Abstract
Rett syndrome (RTT) is a postnatal neurodevelopmental disorder characterized by the loss of acquired motor and language skills, autistic features, and unusual stereotyped movements. RTT is caused by mutations in the X-linked gene encoding methyl-CpG binding protein 2 (MeCP2). Mutations in MECP2 cause a variety of neurodevelopmental disorders including X-linked mental retardation, psychiatric disorders, and some cases of autism. Although MeCP2 was identified as a methylation-dependent transcriptional repressor, transcriptional profiling of RNAs from mice lacking MeCP2 did not reveal significant gene expression changes, suggesting that MeCP2 does not simply function as a global repressor. Changes in expression of a few genes have been observed, but these alterations do not explain the full spectrum of Rett-like phenotypes, raising the possibility that additional MeCP2 functions play a role in pathogenesis. In this study, we show that MeCP2 interacts with the RNA-binding protein Y box-binding protein 1 and regulates splicing of reporter minigenes. Importantly, we found aberrant alternative splicing patterns in a mouse model of RTT. Thus, we uncovered a previously uncharacterized function of MeCP2 that involves regulation of splicing, in addition to its role as a transcriptional repressor.
Keywords: Rett syndrome, Y box-binding protein 1
Approximately one of every 10,000 females will develop Rett syndrome (RTT), a severely disabling neurodevelopmental disease. Despite this high incidence and the uniqueness of its characteristics, RTT remained unrecognized by most clinicians as a distinct syndrome until 1983 (1). In fact, the first description of two girls with RTT, by the pediatrician Andreas Rett, was overlooked until Hagberg and colleagues reported (in English, instead of German, as Rett did in 1966) on 35 patients with this distinctive disorder (1, 2). RTT patients appear normal from birth to ≈6–18 months, although detailed examination of “presymptomatic” records uncovered some nonspecific movement abnormalities (3). During this initial period, they achieve the expected intellectual and motor milestones. Depending on their age of onset, they might learn to sit, crawl, talk and interact with others. After this apparent normal development, they fail to acquire new milestones and enter a distinctive period of regression during which language skills are lost and movement abnormalities are developed, including ataxia and gait apraxia. Stereotyped and almost incessant hand-wringing movements replace purposeful use of the hands. These patients also exhibit impairment in social interactions, develop seizures, breathing dysfunction, deceleration of head growth, and scoliosis. After this period of regression, patients stabilize but fail to acquire new skills. The majority of patients survive to adulthood but may also develop additional neurological abnormalities, such as Parkinsonian features (4–6). Atypical presentations of RTT also occur, including milder and more severe forms of the classic presentation. Patients with more severe phenotypes present without the period of normal development and have congenital hypotonia and infantile spasms. Patients with a milder variant may retain some speech and motor functions and do not have seizures (4–6). This vast phenotypic variability is mainly the result of different mutation types and locations and distinct patterns of X chromosome inactivation.
Initially, RTT was considered to be exclusively a girls' disease because of X dominance and lethality in hemizygous males (7). However, since the discovery of disease-causing mutations in MECP2, the X linked gene encoding for methyl-CpG-binding protein 2 (MeCP2) (8), several RTT boys have been described (9–11). The range of phenotypes presented in these males is even broader than those seen in girls. MECP2 mutations that would cause classic RTT in girls produce lethal neonatal encephalopathy in males. Classic RTT in male patients is seen almost exclusively in cases that have aneuploidy such as an XXY karyotype or are mosaics for MECP2 mutations (4, 11). Mutations that cause mild mental retardation or no phenotype in female carriers cause severe mental retardation, seizures, tremors, and spasticity in male patients (9, 12).
In addition, mutations in MECP2 have been linked to a broader class of human developmental disorders, including Angelman-like syndrome and autism (13–17). In these cases, favorable X chromosome inactivation patterns typically explain either partial or milder phenotypes (17, 18). These findings, together with the discovery that MeCP2's abundance during postnatal development correlates with synapse formation, underscore the importance of MeCP2 for neuronal function (19–21). The specific functions of this protein, however, have not been completely elucidated, and it is not clear how MECP2 mutations cause neuronal dysfunction.
MeCP2 was originally identified based on its ability to bind DNA containing methylated CpG dinucleotides (22). MeCP2 localizes to heterochromatin (23) and acts as a methylation-dependent transcriptional repressor (24). In vitro studies identified two functional domains, the methyl-CpG-binding domain that binds methylated DNA and the transcriptional repressor domain (TRD) that induces long-range repression of gene expression. The TRD associates with a corepressor complex containing Sin3A and Brahma and histone deacetylases, indicating that deacetylation of histones (and/or other proteins) is an essential component of its repressive activity (25, 26). Efforts to identify MeCP2 target genes, however, had limited success. Most notably, transcriptional profiling of RNAs from mice lacking Mecp2 and wild-type controls failed to identify significant gene expression changes despite a dramatic phenotype (27). More recently, some targets of MeCP2 regulation have been identified, including BDNF, REST, Dlx5, and several genes regulated by glucocorticoid (28–31). Mechanistically, however, MeCP2 seems to act differently on these targets. BDNF was identified as an activity-dependent target (28, 29), whose transcriptional repression depends on MeCP2's binding directly to one of its cognate promoters, whereas Dlx5 imprinting-related silencing depends on MeCP2 forming a silent chromatin loop (30). Furthermore, in some instances, binding of MeCP2 and its associated corepressors, does not prevent promoter activation. It has been shown, for example, that the thyroid hormone-induced transcriptional activation of carbonic anhydrase II does not require dislodging of the MeCP2–HDAC2 complex from its promoter (32). Thus, it is becoming clear that MeCP2 has the potential to act differently depending on the molecular context, begging a thorough and unbiased functional analysis.
Therefore, we sought to identify proteins that interact with MeCP2 to gain new insight about its molecular functions and as an attempt to reveal mechanisms of pathogenesis in RTT. Through coimmunoprecipitation and mass spectrometry analysis, we identified the protein Y box-binding protein 1 (YB-1, also known as p50, dbpB, MSY-1, Nsep1, and EF1A) as a MeCP2 partner. YB-1 is involved in many DNA- and RNA-dependent events and is one of the most evolutionarily conserved nucleic acid-binding proteins. It has many cellular functions including regulation of transcription, regulation of translation, DNA repair, and response to stress (33). We investigated the functional significance of this interaction and discuss the possible consequences for RTT pathogenesis.
Materials and Methods
Plasmids. We cloned various domains of MeCP2 into the pcDNA3.1 vector (Invitrogen) by PCR with appropriate sets of primers. The minigene splicing reporters used include a cytomegalovirus (CMV), herpes simplex virus (HSV), or progesterone responsive (PRE2-TATA) promoter and have been previously described: (CMV)-CD44 (34), (CMV)-CD44 ACE3 (34), PRE-CD44 (35), and HSV-CT/CTGRP (35). The CD44-ACE3 construct contains several point mutations in the third A/C-rich exon enhancer (ACE) element of CD44 (34). Other plasmids used for transfections include pcDNA3.1-YB-1 (34) and PCR3.1-PR (35).
Immunoprecipitations and Western Blot Analysis. We transiently transfected HeLa or Neuro2A cells in culture dishes (10-cm diameter) with MeCP2 constructs or pcDNA3.1 (20 μg) and collected cells 36–48 h after transfection. We lysed cells in IPH buffer (1 ml; 50 mM Tris·HCl, pH 8.0/150 mM NaCl/5 mM EDTA/0.5% Nonidet P-40/0.1 mM PMSF) at 4°C for 30 min and removed debris by centrifugation. The cleared lysate was then subjected to preclearance for 2 h with unrelated antibodies, followed by immunoprecipitation for 2 h at 4°C by using 30 μl of a 50% slurry of anti-FLAG antibody attached to agarose beads (ANTI-FLAG M2 Affinity Gel, Sigma) or ANTI-HA agarose conjugate (Clone HA-7, Sigma). Precipitates were washed eight times with IPH buffer (1 ml), eluted into Laemmli buffer, separated on 4–15% gradient SDS/PAGE and stained with Coomassie blue (BioSafe Coomassie, Bio-Rad) or blotted onto nitrocellulose.
The primary antibodies used in this work were as follows: rabbit polyclonal raised against YB-1 (a generous gift from T. A. Cooper, Baylor College of Medicine, 1:2,000 for Western blot); rabbit polyclonal against MeCP2 (C terminus) (Upstate Biotechnology, 1:1,000 for Western blot); rabbit polyclonal against MeCP2 (N terminus) (a generous gift from M. Esteller, Spanish National Cancer Centre, Madrid, 1:40 for immunoprecipitation); and monoclonal anti-Flag antibody (1:5,000 M2, Kodak).
Identification of Proteins by Mass Spectrometry. Coomassie blue-stained protein bands were in-gel digested with trypsin, and the recovered peptides were analyzed by using an Applied Biosystems Voyager DE-STR MALDI-TOF to acquire tandem MS (MS/MS) spectra. Data derived from the MS/MS spectra were used to search a compiled protein database.
Size Exclusion Chromatography. Nuclear extracts were prepared by adapting the method of Franke et al. (36). Details of nuclear extract preparation and Superose 6 fractionation are in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
DNase and RNase Treatments. Protein lysates were treated with 200 units of DNase I (New England Biolabs) or with 25 μl of RNase A (500 units/ml)/T1 (20,000 units/ml) (Ambion) for 2 h at 23°C.
Northern Blot. Total RNA isolated by using TRIzol (Invitrogen) was hybridized against PCR-amplified DNA probes for the NR1 subunit (37). The primers used to amplify the fragments to be used as probes are in Supporting Materials and Methods.
Splicing Assays. For in vivo splicing assays, HeLa cells were plated in six-well plates and cultured 24 h in DMEM plus 10% FBS before plasmid transfection. Cells were grown to 60–70% confluence and then transiently cotransfected by using Lipofectamine 2000 Reagent (Invitrogen) according to manufacturer's instructions, with 200 ng of splicing reporter, 200 ng of effector (pcDNA3.1, MeCP2, or MeCP2 J3–1), and 100 ng of either YB-1 or pcDNA3.1. After 16 h, the cells were washed and maintained in fresh DMEM plus 10% FBS and then harvested 24–36 h later. The splicing assays with the steroid responsive PRE-CD44 minigene included the addition of 10 ng of progesterone receptor to transfection, and the cell culture media substituted with DMEM without phenol red, supplemented with 10% charcoal-stripped FBS. The media was supplemented with 10–8 M progesterone or vehicle (ethanol). All transfections were performed in triplicate, and each experiment was repeated four times. For splicing analysis on reporter minigenes, total RNA was treated with DNase, heat inactivated, and an aliquot was subjected to single-step low-cycle RT-PCR by using the Access RT-PCR system (Promega), according to the manufacturer's instructions. Primers and RT-PCR conditions have been described (34, 35). PCR cycles are limited to 20 cycles, previously determined to be within the linear detection range for all splicing reporters used. Primers used were radiolabeled with γ-32P ATP and T4 kinase (Invitrogen), according to the manufacturer's instructions. Products of the RT-PCR were separated by using a 5% nondenaturing polyacrylamide gel, dried, and then exposed to film or quantified by using a PhosphorImager system (Molecular Dynamics).
UV Cross-Linking of Ribonucleoprotein Complexes. Synthesis of RNA and UV cross-linking have been described in ref. 34. In short, nuclear extracts were prepared from 4-month-old mice brains and then incubated with CD44 precursor RNA that was uniformly radiolabeled by using α-32P UTP (50,000 cpm per reaction) for 5 min at 30°C, spiked with 3 μg of heparin, then UV cross-linked (4 cm from Philips G15T8 lamp) for 20 min at 4°C. After cross-linking, the extract was treated with RNase A for 45 min at 37°C, the reaction was diluted in PXL (1× PBS/0.1% SDS/0.5% deoxycholate/0.5% Nonidet P-40) and loaded into a 10% polyacrylamide gel (NuPAGE Bis-Tris with Mops running buffer, Invitrogen) in 1× LDS loading buffer (Invitrogen)
Splicing Microarray. Custom oligonucleotide microarrays were purchased from Agilent Technologies. We designed these arrays to monitor the expression of 19,763 genes (one probe each), 10,492 alternative 5′ and 3′ ends (one probe each), and 20,383 internal alternative splicing events (two probes per event, one reporting on the inclusion and one on the exclusion), and additional probes reporting on all exons and junctions of prioritized genes (e.g., Mecp2). Alternative splicing was defined by aligning mouse transcripts to the genome. Genomic sequence was used to create exon (60-mer) and exon-exon junction (36-mer on 10-nt T stilts) probes (38, 39). PolyA-purified mRNA was amplified by using a full-length amplification method by using random-priming sequences to reproduce the entire transcript (38). Fluorescent dye-labeling, Cy3 and Cy5, hybridization conditions, and scanning were as described in refs. 38–40. Each amplified sample was hybridized twice in a dye-swap experiment. Log10 ratio-to-pool values and error estimates were calculated as described in ref. 41. Pooled mutant and wild-type cerebral cortex samples were hybridized against each other. Five mutant and five wild-type mice brain samples were individually hybridized against a common reference composed of pooled wild-type mouse brain tissue.
Real-Time PCR. Reverse transcription was performed with random hexamers. PCR were performed at melting temperature 58°C in a Applied Biosystems 7300 Real-Time PCR System, with FAM-labeled primers and TAMRA- or BlackHole-labeled probes. A different set of primers/probes was designed to detect the individual isoforms considered.
Results
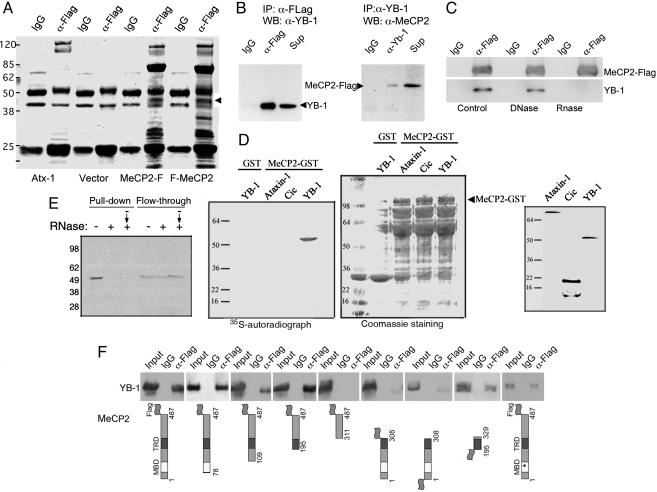
MeCP2 Interacts with YB-1. We purified MeCP2-associated proteins from total cell extracts by one-step coimmunoprecipitation (co-IP). HeLa (human epithelial cell line) and Neuro2A (mouse neuroblastoma cell line) cells transiently expressing tagged versions of human MeCP2 were subjected to co-IP with anti-tag antibodies. FLAG or HA tags were fused either to the amino or carboxyl termini of hMeCP2. To identify proteins that interact with MeCP2 specifically, we determined which proteins coimmunoprecipitated with MeCP2 independent of the type or position of tag or origin of cell line. Proteins fulfilling these criteria and detected by Coomassie blue staining (Fig. 1A) were analyzed by mass spectrometry (42). Western blot analysis of the immunoprecipitate confirmed the presence of Sin3A (data not shown), thus validating the procedure. We focused our effort on characterization of a 50-kDa band, identified as YB-1. Immunoblotting of the immunoprecipitates with anti-YB-1 antibodies confirmed its identity (34). To further confirm the MeCP2–YB-1 interaction, we carried out reciprocal co-IPs: immunoprecipitation of YB-1 could coprecipitate FLAG-tagged MeCP2 in Neuro2A cells and anti-FLAG antibodies immunoprecipitated endogenous YB-1 (Fig. 1B). DNaseI treatment did not modify the MeCP2–YB-1 association, excluding the possibility that this interaction is mediated by DNA (Fig. 1C). However, MeCP2 was unable to co-IP YB-1 after RNase A/T1 treatment, indicating that the MeCP2–YB-1 complex requires RNA for its formation or stabilization (Fig. 1C). The interaction was confirmed by GST pull-down assays by using a GST-MeCP2 fusion protein and in vitro-synthesized YB-1. As shown in Fig. 1D, full-length MeCP2 expressed as a GST fusion pulled down YB-1. Interestingly, GST-MeCP2 was unable to pull down either ataxin-1, a protein that can bind RNA (43, 44), or capicua, a transcriptional repressor (45) (Fig. 1D), despite comparable protein loading, visualized by Coomassie blue staining (Fig. 1D) and comparable input of in vitro-transcribed and -translated (IVTT) products (Fig. 1D Right). This result indicates that the ability of MeCP2 to interact with RNA binding proteins is not generalized but specific for YB-1. The RNA dependence of the complex was also observed in this assay; pretreatment of the IVTT YB-1 with RNase prevented MeCP2-GST's pull down of YB-1 (Fig. 1E). This result also suggests that the intervening RNA molecule should be present in the reaction mixture. Addition of RNase after the complex was allowed to form, by incubating MeCP2-GST and IVTT YB-1 before treatment, was enough to disrupt the interaction (Fig. 1E), indicating a requirement of RNA for the maintenance of the interaction.
Fig. 1.
YB-1 is a MeCP2-associated protein. (A) Coomassie blue staining of anti-FLAG immunoprecipitates from Neuro2A cells transiently transfected with C- or N-terminally FLAG-tagged MeCP2, MeCP2-F, and F-MeCP2, respectively. The arrowhead indicates the 50-kDa band corresponding to YB-1 that is not immunoprecipitated from cells transfected with empty vector or with a plasmid expressing an unrelated nuclear protein (Atx-1, ataxin-1). (B) FLAG-tagged MeCP2 was transfected into Neuro2A cells, immunoprecipitated with unrelated (IgG, anti-HA), anti-FLAG (Left), or anti-YB-1 (Right) antibodies. The immunoprecipitates were analyzed by Western blot for YB-1 (Left) or MeCP2 (Right). Sup, supernatant. (C) Western blot analysis for the identification of MeCP2-FLAG or YB-1, in anti-FLAG immunoprecipitates from Neuro2A cells transfected with MeCP2-FLAG, with or without DNase or RNase treatment of the extracts. RNase treatment disrupts the MeCP2–YB-1 complex. (D) MeCP2-GST was incubated with IVTT, YB-1, or the unrelated proteins ataxin-1 and capicua. MeCP2 interacts specifically only with YB-1, despite comparable protein loading, visualized by Coomassie blue staining (GST-MeCP2 runs at ≈100 kDa) and comparable input of IVTT products (Right). (E) Pretreatment of the IVTT YB-1 with RNase before incubation with MeCP2-GST (+) and treatment of the incubated mixture, including MeCP2-GST and IVTT YB-1, (– → +) prevents YB-1's pull down by MeCP2-GST. (F) The region of MeCP2 that mediates the interaction with YB-1 was identified by anti-FLAG immunoprecipitation from Neuro2A cells transfected with FLAG-tagged MeCP2 deletion constructs.
To determine whether the MeCP2–YB-1 complex requires previous binding to methylated DNA to be established, we explored the effects of a mutation that completely disrupts MeCP2's ability to bind methylated DNA (46). MeCP2 containing a missense mutation in the methyl-CpG-binding domain (R106W) coimmunoprecipitated YB-1 and wild-type MeCP2, suggesting that the interaction is independent of methylated DNA binding (Fig. 1F, rightmost image). We next examined which domains of MeCP2 are required for its interaction with YB-1 (Fig. 1F). A series of N-terminally deleted MeCP2 mutants up to amino acid residue 195, interacted with YB-1 similarly to wild-type MeCP2. No interaction was observed when the N-terminal deletion was extended to amino acid 310, suggesting that the interaction domain resides between amino acid 195 and 310, which corresponds to the TRD domain of MeCP2. The amount of YB-1 coimmunoprecipitated by a C-terminal truncation of MeCP2 (MeCP2-308), which retains the TRD, is only 30–50% of wild-type MeCP2, indicating that sequences beyond the TRD are important for the interaction with YB-1. We therefore tested a fragment of MeCP2 that includes the TRD and surrounding sequences (195–329), which interacted with YB-1 with similar affinity as wild-type MeCP2. These results demonstrate that the region of MeCP2 between amino acid 195 and 329 is required for its interaction with YB-1.
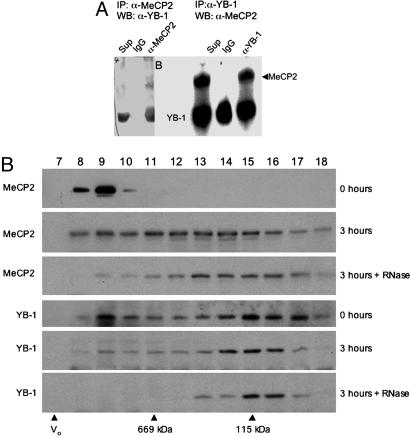
To establish whether YB-1 interacts with endogenous MeCP2, we performed reciprocal co-IP experiments in untransfected CEM-CCRF (human leukemia cell line) cells, which have detectable levels of MeCP2 by Western analysis; MeCP2 is almost undetectable in HeLa or undifferentiated Neuro2A cells (47). Anti-MeCP2 antibody (48) immunoprecipitated endogenous YB-1, and anti-YB-1 antibody immunoprecipitated endogenous MeCP2 (Fig. 2A). Furthermore, by using mouse brain extracts, we found that YB-1 and MeCP2 cofractionate through Superose 6 sizing column as a large complex (>1 MDa), suggesting that this interaction occurs in vivo. Notably, RNase treatment of the brain nuclear extracts before size fractionation results in a dramatic shift of both MeCP2 and YB-1 to smaller sizes, suggesting both proteins are associated with RNA in vivo. After RNase treatment, YB-1 peaks at ≈100 kDa, whereas MeCP2 peaks at ≈400 kDa, indicating that RNA is required for the cofractionation of these proteins (Fig. 2B).
Fig. 2.
MeCP2 and YB-1 interact in vivo in an RNA-dependent manner. (A) Endogenous co-IP of YB-1 and MeCP2 from CEM-CCRF cells. Immunoprecipitation with an unrelated antibody (IgG, anti-HA), anti-N terminus MeCP2 antibody (Left), or anti-YB-1 (Right) antibodies was followed by Western blotting with anti-YB1 (Left) or anti-C terminus MeCP2 (Right) antibody. (B) Western blot analysis for MeCP2 and YB-1 of fractions 7–18 from Superose 6 size exclusion chromatography of mouse brain nuclear extracts. Fraction numbers are indicated on top; void volume and standards are indicated at the bottom. Extracts were subjected to chromatography immediately upon thawing (0 h) or after incubation for 3 h on ice either with (3 h + RNase) or without (3 h) the addition of RNases A and T1, indicated on the right. Endogenous RNase activity in the 3-h incubation likely resulted in some RNA degradation and, thus, a shift in MeCP2 and YB-1 compared with extracts loaded immediately (0 h). This shift is much more dramatic and tight when exogenous RNase is added to the extracts.
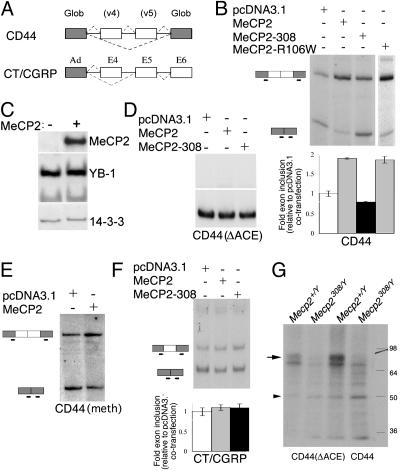
MeCP2 Affects Splicing of Reporter Minigenes. The interaction between MeCP2 and YB-1 suggested a potential functional role for this complex. YB-1 has been shown to regulate alternative splicing of the CD44 minigene through binding to an ACE element and promote the inclusion of the variably spliced exons 4 and 5 (34). Thus, a possible role of the MeCP2–YB-1 complex is to coordinate alternative splicing with gene transcription by recruiting YB-1 to nascent transcripts just as MeCP2 is released from gene promoters. To investigate the functional significance of the MeCP2–YB-1 interaction, we performed minigene-splicing assays in transiently transfected cells (35). In vivo splicing was monitored by low-cycle RT-PCR analysis of total RNA collected from cells transiently transfected with the reporter minigenes and a combination of MeCP2 and/or YB-1 expression vectors. HeLa cells were used for these experiments because of their high efficiency of transfection and the lack of cell-specific differences in the YB-1/MeCP2 interaction (data not shown). We chose to start testing the ability of MeCP2 to modify the splicing patterns of the CD44 minigene, a model system in which concentration-dependent changes of splicing by YB-1 have been established (34). The CD44 minigene used here contains two of the 10 variable exons (v4 and v5) of CD44, which can be either excluded (skipping variant) or included (inclusion variant) during the splicing process, resulting in three major isoforms (Fig. 3A). Cotransfection of MeCP2 resulted in increased inclusion of the variable exons 4 and 5, without observable alterations in transcription (Figs. 3B and 4). Inclusion of both exons increased ≈2-fold (1.90 ± 0.02) compared with the control vector. In contrast, the MeCP2-308 mutant, thereafter referred as MeCP2-308, demonstrated a minimal effect (fold inclusion: 0.79 ± 0.02, Figs. 3B and 4), indicating that the observed effect of wild-type MeCP2 on alternative splicing requires its C-terminal domain. Changes in splicing could be explained either by the modification of the stability of one of the splice variants or by the preferential utilization of splice sites. We ruled out the former by cotransfecting cDNAs for both the inclusion and skipping variants at a fixed ratio. Addition of MeCP2 to the transfection mixture did not alter the ratio of variant production, indicating that the stability of the splice variants remained unchanged in conditions of MeCP2 overexpression (data not shown). Importantly, nuclear levels of YB-1 remained unchanged in HeLa cells overexpressing MeCP2. Subcellular fractionation of transiently transfected cells, followed by Western analysis for the detection of transfected MeCP2 and endogenous YB-1, indicates that nuclear levels of YB-1 are not altered by overexpression of MeCP2. This result rules out the possibility of MeCP2 affecting splicing by inducing degradation or nuclear export of YB-1.
Fig. 3.
MeCP2 regulates splicing of alternative exons. (A) Schematic of the splicing reporter minigenes. (B) HeLa cells were cotransfected with a CD44-derived splicing reporter, together with vector (pcDNA3.1), MeCP2, MeCP2-308, or MeCP2-R106W plasmids. Radiolabeled RT-PCR was performed to detect spliced transcripts. (Upper) Representative RT-PCR autoradiographs. (Lower) Mean (± SD, n ≥ 3) quantifications of the fold exon inclusion, relative to the control DNA. (C) Subcellular fractionation, followed by Western analysis of MeCP2 and endogenous YB-1, indicates that nuclear levels of YB-1 are not altered in MeCP2-transfected cells. (D) HeLa cells were cotransfected with a CD44 minigene deleted for the YB-1 binding element (ΔACE), along with YB-1 and pcDNA3.1, MeCP2, or MeCP2-308. Deletion of the AC-rich element turned the minigene insensitive to cotransfection of wild-type or mutant MeCP2. (E) HeLa cells were cotransfected with an in vitro methylated CD44-derived splicing reporter, together with a vector (pcDNA3.1) or MeCP2. Shown are RT-PCR autoradiographs. In vitro methylation of the CD44 splicing reporter resulted in transcriptional repression by MeCP2, therefore, the amount of the CD44 splicing reporter used in this particular experiments was 10 times the amount used in experiments shown in B and D. The MeCP2-dependent increase in exon inclusion was quantitatively similar to that of the unmethylated CD44 splicing reporter. (F) HeLa cells were cotransfected with a CT/CGRP-derived minigene, along with pcDNA3.1, MeCP2, or MeCP2-308. Splicing of this minigene was not modified by overexpression of MeCP2. (G) 32P-labeled CD44 or CD44(ΔACE) mRNA were incubated with nuclear brain extracts prepared from wild-type or Mecp2308/Y mutant mice, UV irradiated, fractionated by SDS/PAGE, and exposed to x-ray film. Both CD44 mRNAs, wild type and mutant, cross linked to two bands of ≈50 (arrowhead) and 80 (arrow) kDa. The labeled 80-kDa band is absent from the mutant extracts. In addition, the mutation of the ACE element in CD44 decreases its affinity for YB-1 and MeCP2.
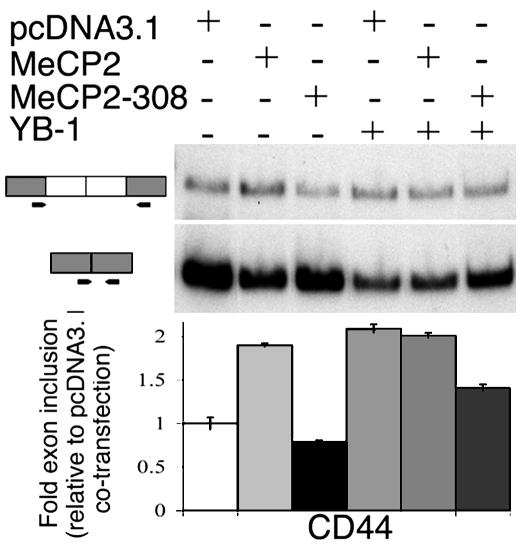
Fig. 4.
HeLa cells cotransfected a CD44 minigene, along with YB-1 and pcDNA3.1, MeCP2, or MeCP2-308. Mutant MeCP2 (MeCP2-308) antagonizes YB-1 exon-inclusion promoting activity. (Upper) Representative RT-PCR autoradiographs. (Lower) Mean (±SD, n ≥ 3) quantifications of the fold exon inclusion, relative to the plasmid control DNA cotransfection.
To determine whether MeCP2's ability to modulate CD44 alternative splicing required DNA binding, we cotransfected a MeCP2 R106W mutant and the CD44 reporter construct. MeCP2 R106W favored exon inclusion to the same extent as the wild-type protein (Fig. 3B). These data, together with the co-IP results, suggested that the effects of MeCP2 on splicing could be mediated by its interaction with YB-1. Deletion of the main YB-1 binding site (ACE) from the CD44 splicing reporter alters its basal splicing (Fig. 3D and ref. 34). This effect of the ACE element deletion is most probably due to the lack of sensitivity of the reporter to the exon inclusion-inducing activity of YB-1, as suggested by the fact that YB-1 cross-linking to the mutated minigene was significantly reduced, whereas overexpression of YB-1 was able to induce exon inclusion to some extent (34). The exon inclusion-promoting activity of MeCP2 on the CD44 minigene was nearly eliminated by deleting the ACE element (Fig. 3D), supporting the notion that YB-1 mediates this activity.
Because the processes of transcription and pre-mRNA splicing are coordinated temporally and spatially (49), we sought to determine whether MeCP2 affected the transcription of the reporter minigene. In vitro methylation of the CD44 splicing reporter resulted in a dose-dependent transcriptional repression by MeCP2. However, the MeCP2-dependent increase in exon inclusion was quantitatively similar to that of the unmethylated CD44 splicing reporter (Fig. 3E). In addition, MeCP2 promoted exon inclusion in a CD44 splicing reporter driven by a steroid-inducible promoter (34) instead of the CMV promoter of the above-mentioned CD44 minigene, suggesting promoter-independent splicing activities of MeCP2 (data not shown).
Alternative splicing decisions are often modulated by the combinatorial binding of both ubiquitous and specific splicing factors to pre-mRNA sequences. Furthermore, splicing factors may show preferential activity toward either alternative cassette exons or alternative terminal exons (50). To determine whether MeCP2 exhibited specificity in its regulation of alternative splicing, we examined other splicing reporters. To examine alternative splicing of an alternative terminal exon cassette, we conducted splicing assays on a CT/CGRP-derived minigene reporter (51). Overexpression of MeCP2 had little effect on altering alternative terminal exon preferences (1.11 ± 0.07 fold, Fig. 3F).
The RNA dependence of the MeCP2–YB-1 complex and the effect of MeCP2 overexpression on the CD44-derived minigene splicing raised the possibility that this complex might include the minigene precursor RNA. We UV cross-linked brain nuclear extracts to radioactively labeled in vitro-transcribed CD44 precursor RNA, subjected the extracts to RNase A treatment, and conducted electrophoretic analysis of the cross-linked proteins. Remarkably, the cross-linked RNA comigrated with proteins of molecular masses (≈50 and 80 kDa) corresponding to those of YB-1 and MeCP2, among others. Under the same conditions, cross-linking of the CD44 precursor RNA with brain extracts obtained from mice carrying a truncating mutation (Mecp2308/Y), form the ≈50-kDa, but not the ≈80-kDa ribonucleoprotein complex, strongly suggesting that the ≈80-kDa complex included MeCP2 (Fig. 3G). To confirm this suggestion, we subjected the RNase treated, cross-linked CD44 mRNA-nuclear extracts to immunoprecipitation by using anti-MeCP2 antibodies (Fig. 7, which is published as supporting information on the PNAS web site). MeCP2 antibodies immunoprecipitated a ribonucleoprotein complex of ≈80 kDa, specifically from Mecp2+/Y but not Mecp2308/Y extracts. This result suggests that MeCP2 and YB-1 reside in the same complex formed on a splicing precursor RNA to influence splicing.
We observed that the MeCP2-308 mutant interacted less efficiently with YB-1 than wild-type MeCP2 (see Fig. 1F); thus, we wanted to determine whether expression of this mutant would affect YB-1 regulation of alternative splicing of the CD44 minigene. As expected, transfection of YB-1 stimulated exon inclusion (2.09 ± 0.05 fold, Fig. 4). However, addition of MeCP2 did not result in a further increase in exon inclusion (2.01 ± 0.03 fold, Fig. 4). The lack of an additive or synergistic effect may be attributed to saturation of exon inclusion by YB-1. Interestingly, MeCP2-308 antagonized YB-1's effect on splicing (1.47 ± 0.04 fold inclusion, Fig. 4), suggesting that the MeCP2-308–YB-1 interaction could generate alterations in the repertoire of alternative transcripts present in cells expressing this mutant. We sought to test this hypothesis by using candidate gene analysis and unbiased screen.
MeCP2 Affects Splicing of NR1. The rationale for selecting a candidate pre-mRNA target of MeCP2 splicing regulation was based on the idea that the regulation of splicing by MeCP2 might be sensitive to neuronal activity (28, 29). We chose the NMDA receptor subunit NR1 as a candidate because an alternative splice site in exon 22 generates variants that express either the C2 or C2′ domains of NR1. Importantly, activity regulates this alternative splicing in cultured cortical neurons (37).
To test whether MeCP2 is involved in splicing of NR1 C-terminal domains, we compared the expression of NR1 mRNA variants differing in the presence of the C2 encoding exon by Northern blot analysis with mRNA obtained from wild-type and MeCP2 null mice (Mecp2–/Y) brain regions. The relative abundance of the NR1 mRNA variants was similar in cerebral cortices of wild-type and Mecp2–/Y mice. However, differential inclusion of NR1 exon 22 was observed in mRNA obtained from subcortical brain tissue (Fig. 5). These data suggest that NR1 pre-mRNA might be a tissue-specific target of MeCP2 splicing regulation.
Fig. 5.
MeCP2 regulates NR1 mRNA splicing. Total RNA from brain regions derived from wild-type or Mecp2–[supi]/Y mice was subjected to Northern analysis. Hybridization with an NR1 probe detects both the C2 and C2′ containing mRNA variants (arrows) and shows different ratios of the two variants in Mecp2–/Y vs. wild type (Mecp2+/Y) specifically in the subcortical samples. Note that both isoforms are equally abundant in wild-type samples, whereas the C2 isoform is more abundant in the mutant samples. No differences were observed in cortex-derived mRNA.
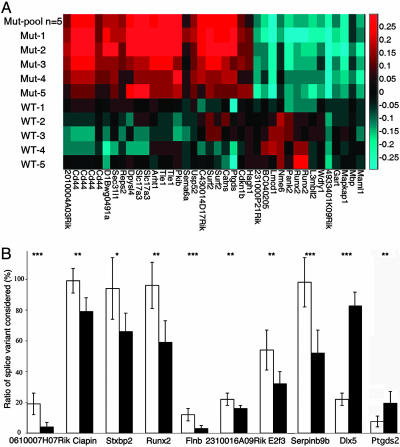
Altered RNA Splicing in a Mouse Model of RTT. To carry out an unbiased search for splicing alterations in endogenous genes, we performed a microarray-based genomewide survey of splicing in cerebral cortex mRNA obtained from wild-type and Mecp2308/Y mice. The Mecp2308/Y mice carry a truncating mutation and reproduce most classical features of RTT (52, 53). Using a full-length mRNA amplification and labeling protocol and custom-designed Agilent microarrays composed of oligonucleotide probes specific to individual exons and exon-exon junctions, we monitored alternative splicing of >14,000 individual variants and >10,000 alternative 5′ and 3′ terminal exons (39). We found alternative splicing to be changed in a significant number of genes in the mutant samples (Fig. 8 and Table 1, which are published as supporting information on the PNAS web site). In addition, supervised clustering with significant splicing changes, including CD44, resulted in a correct classification of the samples according to their genotypes (Fig. 6A). Analysis of the data indicates that most of the splicing changes corresponded to cassette exon changes (99% of splicing changes with a P < 0.0005), which correlates with the cell based-assay results in which MeCP2 promoted exon inclusion. We searched for putative ACE elements in the top 10 genes identified as abnormally spliced. We defined ACE elements as YYYYYYYY, where Y = A or C, and up to two G/T may interrupt the sequence (54). Only ACE sequences within 50 nucleotides from the alternative exon were considered. Interestingly, we found that the alternative exons in the top 10 genes (according to their degree of statistical significance) were enriched by ≈1.5 fold for YB-1 binding sites (ACE) in comparison with their flanking exons (Table 2, which is published as supporting information on the PNAS web site).
Fig. 6.
Aberrant splicing in a mouse model of RTT. (A) Supervised clustering of individual mouse samples based on splice monitoring. For every mutually exclusive splicing event identified, we designed a pair of probes to monitor the inclusion and the exclusion of the event. As an example, a cassette exon would have one probe to monitor the exon and one probe to monitor the junction between adjacent exons. To isolate splicing changes beyond gene expression changes, relative gene expression, as monitored by separate probes, was subtracted from each probe. Plotted is the log10-ratio-to-pool difference between probe pairs. Splicing events were selected that distinguish mutant from wild-type (wt) mouse brain samples with a t test (P < 0.0005). The CD44 variants (second to fifth from left) can distinguish samples. (B) Relative quantification of splice variant-specific mRNA by real-time PCR. The relative abundance of a set of splice variants found to be differentially expressed by microarray analysis were analyzed in cerebral cortices of six mice of wild-type (open bars) or mutant (filled bars) genotypes. Depicted are means of triplicate experiments for six mice of each genotype (±SD). *, P < 0.05; **, P < 0.03; ***, P < 0.01.
Real-time PCR was used to validate candidate transcripts identified by the array. Approximately 35% of the microarray-derived candidate transcripts tested exhibited abnormal splicing in cerebral cortex obtained from a different set of animals than those used to prepare the RNA for the array experiments (Fig. 6B). Interestingly, one of the genes identified as being abnormally spliced in Mecp2308/Y was the homeobox gene Dlx5, a direct target of MeCP2 transcriptional silencing. Dlx5 has at least seven alternative transcripts, some lacking the exon 2-encoded homeodomain (University of California, Santa Cruz, Genome Browser). The splicing event deregulated in brains from Mecp2308/Y mice involves the removal of an alternative intron in the 5′ UTR of the first exon (Fig. 9, which is published as supporting information on the PNAS web site).
Discussion
The involvement of MeCP2 in the etiology of several neurodevelopmental disorders, including Rett and autism, underscores the importance of this protein in neuronal function. However, its function in vivo is far from being entirely understood, precluding a complete understanding of its role in RTT pathogenesis. Identifying interacting partners is a key step to understanding protein function. In fact, several MeCP2 functions have been ascribed based on its identified interacting partners (25, 55, 56).
We discovered that MeCP2 interacts with YB-1, a major component of messenger ribonucleoprotein particles (mRNPs) that belongs to the multifunctional family of DNA/RNA binding proteins containing a highly conserved nucleic acid binding domain (57). In addition to its participation in alternative splicing of mRNA (34, 58), YB-1 functions in the cytoplasm as the main mRNA packaging protein (57), regulates half-life (59) and mRNA template activity in protein synthesis (60, 61). YB-1 has opposite effects on protein synthesis depending on the amount of YB-1 on mRNA, i.e., the YB-1/mRNA ratio (61–64). Interestingly, YB-1 interacts with FMRP, a regulator of mRNA transport and translation that, when absent or defective, gives rise to Fragile X Syndrome (65–67). YB-1 also functions as a transcription factor and regulates expression of genes by binding to a Y box in their promoters (68). In addition, YB-1 is involved in repair and replication of DNA (69–71).
The cofractionation of MeCP2 and YB-1 observed in the size exclusion chromatography from brain nuclear extracts suggests that this interaction occurs in neurons in vivo. Furthermore, both proteins fractionate as belonging to RNA-dependent large molecular weight complexes, consistent with the presence of RNA intervening molecules. The finding that YB-1 is an RNA-dependent MeCP2-interacting protein is interesting in light of findings that several proteins involved in RNA processing are associated with human neurological disorders (58, 72–77). YB-1 controls multiple steps of mRNA processing, including alternative splice site selection (78). Thus, MeCP2, through its association with YB-1, might have RNA splicing-related activities. We found that MeCP2 binds to mRNA from a YB-1-responsive CD44-splicing reporter and promotes exon inclusion. Removal of the ACE element from the CD44 minigene resulted in an abrogation of the MeCP2 exon inclusion enhancement, indicating a requirement of YB-1 for this MeCP2 activity. Notably, aberrant splicing of the endogenous CD44 gene was detected in the splicing-microarray experiments in both the brain (Fig. 6A) and spleen (data not shown) of MeCP2 mutant mice (Mecp2308/Y). This finding suggests a role for MeCP2 in the regulation of the CD44 mRNA splicing in vivo.
Nowhere is the importance of alternative splicing more evident than in the nervous system. Almost all neurotransmitter receptors and channels undergo alternative splicing, and this process represents a key regulatory step in neuronal signaling (79). We identified several genes that are abnormally spliced in brains of Mecp2308/Y mice. The relevance of this finding is twofold: it suggests that MeCP2 has a critical role in regulating alternative splicing in vivo, and it raises the possibility that RTT (and other syndromes associated with MECP2 mutations) might be the result of misregulation of both transcription and splicing. Moreover, splicing alterations could be more consequential than changes in gene expression as defective splicing has the potential to generate proteins with different activities and modify their levels. It is plausible that some alterations in splicing observed in the Mecp2308/Y mice could be due to changes in the expression of splicing factors. However, our findings that MeCP2 alters splicing of a CD44 reporter gene in a sequence-dependent manner, binds the CD44 minigene mRNA, has RNA binding activity (80), and interacts with splicing regulators (YB-1, FBP11, and HYPC) strongly suggest that MeCP2 has a direct role in splicing regulation. Because the R106W mutant, which causes classic RTT but lacks DNA binding, is indistinguishable from wild-type MeCP2 in its interaction with YB-1 and affects splicing of reporter minigenes, it appears that the splicing activity of MeCP2 depends on the genomic context of its microenvironment. For example, MeCP2 and its target minigene are overexpressed in our in vitro assays. The high levels of MeCP2 may permit a spatial relationship between MeCP2 and its splicing targets to be bypassed and allow for binding and splicing modulation to occur in the absence of DNA binding. However, in vivo, the likelihood of MeCP2 encountering its target mRNAs is probably a direct consequence of MeCP2's intranuclear localization. It is well known that the nucleus is functionally compartmentalized (81), and the localization of MeCP2 likely depends on its binding to DNA. We propose that MeCP2 (R106W) does not bind its target DNA sequences, which results in abnormal intranuclear localization, and, thus, its in vivo coordinated regulation of splicing is compromised by this lack of vicinity to the nascent mRNA targets. Our data demonstrating that methylation of the CD44 minigene does not modify MeCP2's modulation of splicing does not disprove the proposed methylation-splicing connection; MeCP2 and its artificial substrates are overabundant in these assays; hence, the putative MeCP2-engaging function of cytosine methylation is not necessary.
Based on the data in this study, we propose that MeCP2 is a multifunctional protein that, in addition to its role as a transcriptional repressor, acts as a splicing regulator. It is tempting to propose that the two activities are economically coordinated, such that when a gene becomes reactivated by releasing MeCP2 from its promoter (through posttranslational modifications, for example), splicing of the nascent transcript is modulated by MeCP2. This proposal is consistent with the recent finding that MeCP2 binds RNA as avidly as it binds methylated DNA and that these two activities are mutually exclusive (80). Moreover, the identification of aberrantly spliced p16INK4a transcripts due to promoter methylation (82, 83) supports a link between DNA methylation and splicing. Interestingly, a recently identified transcriptional target of MeCP2, Dlx5, was identified in our microarray experiments as an in vivo target of MeCP2-dependent splicing regulation. Because MeCP2 binds preferentially methylated CpGs, this hypothesis adds an extra level of complexity, posttranscriptional splicing modulation, for epigenetic control of gene expression.
Supplementary Material
Acknowledgments
We thank Didier Aboeuf and Bert O'Malley for initial splicing experiments and minigene constructs and Tom Cooper and members of the H.Y.Z. laboratory for their critical reading of the manuscript. This work was supported by a Rett Syndrome Research Foundation Research Grant (to J.I.Y.), National Institutes of Health Grants P01 HD40301 and HD24064, and funds from Cure Autism Now (to H.Y.Z.). A.B.B. is a postdoctoral Fellow of the Hereditary Disease Foundation, and H.Y.Z. is an Investigator with the Howard Hughes Medical Institute.
Author contributions: J.I.Y. and H.Y.Z. designed research; J.I.Y., E.P.H., J.C.C., J.C.-B., A.B.B., M.F.R., D.K., R.R., and J.M.J. performed research; J.C.C. and J.M.J. contributed new reagents/analytic tools; J.I.Y., J.C.C., A.B.B., J.M.J., S.B., and H.Y.Z. analyzed data; J.I.Y. wrote the paper; and H.Y.Z. wrote portions of the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: ACE, A/C-rich exon enhancer; co-IP, coimmunoprecipitation; IVTT, in vitro-transcribed and -translated; MeCP2, methyl-CpG binding protein 2; RTT, Rett syndrome; TRD, transcriptional repressor domain; YB-1, Y box-binding protein 1.
References
- 1.Hagberg, B., Aicardi, J., Dias, K. & Ramos, O. (1983) Ann. Neurol., 14, 471–479. [DOI] [PubMed] [Google Scholar]
- 2.Rett, A. (1966) Wien. Med. Wochenschr., 116, 723–726. [PubMed] [Google Scholar]
- 3.Einspieler, C., Kerr, A. M. & Prechtl, H. F. (2004) Pediatr. Res., 57, 696–700. [DOI] [PubMed] [Google Scholar]
- 4.Neul, J. L. & Zoghbi, H. Y. (2004) Neuroscientist 10, 118–128. [DOI] [PubMed] [Google Scholar]
- 5.Weaving, L. S., Ellaway, C. J., Gecz, J. & Christodoulou, J. (2005) J. Med. Genet. 42, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaze, D. G. (2004) Ment. Retard. Dev. Disabil. Res. Rev. 10, 154–158. [DOI] [PubMed] [Google Scholar]
- 7.Zoghbi, H. (1988) J. Child Neurol. 3, S76–S78. [DOI] [PubMed] [Google Scholar]
- 8.Amir, R. E., Van den Veyver, I. B., Wan, M., Tran, C. Q., Francke, U. & Zoghbi, H. Y. (1999) Nat. Genet. 23, 185–188. [DOI] [PubMed] [Google Scholar]
- 9.Meloni, I., Bruttini, M., Longo, I., Mari, F., Rizzolio, F., D'Adamo, P., Denvriendt, K., Fryns, J. P., Toniolo, D. & Renieri, A. (2000) Am. J. Hum. Genet. 67, 982–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orrico, A., Lam, C., Galli, L., Dotti, M. T., Hayek, G., Tong, S. F., Poon, P. M., Zappella, M., Federico A. & Sorrentino, V. (2000) FEBS Lett. 481, 285–288. [DOI] [PubMed] [Google Scholar]
- 11.Van Esch, H., Bauters, M., Ignatius, J., Jansen, M., Raynaud, M., Hollanders, K., Lugtenberg, D., Bienvenu, T., Jensen, L. R., Gecz, J., et al. (2005) Am. J. Hum. Genet. 77, 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couvert, P., Bienvenu, T., Aquaviva, C., Poirier, K., Moraine, C., Gendrot, C., Verloes, A., Andres, C., Le Fevre, A. C., Souville, I., et al. (2001) Hum. Mol. Genet. 10, 941–946. [DOI] [PubMed] [Google Scholar]
- 13.Shibayama, A., Cook, E. H., Jr., Feng, J., Glanzmann, C., Yan, J., Craddock, N., Jones, I. R., Goldman, D., Heston, L. L. & Sommer, S. S. Am. J. Med. Genet. (2004) 128, 50–53. [DOI] [PubMed] [Google Scholar]
- 14.Lynch, S. A., Whatley, S. D., Ramesh, V., Sinha, S. & Ravine, D. (2003) Arch. Dis. Child Fetal Neonatal Ed. 88, F250–F252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carney, R. M., Wolpert, C. M., Ravan, S. A., Shahbazian, M., Ashley-Koch, A., Cuccaro, M. L., Vance, J. M. & Pericak-Vance, M. A. (2003) Pediatr. Neurol. 28, 205–211. [DOI] [PubMed] [Google Scholar]
- 16.Watson, P., Black, G., Ramsden, S., Barrow, M., Super, M., Kerr, B. & Clayton-Smith, J. J. (2001) J. Med. Genet. 38, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammer, S., Dorrani, N., Dragich, J., Kudo, S. & Schanen, C. (2002) Ment. Retard. Dev. Disabil. Res. Rev. 8, 94–98. [DOI] [PubMed] [Google Scholar]
- 18.Young, J. I. & Zoghbi, H. Y. (2004) Am. J. Hum. Genet. 74, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shahbazian, M. D., Antalffy, B., Armstrong, D. L. & Zoghbi, H. Y. (2002) Hum. Mol. Genet. 11, 115–124. [DOI] [PubMed] [Google Scholar]
- 20.Zoghbi, H. Y. (2003) Science 302, 826–830. [DOI] [PubMed] [Google Scholar]
- 21.Kishi, N. & Macklis, J. D. (2004) Mol. Cell. Neurosci. 27, 306–321. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, J. D., Meehan, R. R., Henzel, W. J., Maurer-Fogy, I., Jeppesen, P., Klein, F. & Bird, A. (1992) Cell 69, 905–914. [DOI] [PubMed] [Google Scholar]
- 23.Nan, X., Tate, P., Li, E. & Bird, A. (1996) Mol. Cell. Biol. 16, 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nan, X., Campoy, F. J. & Bird, A. (1997) Cell 88, 471–481. [DOI] [PubMed] [Google Scholar]
- 25.Nan, X, Ng, H. H., Johnson, C. A., Laherty, C. D., Turner, B. M., Eisenman, R. N. & Bird, A. (1998) Nature 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 26.Harikrishnan, K. N., Chow, M. Z., Baker, E. K., Pal, S., Bassal, S., Brasacchio, D., Wang, L., Craig, J. M., Jones, P. L., Sif, S. & El-Osta, A. (2005) Nat. Genet. 37, 254–264. [DOI] [PubMed] [Google Scholar]
- 27.Tudor, M., Akbarian, S., Chen, R. Z. & Jaenisch, R. (2002) Proc. Natl. Acad. Sci. USA 99, 15536–15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, W. G., Chang, Q., Lin, Y., Meissner, A., West, A. E., Griffith, E. C., Jaenisch, R. & Greenberg, M. E. (2003) Science 302, 885–889. [DOI] [PubMed] [Google Scholar]
- 29.Martinowich, K., Hattori, D., Wu, H., Fouse, S., He, F., Hu, Y., Fan, G. & Sun, Y. E. (2003) Science 302, 890–893. [DOI] [PubMed] [Google Scholar]
- 30.Horike, S., Cai, S., Miyano, M., Cheng, J. F. & Kohwi-Shigematsu, T. (2005) Nat. Genet. 37, 31–40. [DOI] [PubMed] [Google Scholar]
- 31.Nuber, U. A., Kriaucionis, S., Roloff, T. C., Guy, J., Selfridge, J., Steinhoff, C., Schulz, R., Lipkowitz, B., Ropers, H. H., Holmes, M. C. & Bird, A. (2005) Hum. Mol. Genet. 14, 2247–2256. [DOI] [PubMed] [Google Scholar]
- 32.Rietveld, L. E., Caldenhoven, E. & Stunnenberg, H. G. (2002) EMBO J. 21, 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohno, K., Izumi, H., Uchiumi, T., Ashizuka, M. & Kuwano, M. (2003) BioEssays 25, 691–698. [DOI] [PubMed] [Google Scholar]
- 34.Stickeler, E., Fraser, S. D., Honig, A., Chen, A. L., Berget, S. M. & Cooper, T. A. (2001) EMBO J. 20, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auboeuf, D., Honig, A., Berget, S. M. & O'Malley B. W. (2002) Science 298, 416–419. [DOI] [PubMed] [Google Scholar]
- 36.Franke, A., DeCamillis, M., Zink, D., Cheng, N., Brock, H. W. & Paro, R. (1992) EMBO J. 11, 2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mu, Y., Otsuka, T., Horton, A., Scott, D. & Ehlers, M. (2003) Neuron 40, 581–594. [DOI] [PubMed] [Google Scholar]
- 38.Schadt, E. E., Edwards, S. W., GuhaThakurta, D., Holder, D., Ying, L., Svetnik, V., Leonardson, A., Hart, K. W., Russell, A., Li, G., et al. (2004) Genome Biol. 5, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R. & Shoemaker, D. D. (2003) Science 302, 2141–2144. [DOI] [PubMed] [Google Scholar]
- 40.Hughes, T. R., Mao, M., Jones, A. R., Burchard, J., Marton, M. J., Shannon, K. W., Lefkowitz, S. M., Ziman, M., Schelter, J. M., Meyer, M. R., et al. (2001) Nat. Biotechnol. 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 41.He, Y. D., Dai, H., Schadt, E. E., Cavet, G., Edwards, S. W., Stepaniants, S. B., Duenwald, S., Kleinhanz, R., Jones, A. R., Shoemaker, D. D. & Stoughton, R. B. (2003) Bioinformatics 19, 956–965. [DOI] [PubMed] [Google Scholar]
- 42.Ogryzko, V. V., Kotani, T., Zhang, X., Schiltz, R. L., Howard, T., Yang, X. J., Howard, B. H., Qin, J. & Nakatani, Y. (1998) Cell 94, 35–44. [DOI] [PubMed] [Google Scholar]
- 43.Yue, S., Serra, H. G., Zoghbi, H. Y. & Orr, H. T. (2001) Hum. Mol. Genet. 10, 25–30. [DOI] [PubMed] [Google Scholar]
- 44.Irwin, S., Vandelft, M., Pinchev, D., Howell, J. L., Graczyk, J., Orr, H. T. & Truant, R. (2005) J. Cell Sci. 118, 233–242. [DOI] [PubMed] [Google Scholar]
- 45.Jimenez, G., Guichet, A., Ephrussi, A. & Casanova, J. (2000) Genes Dev. 14, 224–231. [PMC free article] [PubMed] [Google Scholar]
- 46.Kudo, S., Nomura, Y., Segawa, M., Fujita, N., Nakao, M., Dragich, J., Schanen, C. & Tamura, M. (2001) Brain Dev. 23, S165–S173. [DOI] [PubMed] [Google Scholar]
- 47.El-Osta, A., Kantharidis, P., Zalcberg, J. R. & Wolffe, A. P. (2002) Mol. Cell. Biol. 22, 1844–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballestar, E., Paz, M. F., Valle, L., Wei, S., Fraga, M. F., Espada, J., Cigudosa, J. C., Huang, T. H. & Esteller, M. (2003) EMBO J. 22, 6335–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kornblihtt, A. R., de la Mata, M., Fededa, J. P., Munoz, M. J. & Nogues, G. (2004) RNA 10, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Black, D. L. (2003) Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 51.Lou, H., Helfman, D. M., Gagel, R. F. & Berget, S. M. (1999) Mol. Cell. Biol. 19, 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shahbazian, M., Young, J. I., Yuva-Paylor, L., Spencer, C., Antalffy, B., Noebels, J., Armstrong, D., Paylor, R. & Zoghbi, H. Y. (2002) Neuron 35, 243–254. [DOI] [PubMed] [Google Scholar]
- 53.Moretti, P., Bouwknecht, J. A., Teague, R., Paylor, R. & Zoghbi, H. Y. (2005) Hum. Mol. Genet. 14, 205–220. [DOI] [PubMed] [Google Scholar]
- 54.Coulter, L. R., Landree, M. A. & Cooper, T. A. (1997) Mol. Cell. Biol. 17, 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones, P. L., Veenstra, G. J., Wade, P. A., Vermaak, D., Kass, S. U., Landsberger, N., Strouboulis, J. & Wolffe, A. P. (1998) Nat. Genet. 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 56.Fuks, F., Hurd, P. J., Wolf, D., Nan, X., Bird, A. P. & Kouzarides, T. (2003) J. Biol. Chem. 278, 4035–4040. [DOI] [PubMed] [Google Scholar]
- 57.Skabkin, M. A., Kiselyova, O. I., Chernov, K. G., Sorokin, A. V., Dubrovin, E. V., Yaminsky, I. V., Vasiliev, V. D. & Ovchinnikov, L. P. (2004) Nucleic Acids Res. 32, 5621–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Philips, A. V., Timchenko, L. T. & Cooper, T. A. (1998) Science 280, 737–741. [DOI] [PubMed] [Google Scholar]
- 59.Evdokimova, V., Ruzanov, P., Imataka, H., Raught, B., Svitkin, Y., Ovchinnikov, L. P. & Sonenberg, N. (2001) EMBO J. 20, 5491–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evdokimova, V. M. & Ovchinnikov, L. P. (1999) Int. J. Biochem. Cell Biol. 31, 139–149. [DOI] [PubMed] [Google Scholar]
- 61.Minich, W. B. & Ovchinnikov, L. P. (1992) Biochimie 74, 477–483. [DOI] [PubMed] [Google Scholar]
- 62.Evdokimova, V. M., Kovrigina, E. A., Nashchekin, D. V., Davydova, E. K., Hershey, J. W. & Ovchinnikov, L. P. (1998) J. Biol. Chem. 273, 3574–3581. [DOI] [PubMed] [Google Scholar]
- 63.Nekrasov, M. P., Ivshina, M. P., Chernov, K. G., Kovrigina, E. A., Evdokimova, V. M., Thomas, A. A., Hershey, J. W. & Ovchinnikov, L. P. (2003) J. Biol. Chem. 278, 13936–13943. [DOI] [PubMed] [Google Scholar]
- 64.Davydova, E. K., Evdokimova, V. M., Ovchinnikov, L. P. & Hershey, J. W. (1997) Nucleic Acids Res. 25, 2911–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ceman, S., Nelson, R. & Warren, S. T. (2000) Biochem. Biophys. Res. Commun. 279, 904–908. [DOI] [PubMed] [Google Scholar]
- 66.Khandjian, E. W., Huot, M.-E., Tremblay, S., Davidovic, L., Mazroui, R. & Bardoni, B. (2004) Proc. Natl. Acad. Sci. USA 101, 13357–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagne, J. P., Bonicalzi, M. E., Gagne, P., Ouellet, M. E., Hendzel, M. J. & Poirier, G. G. (August 23, 2005) Biochem. J., 10.1042/BJ20050792. [DOI] [PMC free article] [PubMed]
- 68.Swamynathan, S. K., Nambiar, A. & Guntaka, R. V. (1998) FASEB J. 12, 515–522. [DOI] [PubMed] [Google Scholar]
- 69.Ise, T., Nagatani, G., Imamura, T., Kato, K., Takano, H., Nomoto, M., Izumi, H., Ohmori, H., Okamoto, T., Ohga, T., et al. (1999) Cancer Res. 59, 342–346. [PubMed] [Google Scholar]
- 70.Marenstein, D. R., Ocampo, M. T., Chan, M. K., Altamirano, A., Basu, A. K. Boorstein, R. J., Cunningham, R. P. & Teebor, G. W. (2001) J. Biol. Chem. 276, 21242–21249. [DOI] [PubMed] [Google Scholar]
- 71.Holm, P. S., Bergmann, S., Jurchott, K., Lage, H., Brand, K., Ladhoff, A., Mantwill, K., Curiel, D. T., Dobbelstein, M., Dietel, M., et al. (2002) J. Biol. Chem. 277, 10427–10434. [DOI] [PubMed] [Google Scholar]
- 72.Dansithong, W., Paul, S., Comai, L. & Reddy, S. (2004) J. Biol. Chem. 280, 5773–5780. [DOI] [PubMed] [Google Scholar]
- 73.Buckanovich, R. J., Yang, Y. Y. & Darnell, R. B. (1996) J. Neurosci. 16, 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiledjian, M., Wang, X. & Liebhaber, S. A. (1995) EMBO J. 14, 4357–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Musunuru, K. & Darnell, R. B. (2001) Annu. Rev. Neurosci. 24, 239–262. [DOI] [PubMed] [Google Scholar]
- 76.Ashley, C. T., Jr., Wilkinson, K. D., Reines, D. & Warren, S. T. (1993) Science 262, 563–566. [DOI] [PubMed] [Google Scholar]
- 77.Frugier, T., Nicole, S., Cifuentes-Diaz, C. & Melki, J. (2002) Curr. Opin. Genet. Dev. 12, 294–298. [DOI] [PubMed] [Google Scholar]
- 78.Raffetseder, U., Frye, B., Rauen, T., Jurchott, K., Royer, H. D., Jansen, P. L. & Mertens, P. R. (2003) J. Biol. Chem. 278, 18241–18248. [DOI] [PubMed] [Google Scholar]
- 79.O'Donovan, K. J. & Darnell, R. B. (2001) Sci STKE, PE2. [DOI] [PubMed]
- 80.Jeffery, L.& Nakielny, S. (2004) J. Biol. Chem. 279, 49479–49487. [DOI] [PubMed] [Google Scholar]
- 81.Grigoryev, S. A., Nikitina, T., Pehrson, J. R., Singh, P. B & Woodcock, C. L. (2004) J. Cell Sci. 117, 6153–6162. [DOI] [PubMed] [Google Scholar]
- 82.Suh, S. I., Pyun, H. Y., Cho, J. W., Baek, W. K., Park, J. B., Kwon, T., Park, J. W., Suh, M. H. & Carson, D. A. (2000) Cancer Lett. 160, 81–88. [DOI] [PubMed] [Google Scholar]
- 83.Suh, S. I., Cho, J. W., Baek, W. K., Suh, M. H. & Carson, D. A. (2000) Cancer Lett. 153, 175–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.