently shown that TRF2 migrates rapidly to specific sites of DNA damage caused by DSBs introduced by laser microbeam irradiation (16).
Reports that the PIKKs, ataxia-telangiectasia-mutated (ATM), and DNA-dependent protein kinase catalytic subunits (DNA-PKcs) are critical for mammalian telomere capping (17–23) led us to test whether specific telomere-associated proteins are targets for these kinases. Here, we report that TRF2 is rapidly phosphorylated in response to DNA damage, likely as a result of an ATM-kinase-mediated pathway. The phosphorylated form of TRF2 is not bound to telomeres, in contrast to the ground form of TRF2, and rapidly localizes to damage sites. Furthermore, we report that the phosphorylated form of TRF2 is found at telomeres in cell types undergoing telomere-based crisis and a recombination-driven, telomerase-independent, alternative lengthening of telomeres (ALT) pathway (24–27).
Materials and Methods
Cells. Human BJ, BJ E6/E7, and VA13 cells were kindly provided by Woodring Wright, University of Texas Southwestern Medical Center, Dallas. Human MO59J and MO59K cells were generous gifts from Janice M. Pluth, Lawrence Berkeley National Laboratory, Berkeley, CA. A-T primary fibroblasts (GM05823 and GM03487) were obtained from Coriell Cell Repository (Camden, NJ).
In Vivo Labeling. In vivo labeling experiments were performed as described in ref. 28.
Induction of DNA Damage. Cells were irradiated with 5 Gy or 20 Gy of 250-kVp x-rays at a dose rate of 45 cGy/min on ice. Cells were harvested after 30-min incubation at 37°C, except in the case of time courses where they were harvested at time points ranging from 30 min to 48 h. Drug treatment of cells was carried out by the addition of 10 μg/ml or 50 μg/ml etoposide (Sigma) to the culture media for 1 h. Mock-irradiated controls were incubated for corresponding times on ice.
Antibodies. Anti-TRF2 Thr-188P polyclonal antibody was generated by immunizing rabbits with KLH-conjugated phosphopeptide CSKDPTT[PO3]QKLR and affinity-purification (AnaSpec, San Jose, CA). The following antibodies were used for immunoblotting: anti-TRF2 (4A794, Imgenex, Sorrento Valley, CA, IMG-124), anti-TRF1 (Sigma, TRF-78), anti-p53 (DO-1) (D0104, Santa Cruz Biotechnology), anti-phospho-p53 (Ser-15) (9284, Cell Signaling Technology, Beverly, MA), anti-H2A (2572, Cell Signaling Technology), anti-phospho-H2AX (Ser-139) (26079 and 26513, Upstate Biotechnology, Lake Placid, NY), and β-tubulin (D-10) (A2104, Santa Cruz Biotechnology).
Two-Dimensional PAGE and Immunoblotting. Cells were lysed by incubation at room temperature for 15 min in a sample buffer (7 M urea/2 M thiourea/1% C7BzO/40 mM Tris, pH 7.5/1 mM Na3VO4/10 mM NaF). Cell lysates were centrifuged at 15,000 × g for 30 min at 4°C. The supernatant was snap-frozen in liquid nitrogen and stored at –80°C. Protein concentrations were determined by using a protein-assay system (Bio-Rad), with BSA as a standard.
For two-dimensional PAGE, cell extracts (150 μg) were run on ReadyStrip immobilized pH gradient (IPG) strip (7 cm), ranging from pH 7 to 10, by using a PROTEAN IEF Cell (Bio-Rad). After focusing, the strip was equilibrated and placed on top of a 4–20% Tris-Glycine ZOOM gel (Invitrogen), electrophoresed for protein separation, and blotted to membrane for immunoblot analysis. For immunoblotting, proteins (30 μg) were separated by SDS/PAGE and transferred to a PVDF membrane (Bio-Rad). Blots were blocked in 1% blocking solution (Roche Diagnostics) for 30 min and incubated with primary antibodies overnight, followed by goat anti-mouse IgG-HRP (1:3,000, Santa Cruz Biotechnology) or goat anti-rabbit IgG-HRP (1:4,000, Santa Cruz Biotechnology) for 1 h. Secondary antibodies were detected by enhanced chemiluminescence (ECL) reagent (Amersham Pharmacia).
Immunofluorescence. Cells were fixed in cold methanol for 20 min, permeabilized for 10 min in 0.5% Triton X-100 on ice, and blocked in 5% BSA for 20 min at room temperature. The slides were incubated with anti-TRF2 (1:200) and anti-TRF2 Thr-188P (1:200) antibodies for 1 h, washed in PBS, and incubated with rhodamine red-X goat anti-mouse secondary antibodies (1:1,000, Molecular Probes) and Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:1,000, Molecular Probes) for 1 h at room temperature. Cells were washed in PBS and mounted by using VECTASHIELD mounting medium with DAPI (Vector Laboratories). Fluorescence images were captured by using a Leica DM5000B microscope equipped with a charge-coupled-device camera and spot software (Diagnostic Instruments, Sterling Heights, MI).
Metaphase Chromosomal Spread and FISH. Preparation of metaphase chromosomes from growing cell cultures was carried out by standard methods. FISH with a Cy3-labeled (CCCTAA)3 probe (Applied Biosystems) was performed as described in ref. 29.
Chromatin Extraction. HT1080 cells were harvested after 1-h etoposide treatment. Whole-cell extracts were prepared with various concentrations of KCl (50, 150, 300, and 450 mM) in lysis buffer [20 mM Hepes-KOH, pH 7.9/25% glycerol/0.1 mM EDTA/5 mM MgCl2/0.25% Nonidet P-40/1 mM DTT/protease-inhibitor mixture (Roche Diagnostics)/1 mM Na3VO4/10 mM NaF]. The crude extract was centrifuged at 14,000 × g for 20 min at 4°C, and 30 μg of the supernatant protein extract was separated by PAGE and used for immunoblot analysis as described above. To isolate chromatin, the pellets were resuspended in 1× SDS sample buffer (2% SDS/10% glycerol/50 mM Tris·HCl, pH 6.8/0.86 M mercaptoethanol/0.01% bromophenol blue), sonicated with 10 2-sec bursts by using the Sonic Dismembrater (model 100, Fisher Scientific) and centrifuged at 14,000 × g for 20 min at 4°C; 30 μg of this supernatant protein extract was separated by PAGE and used for immunoblot analysis.
Photoinduction of DNA DSBs. Laser microbeam irradiation was performed as described by Bradshaw et al. (16). Immediately after irradiation, cells were fixed in 2% paraformaldehyde and processed for immunofluorescence. Immunostaining for TRF2-Thr188P and TRF2 was carried out by using primary antibodies at dilutions of 1:400 and 1:100, respectively. DNA breaks in irradiated cells were labeled with Cy3-dCTP by using a published TUNEL protocol (Roche Diagnostics). Images of fixed cells were obtained as described in ref. 16.
Results and Discussion
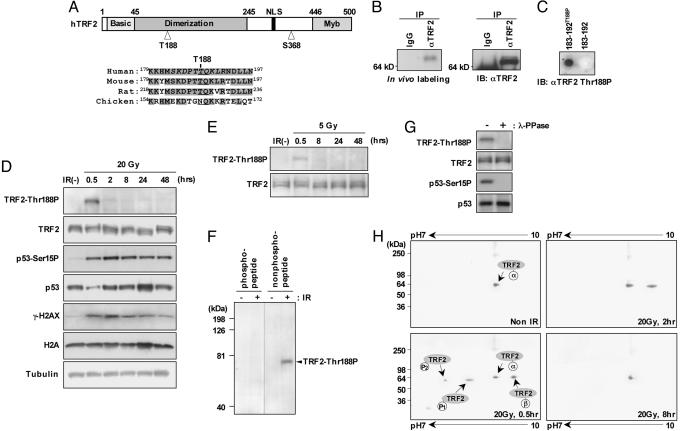
Human TRF2 contains two highly conserved PIKK phosphorylation sites at Thr-188 and Ser-368 (30) (Fig. 1A). In initial in vivo labeling experiments using whole HeLa cells labeled with [32P]orthophosphate, we found a fraction of TRF2 was phosphorylated (Fig. 1B). However, exposure of cells to radioactive [32P]orthophosphate used for conventional in vivo labeling experiments is known to induce DNA damage (31). To investigate whether phosphorylation of TRF2 was DNA damage-dependent, we generated a rabbit polyclonal antibody (anti-TRF2 Thr188P) against a synthetic polypeptide consisting of TRF2 amino acid residues 183–192, with a centrally positioned phosphothreonine-188. The purified anti-TRF2 Thr188P antibody reacted specifically with the immunizing polypeptide phosphorylated at Thr-188 (T-188) but not with the nonphosphorylated peptide (Fig. 1C). When using the anti-TRF2 Thr188P antibody, we did not detect a T-188 phosphorylated form of TRF2 (TRF2-Thr188P) in immunoblots of nuclear extracts from unirradiated HT1080 cells (Fig. 1D). However, TRF2-Thr188P was readily detected 30 min after exposure to 20 Gy of x-rays, as were p53-Ser15P and γ-H2AX, two phosphorylation targets of ATM (Fig. 1D). In a dose-dependent manner, TRF2-Thr188P was also detected at reduced levels 30 min after exposure to lower levels of x-rays (5 Gy) (Fig. 1E). The majority of TRF2-Thr188P was gone by 2 h postirradiation, in marked contrast to the persistence of p53-Ser15P and γ-H2AX (Fig. 1D). We found that the anti-TRF2 Thr188P antibody was immunodepleted during immunoblot analysis when the blotting solution was incubated with the immunizing polypeptide phosphorylated at Thr-188 but not with the nonphosphorylated peptide (Fig. 1F). In addition, phosphatase treatment by λ phosphatase (λ-PPase) eliminated the band detected by the phosphospecific TRF2 antibody (Fig. 1G). Taken together, these results demonstrate that the anti-TRF2 Thr188P antibody is specific for the T-188 phosphorylated form of TRF2.
Fig. 1.
TRF2 is phosphorylated at T-188 after exposure to IR. (A) Schematic diagram of human TRF2 (hTRF2) with conserved PIKK target sequences at T-188. TRF2 T-188, along with adjacent amino acids, is highly conserved in mammalian homologues (shaded). The synthetic phosphorylated polypeptide sequence used as an antigen to generate the anti-TRF2 Thr188P is shown in italics. (B) In vivo labeling. HeLa cells were labeled by [32P]orthophosphate, and TRF2 proteins were immunoprecipitated with anti-TRF2 antibody and separated by SDS/PAGE, followed by autoradiography (Left), and the same membrane was immunoblotted with anti-TRF2 antibody (Right). (C) Anti-TRF2 Thr188P antibody was used to immunoblot synthetic peptides (1 μg) comprised of 10 amino acids of TRF2 with (183–192Thr188P) or without (183–192) phosphorylation at T-188. (D and E) The human fibrosarcoma cell line HT1080 was either mock-irradiated [IR(–)] or irradiated with 20 Gy (D) or 5 Gy(E) of x-rays and harvested at time points ranging from 0.5 to 48 h, as indicated. Whole-cell extracts were prepared and analyzed by immunoblot with anti-TRF2 Thr188P, anti-TRF2 (IMG-124 that recognizes all known forms of TRF2), anti-p53 Ser15P, anti-p53, anti-γ-H2AX, and anti-H2A antibodies. For a loading control, the same membranes were stripped and incubated with anti-β tubulin. (F) HT1080 cells were irradiated with (+) or without (–) 20 Gy and harvested. Immunoblots were incubated with anti-TRF2 T188P antibody (1 μg/ml) in the presence of the immunizing polypeptide phosphorylated at T-188 or nonphosphorylated peptide at a final concentration of 0.5 μg/ml. (G) HT1080 cells were irradiated with 20 Gy (+) and harvested after 30 min. Cell lysates were incubated with or without λ phosphatase (λ-PPase), separated by SDS/PAGE, and probed with anti-TRF2 Thr188P, anti-TRF2, anti-p53 Ser15P, and anti-p53. (H) HT1080 cells were untreated (Non IR) or irradiated with 20 Gy and harvested at time points of 0.5, 2, and 8 h, as indicated. Whole-cell extracts were subjected to isoelectric focusing (pH range, 7–10), SDS/PAGE, and immunoblotting with anti-TRF2 antibody. Here, we refer to the ground state of TRF2 as TRF2α, being present as one major spot without treatment or 8 h after IR. Within 0.5 h after IR exposure, the isoelectric points of TRF2-modified forms shifted ≈0.5 pH unit (TRF2-P1) and ≈1 pH unit (TRF2-P2) toward the acidic direction, as analyzed by two-dimensional PAGE. In addition, TRF2β is present 0.5 and 2 h after IR exposure.
To further investigate the nature of the DNA damage-induced modification of TRF2, we performed two-dimensional PAGE with isoelectric point focusing followed by immunoblotting with anti-TRF2 antibody IMG-124, which recognizes all known forms of TRF2. The addition of a single phosphate group theoretically shifts the isoelectric point of a protein ≈0.5 pH units toward the acidic pole (32). Detection of TRF2 from lysates of mock-irradiated HT1080 cells showed a single protein spot, which we refer to here as TRF2α, focusing at an isoelectric point of ≈pH 9 (Fig. 1H, the theoretical isoelectric point of TRF2 is pH 9.22). Confirming our previous immunoblot results (Fig. 1D), we found that two TRF2-modified forms appear postirradiation with isoelectric points shifted ≈0.5 pH unit (TRF2-P1) and ≈1.0 pH unit (TRF2-P2) toward the acidic pole with the same kinetics of appearance (0.5 h; Fig. 1H) and disappearance (2 h; Fig. 1H) of TRF2 phosphorylation. TRF2-P1 and TRF2-P2 are eliminated after phosphatase treatment by λ phosphatase (λ-PPase), signifying that these TRF2 species are modified by phosphorylation (data not shown). Therefore, one or two phosphate groups are rapidly added to TRF2 in response to ionizing radiation (IR). In addition, an unexpected and unidentified TRF2 species, referred to here as TRF2β, shifted ≈0.5 pH unit toward the basic pole. TRF2β appears rapidly (0.5 h after IR exposure; Fig. 1H) with similar kinetics to the two TRF2-phosphorylated forms but persists past 2 h (Fig. 1H) before disappearing by 8 h (Fig. 1H).
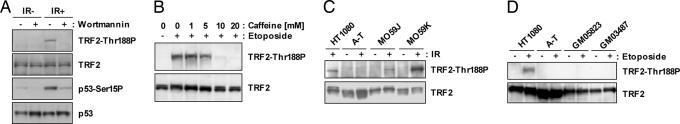
To determine which protein kinase is responsible for DNA damage-induced phosphorylation of TRF2 at T-188, we measured the effect of the PIKK inhibitors wortmannin and caffeine on DNA damage-induced phosphorylation of TRF2. Pretreatment of HT1080 cells with either 20 μM wortmannin or 10 mM caffeine did not affect overall levels of TRF2 protein but blocked measurable TRF2 phosphorylation at Thr-188 by either 20 Gy IR or 10 μg/ml etoposide, a DSB-inducing chemotherapeutic compound (33) (Fig. 2 A and B and data not shown). Because 20 μM wortmannin selectively inhibits DNA-PKcs and ATM but not ATR (34–36), whereas 10 mM caffeine inhibits ATM and ATR but not DNA-PKcs (37), these results suggest that DNA damage-induced phosphorylation of TRF2 at residue T-188 is primarily mediated by ATM. To test this hypothesis, we studied DNA damage-induced phosphorylation of TRF2 in ataxia-telangiectasia fibroblast cells (A-T) deficient in ATM but not DNA-PKcs. We found both IR- and etoposide-induced TRF2 phosphorylation at T-188 to be absent in an A-T cell line (Fig. 2 C and D; data not shown) and two primary A-T cells (Fig. 2D). We also examined the human glioma cell line MO59J, which lacks DNA-PKcs and has reduced activity of ATM and MO59K, a cell line derived from the same tumor that expresses wild-type levels of DNA-PKcs and ATM proteins (38). MO59J showed reduced IR- or etoposide-induced phosphorylation of TRF2 at Thr-188, whereas MO59K showed robust IR-induced T-188 TRF2 phosphorylation (Fig. 2C and data not shown). Taken together, our findings suggest that, under these particular DNA damage-induction conditions, phosphorylation of TRF2 at T-188 by the DSB-inducing agents IR and etoposide depends primarily on the ATM damage-response pathway.
Fig. 2.
Phosphorylation at T-188 is mediated by an ATM kinase signal-transduction pathway. (A) HT1080 cells were preincubated with and without 20 μM wortmannin for 30 min before treatment with 20 Gy of IR (IR+) or without IR (IR–). Whole-cell extracts were prepared 30 min after IR and subsequently analyzed with anti-TRF2 Thr188P, anti-TRF2, anti-p53 Ser15P, and anti-p53 antibodies. (B) HT1080 cells were treated for 1 h with 10 μg/ml etoposide, which induces DSBs, with increasing concentrations of caffeine (1–20 mM). Cell extracts were analyzed by immunoblotting with anti-TRF2 Thr188P or anti-TRF2 antibodies. (C) HT1080, A-T (GM05849), MO59J, and MO59K cells were left untreated (–) or irradiated with 20 Gy (+) and harvested after 30 min. Cell lysates were separated by SDS/PAGE and probed with anti-TRF2 Thr188P or anti-TRF2 antibodies. (D) HT1080, A-T (GM05849), primary A-T fibroblast (GM05823 and GM03487) cells were treated for 1 h with 50 μg/ml etoposide, and immunoblotting was performed with anti-TRF2 Thr188P or anti-TRF2 antibodies.
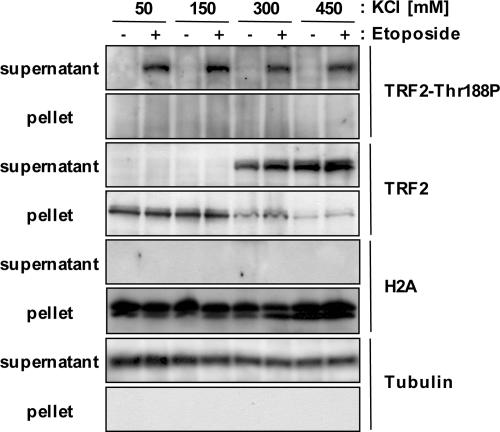
We next tested whether TRF2-Thr188P is bound to telomeric DNA as the ground form of TRF2 (TRF2α) (39, 40). HT1080 cells were either treated with etoposide (50 μg/ml) or untreated, and whole-cell lysates were extracted by using buffers containing 50–450 mM KCl (Fig. 3). Extracts were centrifuged at 14,000 × g to pellet chromatin, and supernatants were resolved by PAGE and examined by immunoblot analysis (Fig. 3). As predicted, β-tubulin fractionates with the supernatant, and the histone protein H2A fractionates with the pellets, both fractionations of these two control proteins being independent of KCl concentration (Fig. 3). As previously shown, TRF2α does not dissociate from chromosomal DNA until buffer concentration reaches 300 mM KCl (41) (Fig. 3). In contrast, TRF2-Thr188P is present in the supernatant regardless of KCl concentration (Fig. 3), indicating that TRF2-Thr188P is not bound to telomeric DNA as the bulk unphosphorylated form of TRF2.
Fig. 3.
TRF2 phosphorylated at T-188 is not bound to telomeric DNA. HT1080 cells were either untreated (–) or treated (+) with 50 μg/ml etoposide. Cells were lysed with various concentrations of KCl (50, 150, 300, and 450 mM), as indicated. After centrifugation, 30 μg of the supernatant was used for immunoblotting (supernatant: total-cell extract fraction). The pellets were resuspended in 1× SDS sample buffer, and 30 μg were used for immunoblot analysis (pellet: chromatin-bound proteins fraction). Antibodies used for immunoblotting were anti-TRF2, anti-TRF2 T188P, anti-H2A, and anti-β tubulin. Note: Bands representing the phosphorylated TRF form at T-188 are present upon longer exposure in eptopside-treated lanes at 50 and 150 mM KCl in the supernatant fraction probed with anti-TRF2 antibody (data not shown).
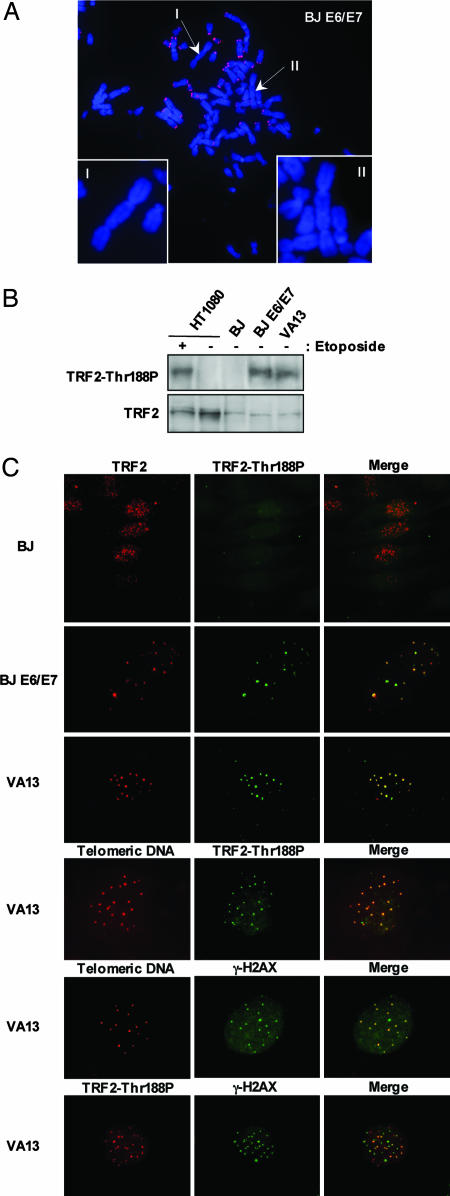
We next studied TRF2-Thr188P expression in untreated primary BJ fibroblasts, late passage BJ E6/E7 fibroblasts (PD86), and VA13 SV40-transformed fibroblasts. Expression of the papilloma virus proteins E6 and E7 allows BJ E6/E7 cells to proliferate in the presence of telomeres that otherwise would be critically short (24, 42), whereas VA13 cells maintain their telomeres by the recombination-driven, telomerase-independent, ALT pathway (25–27). Some telomeres in late-passage BJ E6/E7 cells (Fig. 4A) and in VA13 cells appear to be dysfunctional, because they participate in telomere–telomere fusion and attract γ-H2AX and other DNA damage-response proteins (2, 24, 27, 42)
Fig. 4.
TRF2 phosphorylated at T-188 localizes to telomeres in crisis and ALT cells without DNA-damage induction. (A) Representative chromosome spread of BJ E6/E7 cells (PD 86). Telomeric FISH was performed with Cy3-[CCCTAA]3 PNA probe (red). Chromosomes were stained with DAPI (blue). Arrows and Insets (I and II) show examples of end-to-end fusion without positive Cy3-[CCCTAA]3 PNA telomeric probe staining. (B) Whole-cell lysates of HT1080, BJ (60 PDs), BJ E6/E7 (PD 86), and VA13 cells were analyzed for immunoblotting. Blotting was performed with either anti-TRF2 Thr188P antibody (TRF2-Thr188P) or anti-TRF2 IMG-124 (TRF2). (C) Primary normal fibroblast BJ cells, human papiloma virus E6/E7-infected BJ cells in crisis (BJ E6/E7, PD 86), and SV40-transformed cell-line VA13 cells were stained with a monoclonal antibody recognizing TRF2 (IMG-124, red) or anti-TRF2 Thr188P antibody (green). Colocalization of anti-TRF2 (IMG-124, red) and anti-TRF2 Thr188P (green) antibodies resulting in colocalization is depicted in yellow. VA13 cells were also coimmunostained with anti-TRF2 Thr188P (red) and with a Cy3-[CCCTAA]3 telomere-PNA probe (red) or with an anti-γ-H2AX (Upstate Biotechnology, polyclonal, green) and with a Cy3-[CCCTAA]3 telomere-PNA probe (red). In addition, immunostaining was performed simultaneously with VA13 cells by using anti-TRF2 Thr188P (red) and anti-γ-H2AX (Upstate Biotechnology, monoclonal, green) antibodies.
Immunoblot analysis demonstrated the presence of TRF2-Thr188P in extracts from untreated BJ E6/E7 cells (PD86) and VA13 cells but not in extracts from untreated HT1080 cells, which required exposure to either etoposide or IR to induce detectable amounts of TRF2-Thr-188 (Fig. 4 B and D). Immunostaining with the anti-TRF2 IMG-124 antibody revealed characteristic telomere-associated punctate staining patterns in BJ, BJ E6/E7, and VA13 cells (Fig. 4C, red). In contrast, the majority of untreated BJ and HT1080 cells showed only weak, background immunostaining with anti-TRF2 Thr188P (Fig. 4C, data not shown). The intensity of anti-TRF2 Thr188P staining increased in HT1080 and BJ cells after exposure to IR or etoposide, but the staining lacked a punctate, telomeric pattern (data not shown), consistent with our results that TRF2-Thr188P is not bound to telomeric DNA (Fig. 3). Unlike BJ and HT1080 cells, 2–4% of BJ E6/E7 cells and 5–10% of VA13 cells contained multiple bright anti-TRF2 Thr188P foci (Fig. 4C, green). In VA13 cells, the majority of these bright anti-TRF2 Thr188P foci colocalized with anti-γ-H2AX (both monoclonal and polyclonal forms, Upstate Biotechnology) foci and with a telomeric-repeat-specific PNA probe (Fig. 4C) and with foci of anti-TRF2 IMG-124, anti-TRF1 (TRF-78, Sigma), and anti-Tin2 (12) (data not shown). Taken together, these results suggest that TRF2-Thr188P associates only with dysfunctional telomeres that have induced a localized DNA damage response.
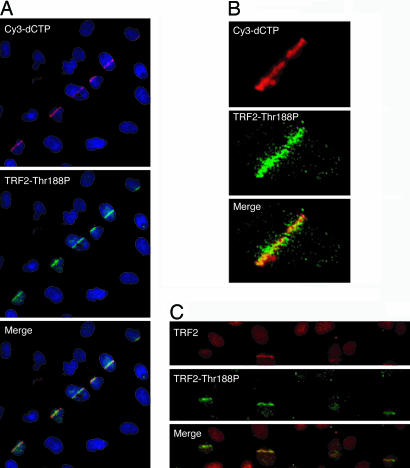
To investigate the possible association of TRF2-Thr188P with induced DSBs in nontelomeric DNA, we induced DSBs in defined nuclear regions of GM0639 human fibroblasts by sequentially exposing the cells to Hoechst 33258, a DNA intercalator, and pulsed 390-nm laser microbeam irradiation (16). Irradiated cells were fixed and analyzed for TRF2-Thr188P and total TRF2 by indirect immunofluorescence. DNA damage was monitored by TUNEL-labeling of DNA breaks with Cy3-conjugated dCTP (16). When examined ≈10 min postirradiation, anti-TRF2-Thr188P foci could be seen colocalizing with Cy3-dCTP-labeled DSBs in irradiated GM0639 fibroblasts (Fig. 5 A and B). Anti-TRF2 Thr188P foci also colocalized with anti-TRF2 IMG-124 foci at sites of photoinduced DSBs in HT1080 cells (Fig. 5C).
Fig. 5.
TRF2-Thr188P associates with photoinduced DSBs. (A) Cy3-dCTP (red) and TRF2-Thr188P (green) form overlapping tracks in nuclei of GM0639 fibroblasts fixed ≈10 min postinduction of DSBs by laser irratiation. (B) A deconvolved image of a single irradiated nucleus demonstrates spatial colocalization between foci of Cy3-dCTP-labeled DNA breaks (red) and TRF2-Thr188P (green). (C) Anti-TRF2 (IMG-124, red) and anti-TRF2 Thr188P (green) antibodies stain overlapping tracks in nuclei of HT1080 cells fixed ≈10 min after photoinduction of DSBs.
Beginning nearly a decade ago, an increasing number of proteins known to play important roles in DNA repair have been found to be critical for telomere maintenance (43). In addition, several recent studies demonstrate that proteins known to play critical roles in maintaining genomic stability via DNA repair pathways localize in cellular-senescence-associated DNA damage foci (8) and telomere-dysfunction-induced foci created by inhibition of TRF2 (10). However, whether the function of these proteins is segregated into separate DNA repair or telomere maintenance functional compartments, depending on their cellular microenvironment, has been the subject of intense investigation. Here, we report that TRF2 is phosphorylated in response to DSBs. In addition, the phosphorylated form of TRF2 is not bound to telomeric DNA, as is the ground form of TRF2 (TRF2α), and rapidly accumulates at DNA damage sites, perhaps analogous to yeast Ku-protein release from telomeres in response to DNA damage and its rapid recruitment to sites of DSBs (44). Like many proteins involved in DSB damage responses, TRF2 is phosphorylated after DSB induction. In cells exposed to etoposide or IR, our results suggest that this phosphorylation depends on ATM, a protein kinase that plays a central role in the major DSB-response network in human cells (7). Recently, it was shown that localization of TRF2 to DNA damage sites induced by photoinduced laser microbeam IR did not require the ATM kinase (16). We propose that the activation of kinases that phosphorylated TRF2 may depend on the specific sources and/or intensities of particular DNA damage agents, as has been demonstrated with other substrates and their corresponding modifying kinases (45, 46). Our results suggest that DNA damage-induced phosphorylation may shift TRF2 spatially and functionally from routine cellular telomere maintenance to a DNA damage response. This shift could occur by altering the range of DSB repair and/or telomere-associated proteins with which TRF2 interacts and/or by changing the nature of those interactions.
Our results implicate two courses of action for TRF2-Thr188P that depend on whether the cell is responding to genomic-wide or telomere-based DNA damage signals. In the case of a genomic-wide DNA repair in response to DSBs occurring throughout the genome by an exogenous source, phosphorylation of TRF2 may cause TRF2 to dissociate from telomeric DNA by disrupting the homodimer form of TRF2, or phosphorylation of TRF2 may be required for function at the damage sites. Alternatively, cells undergoing a telomere-based DNA damage response, such as cells in telomere crisis (BJ E6/E7) or using the telomerase-independent ALT pathway (VA13), we find that phosphorylated TRF2 localizes to telomeres. Phosphorylated TRF2 may be maintained at the telomere by a specific DNA damage-site protein or protein complex that is recruited to the telomere in response to the detection of DNA damage signals at the telomere. Phosphorylated TRF2 may be involved with telomere DNA repair by assisting the recruitment of additional DNA repair proteins to damaged telomeric DNA. In addition, TRF2 phosphorylation may disrupt and/or promote protein–protein interactions with TRF2 protein binding partners to promote a DNA repair response. Moreover, these findings suggest that the phosphorylated form of TRF2 may play a key role in both telomere-based and genomic-wide DNA damage signaling responses.
The exact role TRF2 may play in the DNA damage-response system remains to be determined. However, damage-induced modifications of TRF2 may yield important clues into TRF2 protein's role in a damage response. In addition, continued investigations into whether other telomere-associated protein substrates are modified by PIKKs and/or other kinase families will be critical to determine the extent of direct signaling cross-talk between telomere maintenance and DNA repair-responsive systems. It is likely that additional examples of direct cross-talk between telomere maintenance and DNA repair, two major cellular processes essential for genomic stability, will be discovered with continued work in this active area of research.
Acknowledgments
We dedicate this work to the memory of Anat Krauskopf. We thank Kenneth Cornetta, Brittney-Shea Herbert, Junya Kobayashi, and Elliot Rosen for valuable help during the preparation of this manuscript; Woodring Wright for kindly providing us with BJ, BJ E6/E7, and VA13 cell lines; Janice Pluth for kindly providing us with MO59J/K cell lines; and the Indiana Genomics Initiative (INGEN). This work was supported, in part, by National Institutes of Health/National Cancer Institute Grant R01CA090885-01A2 (to M. S. Mendonca). H.T. was supported by a Japan Society for the Promotion of Science postdoctoral fellowship. INGEN of Indiana University is supported, in part, by the Lilly Endowment. D.G. received funding from the Indiana University Cancer Center, the American Cancer Society, and an Indiana University School of Medicine Biomedical Research grant.
Author contributions: H.T., M. S. Mendonca, L.H.M., M. S. Meyn, and D.G. designed research; H.T., M. S. Mendonca, P.S.B., and D.J.H. performed research; H.T., M. S. Mendonca, P.S.B., D.J.H., L.H.M., M. S. Meyn, and D.G. contributed new reagents/analytic tools; H.T., M. S. Mendonca, P.S.B., D.J.H., L.H.M., M. S. Meyn, and D.G. analyzed data; and H.T., M. S. Meyn, and D.G. wrote the paper
Conflict of interest statement: No conflicts declared.
Abbreviations: ALT, alterative lengthening of telomeres; ATM, ataxia-telangiectasia mutated; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; DSB, double-strand break; IR, ionizing radiation; λ-PPase, λ phosphatase; PIKK, phosphatidylinositol-3 kinase related kinase.
References
- 1.Blackburn, E. H. (2001) Cell 106, 661–673. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska, A. & de Lange, T. (2004) Annu. Rev. Biochem. 73, 177–208. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira, M. G., Miller, K. M. & Cooper, J. P. (2004) Mol. Cell 13, 7–18. [DOI] [PubMed] [Google Scholar]
- 4.Sprung, C. N., Afshar, G., Chavez, E. A., Lansdorp, P., Sabatier, L. & Murnane, J. P. (1999) Mutat. Res. 429, 209–223. [DOI] [PubMed] [Google Scholar]
- 5.DePinho, R. A. (2000) Nature 408, 248–254. [DOI] [PubMed] [Google Scholar]
- 6.Feldser, D. M., Hackett, J. A. & Greider, C. W. (2003) Nat. Rev. Cancer 3, 623–627. [DOI] [PubMed] [Google Scholar]
- 7.d'Adda di Fagagna, F., Teo, S. H. & Jackson, S. P. (2004) Genes Dev. 18, 1781–1799. [DOI] [PubMed] [Google Scholar]
- 8.d'Adda di Fagagna, F., Reaper, P. M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N. P. & Jackson, S. P. (2003) Nature 426, 194–198. [DOI] [PubMed] [Google Scholar]
- 9.Sedelnikova, O. A., Horikawa, I., Zimonjic, D. B., Popescu, N. C., Bonner, W. M. & Barrett, J. C. (2004) Nat. Cell Biol. 6, 168–170. [DOI] [PubMed] [Google Scholar]
- 10.Takai, H., Smogorzewska, A. & de Lange, T. (2003) Curr. Biol. 13, 1549–1556. [DOI] [PubMed] [Google Scholar]
- 11.Greider, C. W. & Blackburn, E. H. (2004) Cell 116, S83–S86. [DOI] [PubMed] [Google Scholar]
- 12.Kim, S. H., Kaminker, P. & Campisi, J. (1999) Nat. Genet. 23, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith, S. & de Lange, T. (2000) Curr. Biol. 10, 1299–1302. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, H. L., Gilley, D., Galande, S. A., Hande, M. P., Allen, B., Kim, S. H., Li, G. C., Campisi, J., Kohwi-Shigematsu, T. & Chen, D. J. (2000) Genes Dev. 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H. & de Lange, T. (1999) Cell 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 16.Bradshaw, P. S., Stavropoulos, D. J. & Meyn, M. S. (2005) Nat. Genet. 37, 193–197. [DOI] [PubMed] [Google Scholar]
- 17.Smilenov, L. B., Morgan, S. E., Mellado, W., Sawant, S. G., Kastan, M. B. & Pandita, T. K. (1997) Oncogene 15, 2659–2665. [DOI] [PubMed] [Google Scholar]
- 18.Bailey, S. M., Meyne, J., Chen, D. J., Kurimasa, A., Li, G. C., Lehnert, B. E. & Goodwin, E. H. (1999) Proc. Natl. Acad. Sci. USA 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilley, D., Tanaka, H., Hande, M. P., Kurimasa, A., Li, G. C., Oshimura, M. & Chen, D. J. (2001) Proc. Natl. Acad. Sci. USA 98, 15084–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goytisolo, F. A., Samper, E., Edmonson, S., Taccioli, G. E. & Blasco, M. A. (2001) Mol. Cell. Biol. 21, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hande, M. P., Balajee, A. S., Tchirkov, A., Wynshaw-Boris, A. & Lansdorp, P. M. (2001) Hum. Mol. Genet. 10, 519–528. [DOI] [PubMed] [Google Scholar]
- 22.Bailey, S. M., Cornforth, M. N., Kurimasa, A., Chen, D. J. & Goodwin, E. H. (2001) Science 293, 2462–2465. [DOI] [PubMed] [Google Scholar]
- 23.Kishi, S., Zhou, X. Z., Ziv, Y., Khoo, C., Hill, D. E., Shiloh, Y. & Lu, K. P. (2001) J. Biol. Chem. 276, 29282–29291. [DOI] [PubMed] [Google Scholar]
- 24.Shay, J. W. & Wright, W. E. (2005) Carcinogenesis 26, 867–874. [DOI] [PubMed] [Google Scholar]
- 25.Murnane, J. P., Sabatier, L., Marder, B. A. & Morgan, W. F. (1994) EMBO J. 13, 4953–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddel, R. R. (2003) Cancer Lett. 194, 155–162. [DOI] [PubMed] [Google Scholar]
- 27.Nabetani, A., Yokoyama, O. & Ishikawa, F. (2004) J. Biol. Chem. 279, 25849–25857. [DOI] [PubMed] [Google Scholar]
- 28.Yannone, S. M., Roy, S., Chan, D. W., Murphy, M. B., Huang, S., Campisi, J. & Chen, D. J. (2001) J. Biol. Chem. 276, 38242–38248. [DOI] [PubMed] [Google Scholar]
- 29.Lansdorp, P. M., Verwoerd, N. P., van de Rijke, F. M., Dragowska, V., Little, M. T., Dirks, R. W., Raap, A. K. & Tanke, H. J. (1996) Hum. Mol. Genet. 5, 685–691. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S. T., Lim, D. S., Canman, C. E. & Kastan, M. B. (1999) J. Biol. Chem. 274, 37538–37543. [DOI] [PubMed] [Google Scholar]
- 31.Bond, J. A., Webley, K., Wyllie, F. S., Jones, C. J., Craig, A., Hupp, T. & Wynford-Thomas, D. (1999) Oncogene 18, 3788–3792. [DOI] [PubMed] [Google Scholar]
- 32.Firulli, B. A., Howard, M. J., McDaid, J. R., McIlreavey, L., Dionne, K. M., Centonze, V. E., Cserjesi, P., Virshup, D. M. & Firulli, A. B. (2003) Mol. Cell 12, 1225–1237. [DOI] [PubMed] [Google Scholar]
- 33.Kurz, E. U. & Lees-Miller, S. P. (2004) DNA Repair (Amsterdam) 3, 889–900. [DOI] [PubMed] [Google Scholar]
- 34.Wymann, M. P., Bulgarelli-Leva, G., Zvelebil, M. J., Pirola, L., Vanhaesebroeck, B., Waterfield, M. D. & Panayotou, G. (1996) Mol. Cell. Biol. 16, 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okayasu, R., Suetomi, K. & Ullrich, R. L. (1998) Radiat. Res. 149, 440–445. [PubMed] [Google Scholar]
- 36.Sarkaria, J. N., Tibbetts, R. S., Busby, E. C., Kennedy, A. P., Hill, D. E. & Abraham, R. T. (1998) Cancer Res. 58, 4375–4382. [PubMed] [Google Scholar]
- 37.Sarkaria, J. N., Busby, E. C., Tibbetts, R. S., Roos, P., Taya, Y., Karnitz, L. M. & Abraham, R. T. (1999) Cancer Res. 59, 4375–4382. [PubMed] [Google Scholar]
- 38.Lees-Miller, S. P., Godbout, R., Chan, D. W., Weinfeld, M., Day, R. S., III, Barron, G. M. & Allalunis-Turner, J. (1995) Science 267, 1183–1185. [DOI] [PubMed] [Google Scholar]
- 39.Broccoli, D., Smogorzewska, A., Chong, L. & de Lange, T. (1997) Nat. Genet. 17, 231–235. [DOI] [PubMed] [Google Scholar]
- 40.Bilaud, T., Brun, C., Ancelin, K., Koering, C. E., Laroche, T. & Gilson, E. (1997) Nat. Genet. 17, 236–239. [DOI] [PubMed] [Google Scholar]
- 41.van Steensel, B., Smogorzewska, A. & de Lange, T. (1998) Cell 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 42.Zou, Y., Sfeir, A., Gryaznov, S. M., Shay, J. W. & Wright, W. E. (2004) Mol. Biol. Cell 15, 3709–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boulton, S. J. & Jackson, S. P. (1996) Nucleic Acids Res. 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin, S. G., Laroche, T., Suka, N., Grunstein, M. & Gasser, S. M. (1999) Cell 97, 621–633. [DOI] [PubMed] [Google Scholar]
- 45.Bode, A. M. & Dong, Z. (2004) Nat. Rev. Cancer 4, 793–805. [DOI] [PubMed] [Google Scholar]
- 46.Kaufman, R. J. (2004) Trends Biochem. Sci. 29, 152–158. [DOI] [PubMed] [Google Scholar]