Abstract
Transcriptional regulation in the archaea involves a mosaic of DNA-binding proteins frequently (although not exclusively) of bacterial type, modulating a eukaryal-type core transcription apparatus. Methanocaldococcus jannaschii (Mja) Ptr2, a homologue of the Lrp/AsnC family of bacterial transcription regulators that are among the most widely disseminated archaeal DNA-binding proteins, has been shown to activate transcription by its conjugate hyperthermophilic RNA polymerase. Here, two in vitro systems have been exploited to show that Ptr2 and a Lrp homologue from the thermophile Methanothermococcus thermolithotrophicus (Mth) activate transcription over a ≈40°C range, in conjunction with their cognate TATA-binding proteins (TBPs) and with heterologous TBPs. A closely related homologue from the mesophile Methanococcus maripaludis (Mma) is nearly inert as a transcriptional activator, but a cluster of mutations that converts a surface patch of Mma Lrp to identity with Ptr2 confers transcriptional activity. Mja, Mth, and Mma TBPs are interchangeable for basal transcription, but their ability to support Lrp-mediated transcriptional activation varies widely, with Mja TBP the most active and Mth TBP the least active partner. The implications of this finding for understanding the roles of TBP paralogues in supporting the gene-regulatory repertoires of archaeal genomes are briefly noted.
Keywords: promoter, Ptr2, RNA polymerase, TATA-binding protein, upstream activating site
Archaeal transcription is a mosaic of components from all three domains of life. The core transcription machinery, the RNA polymerase (RNAP) and its initiation factors, are of eukaryal type: Archaeal promoters consist of a T/A-rich TATA-like element recognized by the archaeal TATA-binding proteins (TBPs), located just downstream of a binding site [the B recognition element (BRE)] for the eukaryotic TFIIB-like initiation factor TFB; TBP and TFB recruit archaeal RNAP to the promoter (just as eukaryotic TBP and TFIIB recruit RNAP II) and place it over a simple initiator element that marks the transcriptional start site (reviewed in ref. 1). For compacting their genomes and constructing their chromatin, different archaea press a great diversity of abundant general DNA-binding proteins into service: homologues of eukaryotic histones H3 and H4, homologues of bacterial HU proteins, and uniquely archaeal proteins, including the widely distributed Alba, among others (reviewed in ref. 2). On the other hand, the great majority of archaeal DNA-binding proteins with known gene-regulatory function or surmised gene-regulatory potential are of bacterial type, with only minor representation of proteins of specific archaeal type and very rare examples of proteins that are homologues of specifically eukaryotic transcriptional regulators (reviewed in ref. 3).
The Lrp/AsnC family of DNA-binding proteins are the most widely represented site-specific DNA-binding proteins in the archaea (4). Whereas Escherichia coli Lrp is the most extensively studied of this bacterial/archaeal protein family, it is the structures of two archaeal Lrp proteins from Pyrococcus species that have been determined (5, 6). The characteristic Lrp family monomer is a two-domain protein with an ≈45-aa N-terminal helix–turn–helix DNA-binding domain joined to an ≈75-aa C-terminal effector domain (with a βαββαβ fold) through an ≈20-aa linker (reviewed in ref. 4). Several of these proteins have been shown to repress transcription by binding to operator sites in such a way as to directly block access by TBP and TFB to the T/A box and BRE or prevent TBP/TFB-mediated RNAP entry to the promoter (7–9). Two Lrp family proteins have been proposed to serve as positive regulators of transcription: Sulfolobus solfataricus LysM (10) and Methanocaldococcus jannaschii (Mja) Ptr2. So far, only Mja Ptr2 has been shown to activate transcription by its conjugate core transcription apparatus in vitro (11–13).
Transcriptional activation by Mja Ptr2 is generated from promoter-proximal upstream activating DNA sites (UAS) by recruiting TBP to the promoter (11). A functional dissection of the architecture of the Mja rb2 promoter's UAS shows that the placement of its two Ptr2-binding sites, relative to each other and to the core promoter's BRE and T/A box, is highly constrained: A pair of optimally placed consensus (strong) Ptr2-binding sites constitutes an especially strong UAS (12).
The preceding analysis has involved just one transcriptional activator, Mja Ptr2, operating on a single in vitro transcription system (its cognate system) at elevated temperature. In this work, we extend our analysis to other archaeal Lrp family proteins closely related to Ptr2. The thermophilic methanogen Methanothermococcus thermolithotrophicus (Mth) and the mesophilic Methanococcus maripaludis (Mma) each encode one Lrp protein that is highly similar to Mja Ptr2 (77% and 76% identity, respectively), with only conservative substitutions in the putative DNA-recognition helix, suggesting that the Mth and Mma orthologues would recognize the Ptr2 consensus DNA-binding site, thus enabling the testing of transcriptional activity by using promoter constructs that have been characterized extensively in conjunction with Mja Ptr2. For this purpose, Mth provides a less thermophilic transcription apparatus that is active at temperatures between 37 and 55°C (14, 15) and allows a comparative analysis of transcriptional activation by these three Lrp family proteins, in conjunction with their cognate and heterologous TBPs. Here, we show that (i) Mja Ptr2 also activates transcription by the heterologous and less thermophilic Mth in vitro transcription system; (ii) the Mth Lrp activates transcription in its conjugate in vitro system and at elevated temperature in the heterologous Mja system; (iii) in contrast, Mma Lrp is almost inert as a transcriptional activator; (iv) mutating a small surface patch in its effector domain to match Mja Ptr2 converts Mma Lrp to a potent transcriptional activator, thus identifying a putative archaeal activation domain; and (v) TBPs from Mja, Mth, and Mma are comparably active in basal transcription but differ widely in their ability to support Ptr2/Lrp-mediated transcriptional activation.
Materials and Methods
Proteins and DNA. Mth RNAP was a generous gift from W. Hausner and M. Thomm (University of Kiel, Germany; currently at the University of Regensburg, Germany), and Mja RNAP was a generous gift from W. Hausner. The preparation of these enzymes has been described in refs. 11, 14, and 15. Mja TBP and TFB were overproduced in E. coli and purified as described in ref. 11. Purification of Mth TFB and Mth and Mma TBP followed the same procedure (the natural Mja TFB primary translation product has an intein that is excised to produce the mature protein, and recombinant Mja TFB was overproduced with an inteinless gene (11); Mth TFB does not have an intein). Mutant genes encoding Mma Lrp mutants 1, 2, and 3 were generated by the Kunkel method of oligonucleotide-directed mutagenesis (16, 17). The purification of N-terminally His6-tagged Mth and Mma Lrp and Mma Lrp mutants 1, 2, and 3 followed the procedure described for Mja Ptr2 (18) but with an additional purification step of size-exclusion chromatography on Superose 12. Protein concentrations were measured by MicroBCA assay, with BSA as standard. Molar concentrations of the Lrp-homologue proteins are expressed in terms of the protein monomer.
DNA for footprinting and transcription was purified as described in ref. 11. DNA was generated by PCR using the previously characterized derivative of the Mja rb2 promoter, with both Ptr2-binding sites of its UAS converted to the in vitro selection (SELEX)-derived consensus (5′-GGACGATTTTCGTCC-3′; the 6-bp inverted repeat is in italics) and a third Ptr2-binding site (site X) centered at base pair +7 (relative to the transcriptional start as +1) mutated (this is the promoter construct designated as cons:cons:mut in figure 5 of ref. 12). A derivative of the Mja ptr1 promoter (18), with increased promoter strength resulting from a mutated BRE, was used to compare the basal transcriptional activities of the three methanococcal TBPs. Complete DNA sequences are available on request.
Hydroxyl Radical (·OH) Footprinting. ·OH footprinting precisely followed the procedure described for Mja Ptr2 at 65°C (12) but was carried out at 55°C with Mth Lrp and at 37°C with Mma Lrp.
In Vitro Transcription. In vitro transcription with the Mja system at 65°C followed (11) and used a transcription buffer [20 mM Na-Hepes, pH 7.8/10 mM MgCl2/1 mM DTT/2.5% (vol/vol) glycerol], containing thermoprotectants [600 mM trehalose/500 mM betaine/5% (wt/vol) polyethylene glycol (PEG3300)], with 400 mM NaCl, 10 nM TBP, 20 nM TFB and ≈30 nM purified RNAP. The same conditions were used for transcription with Mth RNAP and TFB at 55°C. Thermoprotectants were omitted for transcription at 37°C, and the concentration of NaCl was correspondingly reduced to 250 mM in the experiments shown in Fig. 5. Run-off transcription was restricted to a single cycle with poly(dI-dC):poly(dI-dC) as described in ref. 11.
Fig. 5.
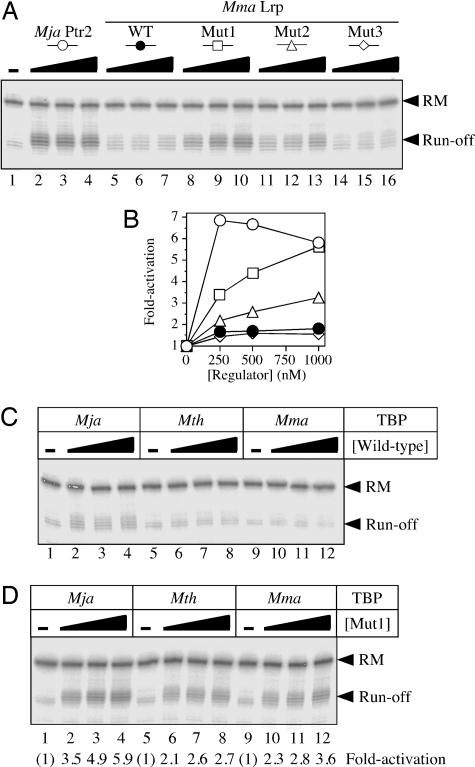
Converting Mma Lrp into a potent transcriptional activator. (A) Comparison of activation levels elicited by 250, 500, or 1,000 nM Mja Ptr2 (lanes 2–4, respectively), wild-type Mma Lrp (lanes 5–7), mutant 1 (lanes 8–10), mutant 2 (lanes 11–13), and mutant 3 (lanes 14–16), in conjunction with Mja TBP, Mth TFB, and Mth RNAP. Single rounds of transcription were carried out at 37°C in the presence of 250 mM NaCl (and in the absence of thermoprotectants). (B) Levels of activation; data from A.(C) Transcriptional response to wild-type Mma Lrp by the three methanococcal TBPs. Single rounds of transcription were carried out as in A. (D) Transcriptional response to Mma Lrp mutant 1 by the three methanococcal TBPs.
Results
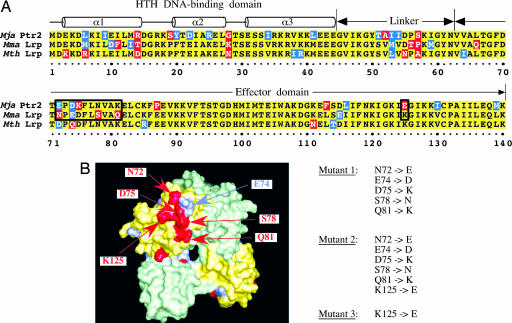
Overlapping Specificities of DNA Binding. Ptr2 and its closely related homologues Mth Lrp and Mma Lrp share a high degree of amino acid sequence identity (Fig. 1A). These methanococcal proteins are also highly similar to pyrococcal Lrp homologues whose structures have been solved, so that their polypeptide chains can be threaded directly into the three-dimensional structures of the pyrococcal proteins (5, 6). Especially notable is the conservation of sequence in the putative presentation helix α3 of the helix–turn–helix DNA-binding domain (amino acids 1–44), which is thought to make base-specific contacts within the DNA major groove (18).
Fig. 1.
Two methanococcal homologues of Ptr2. (A) Sequence alignment of Mja Ptr2, Mth Lrp, and Mma Lrp. Conserved, similar, and differing residues are shaded in yellow, blue, and red, respectively. The putative DNA-binding, linker, and effector domains are indicated above the alignment. (B) Homology model of Mma Lrp, shown in surface representation as a dimer of green and yellow protomers. Positions of sequence divergence from Mja Ptr2 are colored as in A, with putative activation determinants labeled. Also shown are the amino acid substitutions that generate Mma Lrp mutants 1, 2, and 3.
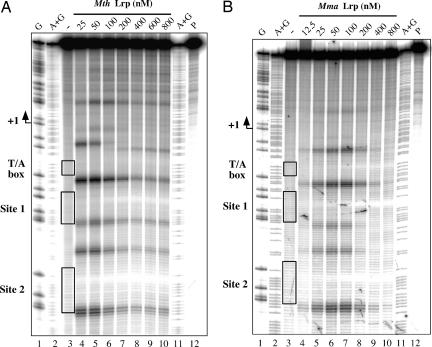
Thus, it was not surprising to find EMSA showing that recombinant Mth Lrp and Mma Lrp (see Materials and Methods) bind to a DNA fragment containing a single consensus Ptr2 site with only slightly lower affinity than does Ptr2 (data not shown). A closer examination, by ·OH footprinting, of DNA binding to the previously analyzed consensus-sequence version of the Mja rb2 promoter UAS (12), at 55°C for Mth Lrp and 37°C for Mma Lrp (Fig. 2), revealed the features that have been shown to characterize the footprint of Mja Ptr2 (at 65°C) (11, 12): protection of DNA from cleavage at the centers of Sites 1 and 2 (the two binding sites of the UAS) flanked by sites of enhanced DNA cleavage, indicating minor-groove widening associated with DNA bending; the changing patterns of this enhanced cleavage at higher protein concentrations suggest additional alterations of DNA structure through formation of higher-order nucleoprotein complexes. The congruence of DNA-binding specificities and properties of these three methanococcal Lrp proteins thus enabled analyses of transcriptional activation by Mth and Mma Lrps to be carried out with promoter constructs that have been characterized extensively in conjunction with analysis of transcriptional activation by Ptr2 (11, 12).
Fig. 2.
Specific binding of Mth and Mma Lrp to the consensus UAS of the rb2 promoter. The DNA probe, 5′-end-labeled on the nontranscribed (nontemplate) strand, was incubated at 55°C (A) or 37°C (B) for 20 min in the absence of Lrp (lane 3 of each panel) or with increasing concentrations of Mth Lrp (A) or Mma Lrp (B) (indicated above each panel and specified throughout for the monomer) and subjected to ·OH cleavage for 30 s at the same temperature. ·OH cleavage products within the T/A box and the two Ptr2 consensus binding sites are boxed in lane 3 and identified at the left of each panel. Also shown are the untreated DNA probe (P) and G and A + G chemical-sequencing ladders.
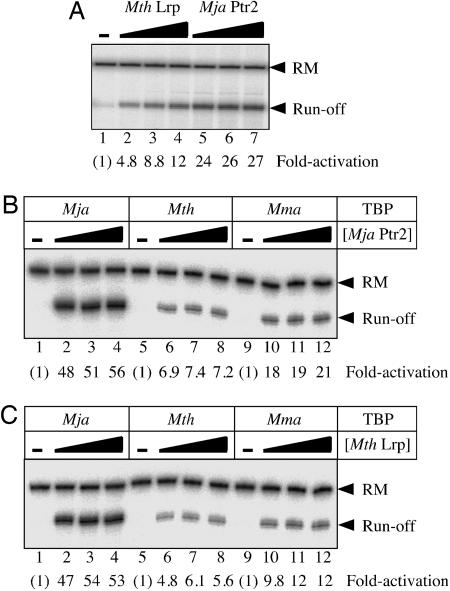
Mth Lrp Is a Transcriptional Activator. Activation of transcription by Mth Lrp was first tested with the Mja in vitro transcription system at the variant of the Mja rb2 promoter that had also been probed by ·OH footprinting (i.e., with a UAS composed of consensus Ptr2-binding sites). Activation was seen to be robust (Fig. 3A, lanes 2–4) but not as strong as with Ptr2 (Fig. 3A, lanes 5–7) under reaction conditions previously optimized for Ptr2-dependent activation, in a reaction medium containing thermoprotectants at relatively high ionic strength (11). In the Mth in vitro system (i.e., Mth TBP, TFB, and RNAP), Ptr2 (Fig. 3B, lanes 6–8), and Mth Lrp (Fig. 3C, lanes 6–8) were seen to elicit similar levels of transcriptional activation at 55°C. Thus, Mth Lrp is the second archaeal regulator so far shown to activate transcription by its cognate machinery in vitro.
Fig. 3.
Mth Lrp is a transcriptional activator. (A) Transcriptional responses to Mth Lrp (lanes 2–4) and to Mja Ptr2 (lanes 5–7) in the Mja in vitro system (i.e., Mja TBP, TFB, and RNAP) are compared. Single rounds of transcription were carried out at 65°C under the conditions previously optimized for Mja Ptr2 (11), by using the consensus rb2 UAS promoter template in the absence (lane 1) or presence of 200 nM (lanes 2 and 5), 400 nM (lanes 3 and 6), or 800 nM (lanes 4 and 7) regulator. The 84-nt Prb2 run-off transcript and the recovery marker DNA (RM) are indicated on the right. Levels of activation are indicated below each lane. (B) Differential response to Mja Ptr2 by the three methanococcal TBPs, Mja TBP (lanes 1–4), Mth TBP (lanes 5–7), and Mma TBP (lanes 8–12), in conjunction with Mth TFB and RNAP. Single rounds of transcription were carried out at 55°C in the presence of 400 mM NaCl and thermoprotectants (see Materials and Methods), in the absence of Ptr2 (lanes 1, 5 and 9) or in the presence of 250 nM (lanes 2, 6, and 10), 500 nM (lanes 3, 7, and 11), or 1,000 nM (lanes 4, 8, and 12) Ptr2. (C) Differential response to Mth Lrp by the three methanococcal TBPs, analyzed as described in B (the low levels of basal transcription in B and C are inapparent in the printed figure, but were readily detectable in the primary data and quantifiable).
Differences Between Methanococcal TBPs That Are Selectively Manifested in Transcriptional Activation. At the rb2 promoter, Ptr2 activates Mja transcription by recruiting TBP, which is structurally and functionally conserved across the archaeal and eukaryal lineages. For example, eukaryotic TBPs (yeast or human) can replace their archaeal counterpart in the Mth in vitro transcription system (19). The rationale for testing the ability of Ptr2 (or that of its homologues) to activate transcription in conjunction with various archaeal TBPs is that it provides a direct comparison of their putative Ptr2-interacting surfaces: Failure to support Ptr2-dependent activation would, in effect, direct attention to candidate sites of interaction based on sequence divergence and location on the TBP surface.
In addition to supporting transcription directed by Mja TBP, the Mth in vitro system (i.e., Mth RNAP and TFB) is transcriptionally active at 37°C and, therefore, suitable for testing the functionality of Mma TBP. Mja, Mth, and Mma TBP (from a hyperthermophile, a thermophile, and a mesophile, respectively) were comparably active in combination with Mth RNAP and TFB at a strong promoter, at 55°C in reaction medium containing thermoprotectants (Fig. 4) and at the weak rb2 promoter, at 37°C in the absence of thermoprotectants (see below; Fig. 5 C and D, lanes 1, 5, and 9). However, these three TBPs differed markedly in their ability to support transcriptional activation by both Mja Ptr2 (Fig. 3B) and Mth Lrp (Fig. 3C). Similar results were also obtained for transcriptional activation by Ptr2 with its homologous RNAP and TFB and with Mja and Mth TBP (data not shown). Mma TBP was not examined at the relatively high temperature of the Mja in vitro system.
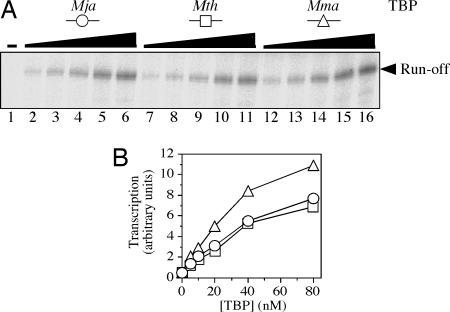
Fig. 4.
The three methanococcal TBPs are equally active in basal transcription. (A) Single rounds of basal transcription at a strong promoter (an up-mutant ptr1 promoter with an improved BRE) were carried out as described for Fig. 3 B and C, in the absence of TBP (lane 1) or in the presence of 5, 10, 20, 40, or 80 nM Mja TBP (lanes 2–6, respectively), Mth TBP (lanes 7–11), or Mma TBP (lanes 12–16). (B) Levels of basal transcription; data from A, quantified.
Thus, it appears that the Mja protein is the most effective of these three TBPs in supporting transcriptional activation by both Mja Ptr2 and its Mth Lrp homologue. Mth TBP is distinctly inferior in this regard; in fact, Mth TBP performed even more poorly (from this perspective) at the optimum temperature of its conjugate in vitro system than did Mma TBP.
Conferring Transcriptional Activation by Mutating Mma Lrp. Although it is optimal for transcription at higher temperature, the Mth in vitro system is also functional at 37°C (as already mentioned) and, therefore, compatible with a test of the mesophilic Mma Lrp as a transcriptional activator. In conjunction with Mja TBP, Ptr2 was found to activate transcription >6-fold at 37°C [Fig. 5 A (lanes 2–4) and B], whereas Mma Lrp was, by comparison, almost inert [Fig. 5 A (lanes 5–7) and B]. The marginal activation by wild-type Mma Lrp was detectable only with Mja TBP and not observed at all with Mth or Mma TBP (Fig. 5C).
The distinctive properties of Mja Ptr2 and Mma Lrp are provocative, in view of their high degree of amino acid sequence identity. Primary sequence alignment of these two proteins, combined with homology-based models derived from the crystal structures of Pyrococcus furiosus LrpA and of a close homologue from another Pyrococcus species (5, 6) drew attention to a clearly defined surface patch of divergences (Fig. 1B) clustering to amino acids 72–81 and to a charge reversal at amino acid 125 (boxed in Fig. 1 A). In Mja Ptr2, a single cysteine substitution within this patch (K75C) strongly diminishes activation while retaining specific DNA-binding (M.O. unpublished observations). Mutagenesis of Mma Lrp to substitute the corresponding Mja Ptr2 residues at amino acids 72, 74, 75, 78, and 81 generated mutant 1; an additional change at residue 125 generated mutant 2; changing only K125 to E generated Mma Lrp mutant 3 (Fig. 1B).
Activation of transcription by these three mutant proteins, wild-type Mma Lrp, and Mja Ptr2 is compared in Fig. 5 A and B, in conjunction with Mja TBP. Mutant 3 was comparable with the wild-type Mma Lrp, in that it was barely capable of activating transcription (Fig. 5A, lanes 14–16). The patch of changes in mutant 1 generated significant transcriptional activation, to levels nearly comparable, at the highest protein concentration, to those elicited by Mja Ptr2 [Fig. 5 A (lanes 8–10) and B], whereas mutant 2, with the additional charge-reversal mutation at amino acid 125, was significantly less transcriptionally active than mutant 1 (Fig. 5B, lanes 11–13). At lower ionic strength, however, the activities of mutants 1 and 2 were comparable, with mutant 2 actually slightly more active (data not shown). As already noted for Mja Ptr2 and Mth Lrp, of the three methanococcal TBPs tested, Mja TBP was the most effective partner for transcriptional activation by Mma Lrp mutant 1 (Fig. 5D, lanes 2–4).
Thus, replacing a patch on the surface of the C-terminal domain of Mma Lrp with its Mja Ptr2 counterpart turns it into a potent transcriptional activator and points to this surface patch as an activation domain.
Discussion
Only one instance of activation of archaeal transcription has been demonstrated in vitro: the Lrp family protein Ptr2, from the hyperthermophile Mja, activates initiation of transcription by its conjugate RNAP at high temperature and recruits TBP to weak T/A-box-containing promoters. Transcriptional activation by Ptr2 is achieved from UASs placed close to the core archaeal promoter.
The experiments that are presented here extend this analysis by examining transcriptional activation by two homologues of Ptr2 from the thermophile Mth and the mesophile Mma. The comparison of the functions of these two proteins is enabled by the Mth in vitro transcription system, which is active at 37°C (14, 15). We show that Mja Ptr2 activates transcription by Mth RNAP at 55°C and even at 37°C. Conversely, Mth Lrp activates transcription by the Mja RNAP at 65°C and by its conjugate RNAP at 55°C.
In contrast, the highly similar Mma Lrp is almost inert as an activator (<2-fold activation; Fig. 5), although its DNA-binding specificity clearly overlaps with that of Mja Ptr2 (Fig. 2). Primary sequence alignment of Mja Ptr2 and Mma Lrp, combined with homology modeling based on the structures of Pyrococcus furiosus LrpA and of a close homologue from another Pyrococcus species (5, 6), points to a cluster of amino acids in the Lrp effector domain as possible determinants of the transcriptional activity of Mja Ptr2. Replacing the corresponding patch of Mma Lrp surface-exposed residues by their Mja Ptr2 counterparts (Fig. 1B) confers a substantial transcriptional-activation capacity on the nearly inactive Mma Lrp, suggesting that this patch of Lrp surface is (at least part of) an activation domain, the first such domain to be defined for an activator of archaeal transcription.
Bringing the Mma Lrp into the fold (as it were) of archaeal activators of transcription is also of special interest because of the recent development of genetic methods for this mesophilic methanogen (reviewed in refs. 20 and 21) and the prospect for developing reporter-gene constructs (e.g., lacZ cassettes under control of various Mja rb2 UAS architectures) for parallel in vivo analysis of Lrp family transcriptional activators and their interactions with the core transcription apparatus.
Because Mja Ptr2 activates transcription at the intrinsically weak Mja rb2 promoter in vitro (at least in part) by recruiting TBP (11), one anticipates that the engineered Mma Lrp mutant 1 functions in the same way, but this remains to be directly established. Whether the Lrp activation surface that is defined here by gain-of-function mutations is a site of direct interaction with TBP (or TFB) or whether it is involved in Lrp dimer interactions that determine the effective presentation of UAS-bound Lrp to the core transcription apparatus also remains to be determined. The crystal structure of an archaeal TBP–TFB–DNA ternary complex (from Pyrococcus woesei) has been determined (22, 23), but the structure of an activator–TBP–TFB–DNA complex remains an important (and possibly arduous) objective. In fact, the determination of crystal structures of promoter-bound transcriptional activators interacting with their target elements in the transcription apparatus is limited to two examples from bacterial transcription: a CAP/RNAP α subunit C-terminal domain/DNA complex (24) and a λcI protein/σ domain 4/DNA complex (25). In each case, interaction involves a strikingly small interface enlisting no more than six residues in each partner. There are no comparable structures for eukaryotic transcription.
Lrp proteins bend DNA (Fig. 2) and form oligomers; DNA wrapping (26) may play an important role in productively aligning the DNA-bound activator with its target(s) in the core transcription apparatus and, possibly, even in postrecruitment steps of initiating transcription. DNA bending by the Lrp activators at close proximity to a T/A-box-bound TBP (or TBP–TFB complex) may be at the heart of the activation mechanism. In this alternative mode of assembly, cooperative DNA binding is determined by structural changes in DNA (rather than purely by protein–protein interaction), as exemplified by the ATF-2 (c-Jun)/interferon regulatory factor 3/IFN β enhancer complex (27). The CAP/α subunit C-terminal domain/DNA complex, on the other hand, provides an example of DNA functioning as a passive scaffold (24).
Finding that the three methanococcal TBPs systematically differ in their ability to support transcriptional activation by all three Lrp homologues (Mja Ptr2, Mth Lrp, and Mma Lrp mutant 1) was unexpected. Mja TBP is clearly the most effective partner for transcriptional activation at all temperatures examined, whether in combination with its conjugate or the heterologous RNAP and TFB. Determining the structural and mechanistic differences that underlie the special effectiveness of Mja TBP is of obvious interest: Many archaea (including some methanogens) encode multiple TBPs that are presumed to enable differential gene expression (28, 29), much as alternative σ factors do in the bacteria. Thus, uncovering differences between homologous methanococcal TBPs that are manifested only in the context of activated transcription provides a framework for thinking about mechanisms of action and function of TBP paralogues. The differential activities of these paralogues might be generated, for instance, by diversifying surface patches for specificity of interaction with regulators. Differences, possibly quite subtle, of DNA bending by the TBP–TFB complex can also change the path of upstream-lying DNA in such a way as to affect activator presentation to the core transcription machinery. These changes may, in turn, be T/A-box-BRE sequence-dependent.
Acknowledgments
We thank G. A. Langham for extensive technical assistance; J. W. Campbell for gene sequence information; G. A. Kassavetis for discussions, advice, and critical comments on the manuscript; W. Hausner and M. Thomm for most generous gifts of materials; I. Porat (University of Georgia, Athens) for kindly providing Methanococcus maripaludis genomic DNA; and the National Institute of General Medical Sciences for support of this research.
Author contributions: M.O. and E.P.G. designed research; M.O. performed research; M.O. contributed new reagents/analytic tools; M.O. analyzed data; and M.O. and E.P.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: BRE, B recognition element; Mja, Methanocaldococcus jannaschii; Mma, Methanococcus maripaludis; Mth, Methanothermococcus thermolithotrophicus; RNAP, RNA polymerase; TBP, TATA-binding protein; UAS, upstream activating DNA site.
References
- 1.Geiduschek, E. P. & Ouhammouch, M. (2005) Mol. Microbiol. 56, 1397-1407. [DOI] [PubMed] [Google Scholar]
- 2.Reeve, J. N. (2003) Mol. Microbiol. 48, 587-598. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L. & Koonin, E. V. (1999) Nucleic Acids Res. 27, 4658-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkman, A. B., Ettema, T. J., de Vos, W. M. & van der Oost, J. (2003) Mol. Microbiol. 48, 287-294. [DOI] [PubMed] [Google Scholar]
- 5.Koike, H., Ishijima, S. A., Clowney, L. & Suzuki, M. (2004) Proc. Natl. Acad. Sci. USA 101, 2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonard, P. M., Smits, S. H., Sedelnikova, S. E., Brinkman, A. B., de Vos, W. M., van der Oost, J., Rice, D. W. & Rafferty, J. B. (2001) EMBO J. 20, 990-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, S. D. & Jackson, S. P. (2000) J. Biol. Chem. 275, 31624-31629. [DOI] [PubMed] [Google Scholar]
- 8.Brinkman, A. B., Dahlke, I., Tuininga, J. E., Lammers, T., Dumay, V., de Heus, E., Lebbink, J. H., Thomm, M., de Vos, W. M. & van Der Oost, J. (2000) J. Biol. Chem. 275, 38160-38169. [DOI] [PubMed] [Google Scholar]
- 9.Dahlke, I. & Thomm, M. (2002) Nucleic Acids Res. 30, 701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinkman, A. B., Bell, S. D., Lebbink, R. J., de Vos, W. M. & van der Oost, J. (2002) J. Biol. Chem. 277, 29537-29549. [DOI] [PubMed] [Google Scholar]
- 11.Ouhammouch, M., Dewhurst, R. E., Hausner, W., Thomm, M. & Geiduschek, E. P. (2003) Proc. Natl. Acad. Sci. USA 100, 5097-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouhammouch, M., Langham, G. E., Hausner, W., Simpson, A. J., El-Sayed, N. M. & Geiduschek, E. P. (2005) Mol. Microbiol. 56, 625-637. [DOI] [PubMed] [Google Scholar]
- 13.Ouhammouch, M., Werner, F., Weinzierl, R. O. & Geiduschek, E. P. (2004) J. Biol. Chem. 279, 51719-51721. [DOI] [PubMed] [Google Scholar]
- 14.Frey, G., Thomm, M., Brudigam, B., Gohl, H. P. & Hausner, W. (1990) Nucleic Acids Res. 18, 1361-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hausner, W. & Thomm, M. (1993) J. Biol. Chem. 268, 24047-24052. [PubMed] [Google Scholar]
- 16.Kunkel, T. A., Bebenek, K. & McClary, J. (1991) Methods Enzymol. 204, 125-139. [DOI] [PubMed] [Google Scholar]
- 17.McClary, J. A., Witney, F. & Geisselsoder, J. (1989) BioTechniques 7, 282-289. [PubMed] [Google Scholar]
- 18.Ouhammouch, M. & Geiduschek, E. P. (2001) EMBO J. 20, 146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wettach, J., Gohl, H. P., Tschochner, H. & Thomm, M. (1995) Proc. Natl. Acad. Sci. USA 92, 472-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allers, T. & Mevarech, M. (2005) Nat. Rev. Genet. 6, 58-73. [DOI] [PubMed] [Google Scholar]
- 21.Whitman, W. B., Tumbula, D. L., Yu, J. P. & Kim, W. (1997) Biofactors. 6, 37-46. [DOI] [PubMed] [Google Scholar]
- 22.Kosa, P. F., Ghosh, G., DeDecker, B. S. & Sigler, P. B. (1997) Proc. Natl. Acad. Sci. USA 94, 6042-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Littlefield, O., Korkhin, Y. & Sigler, P. B. (1999) Proc. Natl. Acad. Sci. USA 96, 13668-13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benoff, B., Yang, H., Lawson, C. L., Parkinson, G., Liu, J., Blatter, E., Ebright, Y. W., Berman, H. M. & Ebright, R. H. (2002) Science. 297, 1562-1566. [DOI] [PubMed] [Google Scholar]
- 25.Jain, D., Nickels, B. E., Sun, L., Hochschild, A. & Darst, S. A. (2004) Mol. Cell 13, 45-53. [DOI] [PubMed] [Google Scholar]
- 26.Jafri, S., Chen, S. & Calvo, J. M. (2002) J. Bacteriol. 184, 5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panne, D., Maniatis, T. & Harrison, S. C. (2004) EMBO J. 23, 4384-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baliga, N. S., Goo, Y. A., Ng, W. V., Hood, L., Daniels, C. J. & DasSarma, S. (2000) Mol. Microbiol. 36, 1184-1185. [DOI] [PubMed] [Google Scholar]
- 29.Galagan, J. E., Nusbaum, C., Roy, A., Endrizzi, M. G., Macdonald, P., FitzHugh, W., Calvo, S., Engels, R., Smirnov, S., Atnoor, D., et al. (2002) Genome Res. 12, 532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]