Abstract
In an attempt to characterize early B cell development including the commitment of progenitor cells to the B cell lineage, we generated and compared genomewide gene expression profiles of human hematopoietic stem cells (HSCs) and pre-B cells (PBCs) by using serial analysis of gene expression. From more than 100,000 serial analysis of gene expression tags collected from human CD34+ HSCs and CD10+ CD19+ PBCs, 42,399 unique transcripts were identified in HSCs but only 16,786 in PBCs, suggesting that more than 60% of transcripts expressed in HSCs were silenced during or after commitment to the B cell lineage. On the other hand, mRNAs of pre-B cell receptor (pre-BCR)-associated genes are virtually missing in HSCs but account for more than 10% of the transcriptome of PBCs, which also show increased expression of apoptosis-related genes. Both concentration of the transcriptional repertoire on pre-BCR-related genes together with marked up-regulation of apoptosis mediators in PBC might reflect selection for the expression of a functional pre-BCR within the bone marrow. Besides known regulator genes of early B cell development such as PAX5, E2A, and EBF, the most abundantly expressed genes in PBCs include ATM, PDGFRA, SIAH1, PIM2, C/EBPB, WNT16, and TCL1, the role of which has not been established yet in early B cell development.
B cells originate from bone marrow hematopoietic progenitor cells (1), which give rise to a common lymphoid progenitor (2). Before B cell differentiation in a sequence defined by the configuration of the Ig gene loci (3), the pluripotent progenitor cells undergo commitment to the B cell lineage (4), which critically relies on expression of the PAX5 transcription factor (5). Developmental progression of a committed B cell precursor including rearrangement of Ig DH to JH gene segments, depends on two transcription factors encoded by the E2A (6) and EBF (7) genes. Pro-B cells typically harbor DH-JH gene rearrangements and pre-B cells (PBCs) carry VH-DHJH gene rearrangements (8), whereas the Igκ and Igλ light chain loci undergo rearrangement only in late PBCs (9). Instead of Ig light chain gene rearrangements, pro-B and early PBC express Vpre-B- and λ5-surrogate light chain genes (10). Concomitant expression of RAG1, RAG2, and TdT genes (11) indicates that the recombination machinery is active at the PBC stage. Although many aspects of early B cell development have been studied in detail, the molecular mechanisms underlying the decision of progenitor cells for a B cell fate remain elusive. In an attempt to identify genes that determine commitment to the B cell lineage, human bone marrow hematopoietic stem cells (HSCs) and PBCs were analyzed by using serial analysis of gene expression (SAGE) (12), which allows for the unbiased quantitative analysis of transcriptomes (i.e., the totality of transcripts within a given population), including novel genes and known genes that have not been implicated in B cell differentiation so far.
Materials and Methods
Isolation of Human HSCs, PBCs, and Mature B Cell Subsets.
HSCs and PBCs were purified from bone marrow and umbilical cord blood. Purification of bone marrow CD34+ HSCs was described in Zhou et al. (13). For verification experiments, cord blood HSCs were isolated by using anti-CD34 immunomagnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). For enrichment of PBCs, mononuclear cells were isolated from four bone marrow samples (Poietics, Gaithersburg, MD) and 28 umbilical cord blood samples by Ficoll density gradient centrifugation. All samples were obtained according to the principle of informed consent. T cells and myeloid cells were depleted by using anti-CD3 and anti-CD15 immunomagnetic beads (Dynal, Oslo). Among the remaining cells, immature CD10low CD19+ CD20+ B cells and CD138+ plasma cells were depleted by using an anti-CD20 IgG1 antibody (BD Biosciences, Heidelberg) together with anti-IgG1 beads and anti-CD138 beads (Miltenyi Biotec), respectively. The PBCs were enriched by using anti-CD19 immunomagnetic multisort beads (Miltenyi Biotec); the beads were released enzymatically from the CD19+ cells. The purified cells were subsequently labeled with a mouse anti-CD10 IgG1 antibody (CALLA; BD Biosciences) and separated by using anti-mouse IgG1 beads (Miltenyi Biotec). Mature B cell subsets (IgD+ CD27− naïve B cells and CD19+ CD27+ memory B cells) were isolated from seven tonsillectomy specimens as described in Müschen et al. (14).
Verification of B Cell Subset Purification.
The identity of the purified B cell subsets was verified genotypically and phenotypically. The genotype of purified PBCs and mature B cell subsets (naïve and memory B cells) was assessed by PCR amplification of rearranged VH1 genes from genomic DNA and subsequent cloning and sequencing of the PCR products as described in Müschen et al. (15). In addition, the phenotype of the purified HSCs, PBCs, and memory B cells was assessed by semiquantitative reverse transcriptase–PCR (RT-PCR) at the mRNA level by using IgH Cμ-, Cγ1-, Ig Cκ-, Cλ-, Vpre-B1-, λ5- and β2-microglobulin-specific primers (Integrated DNA Technologies, Coralville, IA). The phenotype of purified PBCs was verified at the protein level by flow cytometry by using CD10- and CD19-specific antibodies (BD Biosciences).
SAGE Analysis.
RNA isolation, cDNA synthesis, and SAGE analysis were carried out according to Lee et al. (16). The UniGene reference database (March 2001) was obtained at http://www.sagenet.org/SAGEDatabases/unigene.htm. A total of 106,021 SAGE tags collected from HSCs (13) and 110,788 SAGE tags from PBCs were used in the analyses. All SAGE tags were compacted in a unique data set with quantitative information and matched to the SAGE reference database for gene identification.
Verification of SAGE Tags Matching Multiple UniGene Clusters by Generation of Longer 3′ Expressed Sequence Tags from SAGE Tags for Gene Identification (GLGI).
About 15% of the SAGE tags collected from HSCs and PBCs matched to more than one UniGene cluster. The multimatch-SAGE tags were sorted based on the ratio of their frequency in PBCs and HSCs. For 96 multimatch-SAGE tags with the highest ratio (PBC count/HSC count), a longer 3′ expressed sequence tag corresponding to each tag was generated by GLGI (17) by using 3′ cDNAs from HSCs or PBSs to determine the correct match.
Verification of Quantitative Accuracy of SAGE Data.
To corroborate quantitative differences in gene expression between HSCs and PBCs as determined by SAGE, semiquantitative RT-PCR analysis was performed for a set of 45 selected genes by using cDNAs from HSCs and PBCs purified from umbilical cord blood as template. RT-PCR was done for 18, 24, and 30 cycles and normalized for the COX6B gene encoding the cytochrome c oxidase subunit VI.
Functional Clustering of SAGE Data.
For graphic representation of SAGE data, SAGE tags derived from known transcripts were arranged in functional clusters and sorted based on the ratio of their frequency in PBCs and HSCs. For transformation and graphic representation of SAGE tag counts, cluster and treeview software was used (kindly provided by Michael B. Eisen, Lawrence Berkeley National Laboratory, http://rana.lbl.gov/).
Results and Discussion
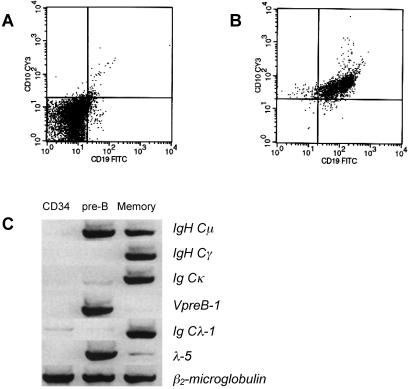
Purified PBCs exhibit a typical phenotype in that they express IgH μ chains together with Vpre-B or λ5 surrogate light chains (Fig. 1C), which in association with the Igα and Igβ signaling chains form the pre-B cell receptor (pre-BCR). Also supporting the immature phenotype of the purified cells, mRNA for the Cγ1 gene (expressed by postgerminal center B cells after class-switch recombination) was not detected. No mRNAs for Ig Cκ or Cλ light chain genes were detected in human PBCs. This finding was unexpected, because in mice, Igκ and Igλ light chain genes are already expressed in late PBCs [small resting pre-BII cells (8)] before the assembly of the BCR, which occurs in immature B cells.
Figure 1.
Phenotype of purified human PBCs. The FACS plots show enrichment of CD10+ CD19+ (CD3− CD15− CD20− CD138−) PBCs from mononuclear bone marrow cells before (A) and after (B) magnetic cell sorting enrichment. Using cDNA from the purified cells together with cDNAs from CD34+ HSC and memory B cells, the phenotype of purified PBCs was confirmed by semiquantitative RT-PCR.
Absence of Ig Cγ1 and Ig Ck or Cλ light chain transcripts, however, is consistent with an immature phenotype of PBCs. Rearranged VH1 genes were amplified from genomic PBC DNA; the PCR products were cloned and sequenced. None of 10 VH1 gene rearrangements harbored somatic mutations, which are a hallmark of antigen-experienced B cells like bone marrow plasma cells. Of note, eight of the amplified rearrangements were not productive, either by loss of the reading frame during V(D)J-recombination or by rearrangement of VH1 pseudogenes (not shown), both of which are common features of an immature B cell repertoire that is not readily selected for the expression of a functional Ig heavy chain within the pre-BCR. The purity of enriched PBCs as assessed by flow cytometry ranged between 92% and 95% CD10+ CD19+ CD20− cells (Fig. 1 A and B).
Detailed features of the HSC SAGE library have been described in Zhou et al. (13). For both HSCs and PBCs, more than 100,000 SAGE tags were collected and matched to UniGene. Although the HSC library contained 42,399 unique tags, only 16,786 unique tags were identified in the PBC library. This finding indicates that the repertoire of gene expression at the mRNA level is far less complex in PBCs and suggests that committed B cell precursors express many fewer genes than uncommitted HSCs (Table 1). Except for the fact that more than 60% of the genes expressed in HSCs were silenced in PBCs, the quantitative distribution of SAGE tags is similar in both SAGE profiles: Around 15% of SAGE tags match to more than one UniGene cluster (ranging between two and 98 matches), 45% matched to one single UniGene cluster, and about 40% of tags do not have a match in UniGene and are likely to be derived from novel genes (Table 1). Novel SAGE tags can be further investigated by GLGI (17) and 5′ rapid amplification of cDNA ends to identify full-length cDNAs from novel candidate genes.
Table 1.
Distribution of SAGE tags in CD34+ HSCs and PBCs
| CD34+HSC | CD10+ CD19+PBC | |
|---|---|---|
| Total tags | 106,021 | 110,788 |
| Unique tags | 42,399 | 16,786 |
| Tags matching multiple genes* | 5,918 (14)† | 2,686 (16) |
| Tags matching single gene | 16,914 (40) | 7,614 (46) |
| Novel tags | 19,567 (46) | 6,486 (39) |
| Abundance classes of SAGE-tags‡ | ||
| 1 | 32,453 (76) | 10,284 (61) |
| 2 to 4 | 7,757 (18) | 4,284 (26) |
| 5 to 9 | 1,328 (3) | 1,348 (8) |
| 10 to 99 | 771 (2) | 676 (4) |
| ≥100 | 91 (<1) | 194 (1) |
| Tags not reflecting 3′ splice variants§ | 10,857 (64) | 5,787 (76) |
| Tags potentially reflecting isoforms§ | 6,057 (36) | 1,827 (24) |
| Number of 3′ splice variants | ||
| 1 | 2,875 (17) | 1,142 (15) |
| 2 | 1,522 (9) | 381 (5) |
| 3 | 846 (5) | 152 (2) |
| 4 | 338 (2) | 77 (1) |
| 5 | 126 (1) | 38 (<1) |
| ≥6 | 350 (2) | 37 (<1) |
Among the tags matching multiple UniGene clusters, 83 tags most frequently found in PBCs were verified by GLGI.
Percentages are given in parentheses.
Abundance classes refer to tag counts in the two SAGE libraries.
If more than one SAGE tag was derived from transcripts being assigned to a single UniGene cluster, these SAGE tags were taken as indication for 3′ splice variants.
The proportions of abundance classes are comparable in both HSCs and PBCs (Table 1). Multiple SAGE tags matching to one UniGene cluster (i.e., indicating the presence of splice variants in the 3′ region of a given gene) tend to be more frequent in HSCs as compared with PBCs and point to another level, at which the pattern of gene expression is more complex in HSCs than in PBCs.
Before the comparison of the SAGE profiles for HSCs and PBCs, SAGE tags matching to multiple UniGene clusters were subjected to GLGI for unambiguous assignment to a single UniGene cluster. GLGI was performed for 96 SAGE tags that were most significantly overpresented in the PBC profile as compared with HSCs. For 83 GLGI reactions, a PCR product was obtained that yielded a reproducible 3′ expressed sequence tag (EST) sequence after sequencing. The length of the ESTs varied between 45 and 560 bp. Among the genes, which were identified as the correct origin of SAGE tags by GLGI, are the CD19 gene (3,601 copies in PBCs, identified among 60 alternative matches in UniGene), the CD84 gene (2,227 copies in PBCs, 98 alternative matches), the ATM gene (1,109 copies, 10 alternative matches), and the MAF gene (445 copies in PBCs, 26 alternative matches; see Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). Given the critical role of the CD19 and ATM genes in normal and malignant B cell development, respectively, their identification as the correct source of SAGE tags demonstrates that GLGI represents an important means to overcome specificity limitations in gene identification by 10-bp SAGE tags (Table 3). After GLGI, the genes that were up- and down-regulated during the development of HSC to PBC were identified (Table 2). Among the known genes that have the highest ratio of tag counts in PBCs and HSCs (up-regulated genes) are many known regulators of early B cell development. This finding is in agreement with recent microarray data confirming that known B cell markers can indeed distinguish B cell lineage populations from other lineages (18). λ5 (1,975 tags), Vpre-B1,3 (287 tags), Ig heavy chain genes (1,090 tags), Igα (58 tags), and Igβ (181 tags) are constituents of the pre-BCR and are among the high abundance class genes in the PBC library (Table 2). The set of up-regulated genes also includes RAG2 (125 tags), TdT (90 tags), and RAG1 (83 tags), which are required for V(D)J gene recombination, as well as proximal components of the pre-BCR signaling cascade like CD19 (3,657 tags), BLK (139 tags), BLNK (81 tags), BTK (71 tags), IKAROS (62 tags), CD72 (26 tags), APS (23 tags), and BAP29 (15 tags) and B lineage-specific transcription factors such as OBF1 (174 tags), E2A (164 tags), EBF (131 tags), PAX5 (67 tags), and OCT2 (53 tags). A rough survey of all SAGE tags matching to a single UniGene cluster in HSCs and PBCs indicates that genes encoding constituents of the pre-BCR or parts of its signaling cascade account for less than 0.2% of the transcriptome in HSCs but more than 10% in PBCs.
Table 2.
Genes differentially expressed in CD34+ HSCs and PBCs
| Up-regulated in PBCs
|
Down-regulated in PBCs
|
||||
|---|---|---|---|---|---|
| Gene | Tag counts in
|
Gene | Tag counts in
|
||
| HSC | PBC | HSC | PBC | ||
| λ-5 | 0 | 1,975 | KIF5C | 115 | 0 |
| CGLA | 0 | 326 | RAB5 | 103 | 0 |
| CD19 | 6 | 3,657 | SOS1 | 47 | 0 |
| CD84 | 4 | 2,227 | TRIAD | 37 | 0 |
| Vpre-B1 | 0 | 177 | HGF | 36 | 0 |
| RAG2 | 0 | 125 | MAGOH | 36 | 0 |
| Ig VH3 | 0 | 97 | DS10 | 145 | 2 |
| BLNK | 0 | 81 | HSP90 | 29 | 0 |
| SIAH1 | 0 | 73 | TMSB10 | 25 | 0 |
| SYNJ1 | 0 | 69 | Topoisomerase II | 24 | 0 |
| IKAROS | 0 | 62 | CST8 | 23 | 0 |
| OCT2 | 0 | 53 | ARHI | 23 | 0 |
| ATM | 9 | 1,109 | PBX3 | 23 | 0 |
| GAP | 0 | 45 | MARP-2 | 22 | 0 |
| TNFR75 | 0 | 44 | NR113 | 51 | 1 |
| GNRHR | 0 | 43 | MSH5 | 20 | 0 |
| TGFBR3 | 0 | 43 | HARP | 143 | 3 |
| PDGRFRA | 6 | 624 | ETS | 19 | 0 |
| IL7R | 0 | 40 | ROR2 | 15 | 0 |
| TdT | 1 | 90 | SHOX2 | 143 | 4 |
| FYNBP | 1 | 87 | MPO | 33 | 1 |
| BCRDS1 | 0 | 34 | HSP70 | 16 | 0 |
| TCL1 | 0 | 34 | HDAC2 | 13 | 0 |
| CGLB | 0 | 31 | p300/CBPAF | 13 | 0 |
| E2A | 2 | 164 | ICAPA1 | 13 | 0 |
| PAX5 | 1 | 67 | NAP1L4 | 13 | 0 |
| EBF | 2 | 131 | AP3 | 24 | 1 |
| CD72 | 0 | 26 | RALABP1 | 12 | 0 |
| Ig Cμ | 15 | 962 | HHOX | 11 | 0 |
| LARD | 0 | 24 | ING4 | 11 | 0 |
| Igα | 1 | 58 | CD164 | 27 | 1 |
| APS | 0 | 23 | DRG1 | 25 | 1 |
| Vpre-B3 | 2 | 110 | LAP18 | 25 | 1 |
| NFKB2 | 0 | 22 | AF4L1 | 9 | 0 |
| FADD | 0 | 21 | ARHGAP4 | 9 | 0 |
| IL1RII | 0 | 21 | NFI/C | 9 | 0 |
| BLK | 3 | 139 | MAGOH | 40 | 2 |
| Igβ | 4 | 181 | CAMKII | 8 | 0 |
| FAF | 0 | 18 | AML3 | 8 | 0 |
| Ig VH1 | 0 | 18 | ABL1 | 8 | 0 |
| LIFR | 0 | 18 | Topoisomerase-IIBP | 8 | 0 |
| GRAF | 2 | 86 | KOX15 | 8 | 0 |
| IL10RA | 0 | 16 | SPTBN2 | 19 | 1 |
| BAP29 | 0 | 15 | KLK8 | 7 | 0 |
| TRAF1 | 0 | 15 | KYNU | 7 | 0 |
| PIM2 | 5 | 181 | ETEFA1 | 208 | 12 |
| OBF1 | 5 | 174 | NUDT4 | 49 | 3 |
| LAG1 | 1 | 33 | HSP60 | 48 | 3 |
| Ig VH4 | 0 | 13 | SCP | 16 | 1 |
| MYB | 0 | 13 | CD46 | 16 | 1 |
| HK3 | 1 | 32 | H2KBF2 | 16 | 1 |
| DLEU1 | 1 | 31 | HSP70IP | 16 | 1 |
| IL4R | 1 | 29 | MAEA | 31 | 2 |
| CASP8 | 3 | 83 | NUP62 | 15 | 1 |
| CD83 | 2 | 55 | p300TED | 6 | 0 |
| E2F | 2 | 39 | CDC2L1 | 6 | 0 |
| PBX1 | 4 | 68 | DNM1L | 6 | 0 |
| C/EBPB | 4 | 66 | ELAV1 | 6 | 0 |
| WNT16 | 4 | 39 | EREG | 6 | 0 |
The receptors for IL-4 (29 tags), IL-10 (16 tags), and notably IL-7 (40 tags), which is indispensable for early B cell development in the bone marrow, are among the most abundantly expressed genes in PBC (Table 2). The genes up-regulated in PBC also include a number of direct apoptosis mediators [Caspase 8 (83 tags), TNFR2 (40 tags), FADD (21 tags), FAF (18 tags), and TRAF1 (15 tags); see Table 2] and the apoptosis-sensitizing gene E2F (39 tags), indicating the propensity of PBCs to apoptosis. Besides these examples, SAGE tag counts for proapoptotic molecules are consistently higher in PBCs as compared with HSCs throughout the SAGE profiles. Increased susceptibility to apoptosis as a feature of committed B cell precursors within the bone marrow is further supported by the finding that in PBCs antiapoptotic genes frequently are silenced. This group of genes includes BCL2 (five tags in HSCs, one tag in PBCs), the FLICE inhibitor FLIP (4 vs. 0 tags), the BCL-2 binding protein BAG1 (3 vs. 0 tags), the NF-κB-activating kinase NAK and the apoptosis-inhibiting BCL2-homologues BCLW (3 vs. 0 tags), and BCLX (7 vs. 0 tags). In the case of BCLX, the SAGE tags found in the libraries could distinguish between the antiapoptotic isoform, termed BCL-XL, and its truncated proapoptotic splice variant, BCL-XS. Concomitant up-regulation of positive mediators of apoptosis with pre-BCR-related molecules within the context of a restricted repertoire of gene expression might reflect a “concentration effect” of selection for a functional pre-BCR within the bone marrow. Indeed, PBCs are thought to enter a readily initiated apoptosis program unless they productively rearrange Ig V region genes on one allele of the IgH locus and are subsequently rescued by survival signals through a functional pre-BCR (1).
For the majority of genes up-regulated in PBCs as compared with HSCs, a role in early B cell development already has been established. However, many other up-regulated genes are known for a specific function, yet unrelated to normal B cell development. These up-regulated genes in PBCs include ATM (nine tags in HSC, 1,109 tags in PBCs), which mediates DNA double-strand break (DSB) repair and whose inactivation by somatic mutation frequently is associated with abnormal rearrangement of Ig and T cell antigen receptor genes and a predisposition to chronic lymphocytic B cell leukemia (19). Comparing the SAGE tag counts for ATM in PBCs to mature naive, germinal center, and memory B cells, the expression of the ATM gene appears to be specific for the PBC stage (N.F., V.S.B., and M.M., unpublished work) and possibly linked to the occurrence of DSBs during V(D)J recombination. Thus, one possible function of stage-specific up-regulation of ATM in normal PBCs could be DSB repair during V(D)J recombination and prevention of aberrant gene rearrangements. SIAH1 (0 tags in HSCs, 73 tags in PBCs), which is known as an apoptosis inducer and tumor suppressor gene (20), was recently shown to degrade OBF-1, a transcriptional activator of the Ig heavy chain locus upon BCR crosslinking (21). Therefore, coexpression of OBF1 (174 tags) with SIAH1 (73 tags) in PBCs might indicate the presence of a negative feedback loop, which could limit the duration of pre-BCR-dependent activation signals. The balance between activation and inhibitory signals may also be affected by PDGFRA (624 tags), which is implicated in tyrosine kinase signaling (22), the MYC-enhancing protooncogene PIM2 (ref. 23; 181 tags), the AKT-inducing protooncogene TCL1 (ref. 24; 34 tags), the IL-6 dependent transcription factor C/EBPB (ref. 25; 66 tags), and WNT16 (ref. 26; 39 tags), a target gene of the B lineage leukemia fusion E2A-PBX1. Differential expression of these genes between HSCs and PBCs has been verified by semiquantitative RT-PCR (Fig. 2). The relatively high levels of expression of these genes in PBCs, as heretofore unrecognized, are potentially meaningful, while their function in early B cell development remains to be clarified.
Figure 2.
Assessment of quantitative accuracy of SAGE data. To corroborate differential gene expression between HSCs and PBCs as assessed by SAGE, semiquantitative RT-PCR was performed for 45 selected genes. The RT-PCR was normalized for 18, 24, and 30 cycles by using a cDNA fragment of the COX6B gene (41 SAGE tags in the HSC and PBC libraries) as a standard. Shown is a comparison of the RT-PCR products (Left) to SAGE tag counts (Center), and the gene names are indicated (Right). For three of 45 genes, no or no specific amplification product was obtained (not shown).
The group of genes that are silenced during or after B cell lineage commitment is much more heterogenous than the genes induced at the PBC stage: transcription factors like HGF (36 copies in HSC), ETS (19 copies), p300/CBPAF (13 copies), and AF4-like (nine copies) are absent in PBCs as well as signaling molecules such as HHOX (11 copies), LAP18 (25 copies), AML3 (eight copies), ABL1 (eight copies), and NUP62 (15 copies). Of note, expression of topoisomerase II (24) and its binding protein (8), the former being implicated in the development of acute myeloid leukemia, can be detected in HSCs but not in PBCs. As expected, typical markers of HSC such as CD34 (19), CD164 (27), and MPO are also missing in the gene expression profile of PBCs.
Within the group of genes that have been silenced during or after commitment to the B cell lineage are also heat shock proteins (HSPs) that are expressed at high abundance in HSCs. This finding not only applies to HSP60 (48 copies in HSCs), HSP90 (29 copies), and HSP70 (16 copies; Table 2) but also to all other members of this family that are expressed in HSCs. We compared the expression levels of HSPs in the PBC SAGE profile to SAGE libraries that have been generated for mature B cell subsets including naïve, germinal center, and memory B cells (N.F., Ines Schwering, V.S.B., Ralf Küppers, and M.M., unpublished work). In naïve, germinal center, and memory B cells, however, expression levels of HSPs are within the same order of magnitude as in HSCs. Silencing of HSP genes, which are implicated in the maintenance of the appropriate conformational protein structure, appears to be specific for PBCs yet unclear in its functional significance.
PBCs as Normal Precursors of B Cell Lineage Leukemia.
Unlike HSCs, PBCs frequently give rise to leukemia, which correlates with the activity of the recombination machinery during B lymphopoiesis. We selected a number of genes, which appear to be relevant in PBCs based on SAGE data and searched for them in recently published microarray profiles on PBC-derived B lineage acute lymphoblastic leukemias (B-ALLs). As expected, the PBX1 gene is strongly expressed in B-ALLs carrying an E2A-PBX1 rearrangement (27) but also up-regulated in normal PBCs (four tags in HSCs, 68 tags in PBCs; Table 2), suggesting that up-regulation of PBX1 resulting from the fusion with E2A as such does not necessarily constitute an oncogenic event. Interestingly, the OBF1 gene, which is strongly expressed in PBCs as compared with HSCs (73 and 0 tags, respectively) is specifically up-regulated in B-ALLs carrying a TEL-AML1 rearrangement and can, thus, distinguish this subentity from B-ALLs harboring other translocations (27). In another study, the OBF1 gene could also discriminate B-ALLs carrying an MLL-AF4 fusion, in which expression of OBF1 was entirely missing, from other types of B-ALLs (28). Indeed, the MLL-AF4 rearrangement confers a mixed-lineage leukemia phenotype, i.e., partial loss of PBC identity of the malignant clone, suggesting that fidelity to the B cell lineage correlates with the degree of OBF1 expression in normal and leukemic PBCs. The ATM gene, which is almost exclusively and at high levels expressed in PBCs, was identified as a marker to differentiate B-ALLs carrying a BCR-ABL1 gene rearrangement from others B-ALLs (27). Contrasting BCR-ABL+ B-ALLs, however, the ABL1 gene, which is strongly expressed in the leukemia cells as a result of the translocation, is silenced in PBCs (eight tags in HSCs, 0 tags in PBCs; Table 2).
To corroborate quantitative differences of gene expression by using a complementary method, we performed semiquantitative RT-PCR for 45 selected genes (Fig. 2). In this experiment, cDNAs from umbilical cord blood HSCs and PBCs (i.e., cell populations that were not subjected to SAGE analysis) were used as templates. Among 45 genes tested, amplified fragments of the expected size were obtained in 42 cases. The intensity of the RT-PCR bands reflected the quantitative differences between HSCs and PBCs (as tag counts in the SAGE analysis) for all 42 informative cases (Fig. 2). We conclude that the vast majority of quantitative differences observed in the comparison of SAGE profiles for HSCs and PBCs are reproducible by using an alternative method.
For graphic representation of quantitative differences of gene expression within groups of genes of similar function, the SAGE data have been arranged within functional clusters and sorted according to the ratio of the frequency of SAGE tags in PBCs and HSCs (Fig. 3). As expected, positive mediators of pre-BCR signaling and costimulatory molecules are exclusively or predominantly expressed in PBCs, with the exception of SOS1 (Fig. 3A), which is part of the proximal signaling cascade after antigen-receptor engagement. Analyzing SAGE libraries from mature naïve and memory B cells, we indeed find high expression levels of SOS1 (N.F., V.S.B., and M.M., unpublished work), suggesting that SOS1 is a constituent of BCR signaling in mature but not of pre-BCR signaling in PBCs. Conversely, many inhibitory molecules or negative regulators of antigen-receptor signaling are not specifically expressed in PBCs and also found in HSCs (Fig. 3B), suggesting that molecules known for their negative regulatory role in antigen receptor signaling of mature B cells are in fact not lineage specific in all instances. To identify molecules that distinguish progenitors of the B cell lineage from the uncommitted hematopoietic progenitor cell, we compared the expression levels of genes that have been implicated in the commitment to the B, T, or myeloid lineages. Intriguingly, most of these lineage-specific genes overexpressed in PBCs are linked to pre-BCR signaling or encode components of the pre-BCR (Fig. 3C), suggesting that signals through the pre-BCR are of particular importance in the maintenance and further development of B cell identity. Markers specific for the T cell (CD2, CD3, CD5) and the myeloid cell (CD33, CD164, MPO) lineages are down-regulated in PBCs, confirming that the expression of cross-lineage markers does not significantly contribute to the PBC phenotype.
Figure 3.
Functional clustering of SAGE data in HSCs and PBCs. Shown is the graphic representation of SAGE data from HSC and PBC SAGE libraries on a color scale from black (no or low expression) to red (high expression levels). The data are arranged within functional clusters for costimulatory molecules and positive regulators of pre-BCR signaling (A), inhibitory molecules and negative regulators of pre-BCR signaling (B), lineage-specific molecules (C), and constituents of pre-BCR and genes related to V(D)J gene recombination (D) and are sorted based on the ratio of SAGE tag counts in PBCs and HSCs.
Seemingly contradicting these observations, known components of the BCR including Igκ and Igλ light chain variable and constant genes, Ig heavy chain α and γ constant genes are underrepresented in the PBC SAGE library (Fig. 3D). The expression of Igκ and Igλ light chains and Ig heavy chain constant genes are regulated in a stage-specific manner: expression of Igκ and Igλ light chain genes defines a late stage of B cell development within the bone marrow (see below), expression of Cγ and Cα genes requires class switch recombination, which takes place during the germinal center reaction of mature B cells. Therefore, absence of expression of Igκ and Igλ light chain and IgCα and IgCγ genes in PBCs was indeed expected.
Rearrangement and Expression of Ig Light Chain Genes.
In mice, several functionally distinct subsets within the PBC pool have been identified (8). Whereas murine pre-BI cells (nomenclature according to Melchers and Rolink; ref. 8) exhibit ongoing rearrangement of Ig heavy chain genes together with surrogate light chain expression, large cycling pre-BII cells (8) express a pre-BCR in the absence of an active recombination machinery. A second wave of recombination acting within the Ig light chain loci is initiated at the small resting pre-BII cell (8) stage with subsequent expression of Ig light chain genes in mice. A recent microarray study on gene expression profiling of early B cell development in mice indeed shows significant expression levels of Igκ light chain genes notably in small resting pre-BII cells and—although to lesser extent—in large cycling pre-BII cells (29). The strong expression of Vpre-B and λ5 surrogate light chain genes in the absence of Igκ and Igα light chain gene expression in the human CD10+ CD19+ CD20− PBCs studied here diverges from the finding in mice (8, 29) and suggests that human PBCs are more homogenous than their murine equivalent. One explanation could be that in human PBCs—unlike in murine small resting pre-BII cells—Igκ and Igλ light chain genes are being rearranged but not expressed before the immature B cell stage. Ig light chain expression might be linked to a phenotype change (i.e., loss of CD10 and/or gain of CD20 expression). In this analysis, CD10low CD19+ CD20+ immature B cells, i.e., the progeny of PBCs and a known source of Ig light chain expression, have been depleted during purification of human PBCs. Thus, further investigation will clarify whether Ig light chain expression in the human occurs first in late PBCs or in immature B cells.
Supplementary Material
Acknowledgments
We thank Klaus Rajewsky and Ralf Küppers for critical discussions and Stefanie Jauch for excellent technical assistance. We thank Stephan Morris for thoughtful review of the manuscript. This work was supported by the Köln Fortune Program/Faculty of Medicine, University of Cologne (to M.M.), the Cancer Research Institute (tumor immunology program; to M.M.), the Emmy-Noether Program of the Deutsche Forschungsgemeinschaft (to M.M.), National Institutes of Health Grants CA 78862-01 (to J.D.R. and S.M.W.) and CA 84405 (to J.D.R.), American Cancer Society Grant IRG-41-40 (to S.M.W.), and the G. Harold and Lelia Y. Mathers Foundation (to S.M.W.).
Abbreviations
- B-ALL
B lineage acute lymphoblastic leukemia
- BCR
B cell receptor
- SAGE
serial analysis of gene expression
- GLGI
generation of longer 3′ expressed sequence tags from SAGE tags for gene identification
- HSP
heat shock protein
- HSC
hematopoietic stem cell
- PBC
pre-B cell
- RT-PCR
reverse transcriptase–PCR
References
- 1.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Kondo M, Weissmann I L, Akashi K. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 3.Tonegawa S. Nature (London) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 4.Allman D, Li J, Hardy R R. J Exp Med. 1999;189:735–740. doi: 10.1084/jem.189.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nutt S L, Heavey B, Rolink A G, Busslinger M. Nature (London) 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 6.Bain G, Robanus Maandag E C, te Riele H P, Feeney A J, Sheehy A, Schlissel M, Shinton S A, Hardy R R, Murre C. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 7.Lin H, Grosschedl R. Nature (London) 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 8.Melchers F, Rolink A. In: Fundamental Immunology. Paul W E, editor. Philadelphia: Lippincott–Raven; 1999. pp. 183–224. [Google Scholar]
- 9.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karasuyama H, Rolink A, Shinkai Y, Young F, Alt F W, Melchers F. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 11.LeBien T W. Blood. 2000;96:9–23. [PubMed] [Google Scholar]
- 12.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 13.Zhou G, Chen J, Lee S, Clark T, Rowley J D, Wang S M. Proc Natl Acad Sci USA. 2001;98:13966–13971. doi: 10.1073/pnas.241526198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müschen M, Re D, Jungnickel B, Diehl V, Rajewsky K, Küppers R. J Exp Med. 2000;192:1833–1840. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müschen M, Rajewsky K, Bräuninger A, Baur A S, Oudejans J J, Roers A, Hansmann M L, Küppers R. J Exp Med. 2000;191:387–394. doi: 10.1084/jem.191.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Zhou G, Clark T, Chen J, Rowley J D, Wang S M. Proc Natl Acad Sci USA. 2001;98:3340–3345. doi: 10.1073/pnas.051013798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J-J, Rowley J D, Wang S M. Proc Natl Acad Sci USA. 2000;97:349–353. doi: 10.1073/pnas.97.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staudt L M. Annu Rev Med. 2002;53:303–318. doi: 10.1146/annurev.med.53.082901.103941. [DOI] [PubMed] [Google Scholar]
- 19.Stankovic T, Weber P, Stewart G, Bedenham T, Murray J, Byrd P J, Moss P A, Taylor A M. Lancet. 1999;353:26–29. doi: 10.1016/S0140-6736(98)10117-4. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Stevens J, Rote C A, Yost H J, Hu Y, Neufeld K L, White R L, Matsunami N. Mol Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 21.Boehm J, He Y, Greiner A, Staudt L M, Wirth T. EMBO J. 2001;20:4153–4162. doi: 10.1093/emboj/20.15.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heidaran M A, Pierce J H, Lombardi D, Ruggiero M, Gutkind J S, Matsui T, Aaronson S A. Mol Cell Biol. 1991;11:134–142. doi: 10.1128/mcb.11.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haupt Y, Alexander W S, Barri G, Klinken S P, Adams J M. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 24.Virgilio L, Narducci M G, Isobe M, Billips L G, Cooper M D, Croce C M, Russo G. Proc Natl Acad Sci USA. 1994;91:12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McWhirter J R, Neuteboom S, T, Wancewicz E V, Monia B P, Downing J R, Murre C. Proc Natl Acad Sci USA. 1999;96:11464–11469. doi: 10.1073/pnas.96.20.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeoh E J, Ross M E, Shurtleff S A, Williams W K, Patel D, Mahfouz R, Behm F G, Raimondi S C, Relling M V, Patel A, et al. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong S A, Staunton J E, Silverman L B, Pieters R, den Boer M L, Minden M D, Sallan S E, Lander E S, Golub T R, Korsmeyer S J. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann R, Seidl T, Neeb M, Rolink A, Melchers F. Genome Res. 2002;12:98–111. doi: 10.1101/gr.201501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.