Abstract
The p53 tumor suppressor protein plays a central role in cell cycle regulation, DNA repair, and apoptosis. Recent studies indicate that DNA damage and somatic mutations in the p53 gene can occur because of genotoxic stress in many tissues, including the skin, colon, and synovium. Although somatic mutations in the p53 gene have been demonstrated in rheumatoid arthritis (RA) synovial tissue and synoviocytes, no information is available on the location or extent of p53 mutations. Using microdissected RA synovial tissue sections, we observed abundant p53 transition mutations, which are characteristic DNA damage caused by oxidative stress. p53 mutations, as well as p53 mRNA expression, were located mainly in the synovial intimal lining rather than the sublining (P < 0.01). Clusters of p53 mutant subclones were observed in some microdissected regions, suggesting oligoclonal expansion. Because IL-6 gene expression is regulated by wild-type p53, IL-6 mRNA expression in microdissected tissues was quantified by using real-time PCR. The regions with high rates of p53 mutations contained significantly greater amounts of IL-6 mRNA compared with the low mutation samples (P < 0.02). The microdissection findings suggest that p53 mutations are induced in RA synovial tissues by inflammatory oxidative stress. This process, as in sun-exposed skin and inflamed colonic epithelium, provides some of the mutant clones with a selective growth advantage. A relatively low percentage of cells containing p53 mutations can potentially affect neighboring cells and enhance inflammation through the elaboration of proinflammatory cytokines.
The p53 tumor suppressor protein plays a central role in cell cycle regulation, DNA repair, and apoptosis (1, 2). Mutations in this gene are known to contribute to the pathogenesis of many neoplastic diseases (3, 4). However, the mechanisms of DNA damage and the contribution of abnormal p53 to non-neoplastic diseases have only recently been appreciated. For instance, somatic mutations can occur because of genotoxic stress in many tissues, including the skin, colon, and synovium (5–7). Oxidative damage caused by inflammation appears to cause p53 mutations in colon and synovium whereas UV light causes mutations in sun-exposed skin. The localization of somatic mutations within non-neoplastic tissues and the potential impact on the pathogenesis of inflammatory disease is only beginning to be appreciated.
Rheumatoid arthritis (RA) is a chronic inflammatory disease characterized by excessive growth and invasion of synovial tissues (8). Although RA has many features of autoimmunity, nonimmunologic factors also play a significant role in chronic disease (9–11). Clinical observations suggest that joint destruction can progress independent of inflammation (12–14), and autonomous proliferation of fibroblast-like synoviocytes might contribute to this phenomenon (10, 11). RA synoviocytes and synovial tissues exhibit some features of transformation that might participate in this process, including loss of contact inhibition, expression of oncogenes, autonomous invasion into cartilage, and insufficient apoptosis (10, 11, 15, 16). Genetic changes caused by local oxidative damage have been proposed as a potential mechanism that permanently alters or imprints RA synovial cells (7, 17). In contrast, mutations in p53 were not observed in osteoarthritis synovial samples. The markedly higher production of reactive oxygen and nitrogen in RA increases the likelihood of mutagenic events in RA as observed in inflammatory bowel disease.
To investigate the aggressive behavior of RA synoviocytes, we have focused our attention on the structure and function of the p53 tumor suppressor gene. Previous studies have demonstrated p53 overexpression in RA synovial tissues (18). Somatic mutations in the p53 gene have also been observed in RA synovium and cultured fibroblast-like synoviocyte cDNA and genomic DNA, although quantitative differences between studies have been noted (7, 19–21). Most of the p53 mutations in RA are characterized by transition base changes (7, 21) and have also been previously identified in human neoplastic tissue (22). Furthermore, certain p53 mutations in RA are dominant negative and can suppress endogenous wild-type p53 function (23). Inactivation of p53 protein can recapitulate many of the phenotypic changes observed in RA, such as increased proliferation and invasion of synovial cells (24, 25).
Despite this wealth of data, there is no information on the location or extent of p53 mutations in synovial tissue. Therefore, we examined for p53 mutations by using microdissected RA synovial tissue sections. These data indicate that p53 mutations are located primarily in the synovial intimal lining and can be associated with increased local expression of IL-6. Even a relatively small percentage of cells within the synovium could increase proinflammatory cytokines and activate neighboring cells in the synovium.
Materials and Methods
Synovial Tissues and Microdissection.
Synovial tissue samples were obtained from RA patients who underwent joint replacement (except one synovectomy sample). The diagnosis of RA conformed to the 1987 American College of Rheumatology revised criteria (26). Tissue samples were embedded in TissueTek OCT compound (Miles Diagnostics, Elkhart, IN) by immersion in methylbutane and stored at −80°C until use. Frozen tissues were cut into 10-μm sections. The first section of each block was stained with hematoxylin and eosin, and histology was noted to define intimal lining and sublining regions. The lining regions and sublining regions were then microdissected by using a sterile scalpel blade with the light microscopy. The area of each region was ≈0.25 mm2.
PCR Amplification and Sequencing.
Total RNA was isolated from microdissected RA synovial tissues by using RNA STAT-60 (Tel-Test, Friendswood, TX). Extracted RNA was reverse-transcribed into cDNA with random primers and Moloney murine leukemia virus reverse transcriptase according to the manufacturer's protocol (ProSTAR First-Strand RT-PCR kit, Stratagene). Exons 5–10 of the p53 gene were amplified from cDNA of microdissected RA synovial tissues by a nested PCR with primers flanking the gene sequences (external primer sense: 5′-ACCTACCAGGGCAGCTACGGTTTC-3′; external primer antisense: 5′-TTTATGGCGGGAGGTAGACGGACC-3′; internal primer sense: 5′-TCATTTAGGTGACACTATAGGACTTCAGGTGGCTGGA-3′; internal primer antisense: 5′-GATAATACGACTCACTATAGGGCTTCTTGCATTCTG-3′) as described (7). The PCR mixture consisted of 1× high-fidelity Advantage-HF2 polymerase mix (CLONTECH), 1× HF2 PCR buffer (CLONTECH), 1× HF2 dNTP mix (CLONTECH), 200 nM forward and reverse primers, and cDNA of samples in a total volume of 50 μl. For each PCR, cDNA was amplified for 45 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and extension at 68°C for 90 sec, with a final extension at 68°C for 3 min. Amplified PCR products were resolved by electrophoresis on 1.5% agarose gel containing ethidium bromide. The p53 bands were excised from the gels with a sterile blade and purified by using a QIAquick Gel extraction kit (Qiagen, Valencia, CA). Purified p53 PCR products were subcloned and sequenced by using the Applied Biosystems PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit on an Applied Biosystems PRISM 3100 Genetic Analyzer.
Real-Time PCR.
Real-time reverse transcriptase–PCR was performed by using the GeneAmp 5700 Sequence Detection System (Perkin–Elmer Applied Biosystems). Predeveloped sequence detection reagents specific for human IL-6 and glyceraldehyde-3-phosophodehydrogenase (GAPDH) (Perkin–Elmer Applied Biosystems) were used. PCR was performed in a TaqMan universal PCR Master Mix by using the following protocol: initial activation of AmpliTaq Gold DNA polymerase at 95°C for 10 min, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. The fluorescent signal was plotted versus cycle number, and the threshold cycle (Ct, the cycle number at which an increase above background fluorescence could be reliably detected) was determined.
Peripheral blood mononuclear cells from healthy controls were isolated with Ficoll-Paque (Amersham Pharmacia) and cultured overnight at 5 × 106 cells per ml in the presence of 1 μg/ml ConA. Peripheral blood mononuclear cell cDNA was prepared and diluted serially. Aliquots of each standard concentration was included in each PCR run, and standard curves for IL-6 and GAPDH were generated by linear regression using log [Ct] versus log (cell number). Sample Ct values were then used to calculate the cell equivalent number for IL-6 and GAPDH in unknown samples, and data were expressed as the ratio between IL-6 Ct and GAPDH Ct (cell equivalent ratio). Each PCR run also included nontemplate controls containing all reagents except cDNA. These controls generated a Ct greater than 40 in all experiments.
Statistical Analysis.
Data are expressed as means ± SEM. Comparisons among two groups were done by Mann–Whitney or Wilcoxon tests. P values less than 0.05 were considered significant.
Results
Microdissection and p53 RNA Expression in Synovial Tissues.
Frozen tissue sections from RA synovium (n = 8) were microdissected, including 65 intimal lining regions and 49 sublining regions. Fig. 1 shows an example of a synovial tissue section stained with hematoxylin and eosin, demonstrating the relative size and location of the microdissected regions. Based on the relative amounts of GAPDH detected in the microdissected samples with quantitative real-time PCR, we estimate that the regions contained approximately 1,000 cells. The yield for detectable p53 mRNA was substantially lower if smaller regions were evaluated. The cellularity and RNA content as determined by GAPDH expression were similar in microdissected samples isolated from the lining and sublining regions.
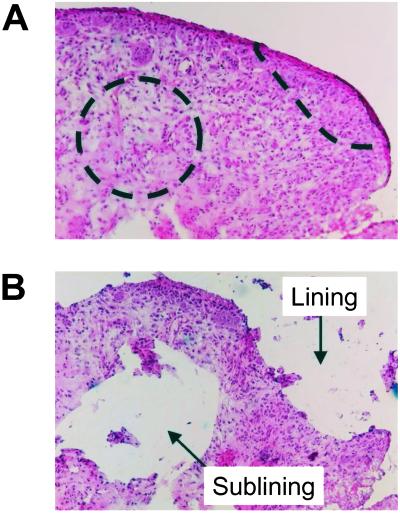
Figure 1.
Microdissection of RA synovial tissues. Frozen synovial tissues were cut into 10-μm sections. Intimal lining and sublining regions were microdissected by using a sterile blade under light microscopy. An example of the size and location of microdissected regions is shown (A: before microdissection, B: after microdissection). The area of each region was ≈0.25 mm2, and the number of the cells in the region was estimated at approximately 1,000 cells. (Magnification: ×100.)
Reverse transcriptase–PCR was then performed on the 114 microdissected regions to determine whether p53 mRNA could be detected. As shown in Fig. 2, the percentage of regions that expressed p53 mRNA as determined by agarose gel electrophoresis was significantly higher in the intimal lining regions compared with the sublining regions (P < 0.01).
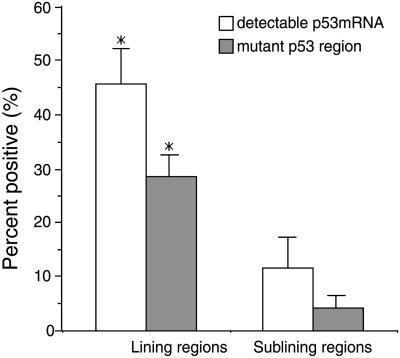
Figure 2.
p53 mRNA expression and p53 mutations in microdissected synovial tissues. Microdissected regions from eight RA patients were analyzed for p53 expression and mutations. The percentage of p53 mRNA positive regions by reverse transcriptase–PCR was significantly higher in the lining compared with the sublining (P < 0.01). The percentage of regions with a high mutation rate (i.e., >15% of subclones) was significantly higher in the lining compared with the sublining (P < 0.01).
p53 Mutations in Microdissected Synovial Tissues.
After PCR amplification, the 34 microdissected regions with detectable p53 mRNA were evaluated for the presence of wild-type or mutant p53. Ten to 12 subclones from each region were sequenced, and a total of 392 p53 subclones were analyzed. Twenty seven percent of the subclones in the p53 mRNA-positive microdissected regions contained missense mutations. Table 1 shows the location and relative frequency of the missense p53 mutations detected in microdissected synovial tissues. Some of the subclones contained more than one mutation. The intimal lining was more likely to contain a high abundance (>15%) of mutant p53 subclones than the sublining (see Fig. 2; P < 0.01). p53 missense mutations were widely distributed in exons 5–10, and multiple copies of individual p53 mutant subclones were found in three of the eight patients (RA1, RA2, and RA8). Sequence analysis after PCR amplification of wild-type p53 plasmid did not demonstrate the introduction of new mutations and confirmed the fidelity of amplification.
Table 1.
p53 mutations in microdissected RA synovial tissue
| Patient | Location | Mutant/total subclones | p53 mutation |
|---|---|---|---|
| RA1 | Lining | 7/9 | 138 GCC>GTC (Ala>Val) |
| Lining | 1/11 | 316 CCC>TCC (Pro>Ser) | |
| Lining | 8/11 | 176 TGC>CGC (Cys>Arg) | |
| 6/11 | 213 CGA>TGA (Arg>*stop) | ||
| 2/11 | 138 GCC>GTC (Ala>Val) | ||
| 1/11 | 128 CCT>ACT (Pro>Thr) | ||
| Sublining | 5/12 | 138 GCC>GTC (Ala>Val) | |
| 1/12 | 149 TCC>TTC (Ser>Phe) | ||
| 1/12 | 202 CGT>TGT (Arg>Cys) | ||
| 1/12 | 318 CCA>TCA (Pro>Ser) | ||
| RA2 | Lining | 1/11 | 365 CAC>TAC (His>Tyr) |
| Lining | 8/12 | 144 CAG>TAG (Gly>*stop) | |
| 1/12 | 340 ATG>ACG (Met>Thr) | ||
| Lining | 1/12 | 317 CAG>CAT (Gln>His) | |
| Lining | 1/12 | 255 ATC>ACC (Ile>Thr) | |
| 1/12 | 282 CGG>CAG (Arg>Gln) | ||
| Lining | 1/11 | 269 AGC>GGC (Ser>Gly) | |
| 1/11 | 342 CGA>TGA (Arg>*stop) | ||
| Sublining | 1/11 | 132 AAG>GAG (Lys>Glu) | |
| RA3 | Sublining | 1/12 | 138 GCC>ACC (Ala>Thr) |
| 1/12 | 152 CCG>CTG (Pro>Leu) | ||
| Lining | 1/12 | 151 CCC>TCC (Pro>Ser) | |
| 1/12 | 156 CGC>CAC (Arg>His) | ||
| 1/12 | 217 GTG>ATG (Val>Met) | ||
| 1/12 | 342 CGA>CAA (Arg>Gln) | ||
| Sublining | 1/11 | 125 ACG>ATG (Thr>Met) | |
| 1/11 | 295 CCT>TCT (Pro>Ser) | ||
| RA4 | Lining | 1/11 | 189 GCC>ACC (Ala>Thr) |
| 1/11 | 259 GAC>GGC (Asp>Gly) | ||
| RA5 | Lining | 1/12 | 244 GGC>GAC (Gly>Asp) |
| 1/12 | 324 GAT>AAT (Asp>Asn) | ||
| Lining | 1/12 | 161 GCC>ACC (Ala>Thr) | |
| 1/12 | 362 AGC>GGC (Ser>Gly) | ||
| RA6 | Lining | 1/12 | 280 AGA>ACA (Arg>Thr) |
| 1/12 | 360 GGG>AGG (Gly>Arg) | ||
| RA7 | Lining | 1/12 | 273 CGT>TGT (Arg>Cys) |
| Lining | 1/12 | 183 TCA>TTA (Ser>Leu) | |
| 1/12 | 306 CGA>TGA (Arg>*stop) | ||
| Lining | 1/12 | 158 CGC>CTC (Arg>Leu) | |
| RA8 | Lining | 1/12 | 121 TCT>CCT (Ser>Pro) |
| 1/12 | 177 CCC>TCC (Pro>Ser) | ||
| 1/12 | 178 CAC>*AC deletion (His>*stop) | ||
| 1/12 | 369 CTG>CCTG insert (Leu>*stop) | ||
| Lining | 1/11 | 157 GTC>ATC (Val>Ile) | |
| 1/11 | 229 TGT>TAT (Cys>Tyr) | ||
| 1/11 | 250 CCC>CTC (Pro>Leu) | ||
| Lining | 12/12 | 337 CGC>AGC (Arg>Ser) | |
| Lining | 2/12 | 300 CCC>TCC (Pro>Ser) | |
| Lining | 12/12 | Deletion 143–220 | |
| 1/12 | 228 GAC>AAC (Asp>Asn) | ||
| Lining | 1/12 | 141 TGC>TAC (Cys>Tyr) | |
| 1/12 | 146 TGG>TGA (Trp>*stop) | ||
| 1/12 | 221 GAG>GCG (Glu>Ala) | ||
| Sublining | 12/12 | Deletion 143–220 | |
| 1/12 | 345 AAT>GAT (Asn>Asp) |
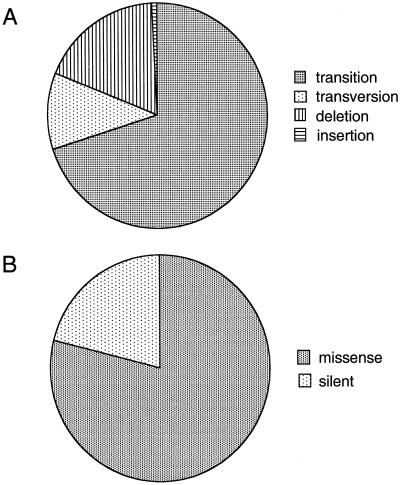
Previous studies have shown an abundance of transition mutations in RA synovium, possibly caused by the effects of oxidative stress (7, 17). The mutations detected in the current study were similar because transitions represented the vast majority of substitutions detected in the microdissected samples (Fig. 3A). Most of the p53 mutations in microdissected synovial tissues were missense mutations (Fig. 3B).
Figure 3.
p53 mutations in microdissected RA synovial tissues. (A) Type of mutation. As previously demonstrated, the majority of abnormalities were transition mutations. (B) Percentage of missense mutations. The majority of p53 abnormalities were missense mutations.
Localization of p53 Mutations in RA Synovial Tissues.
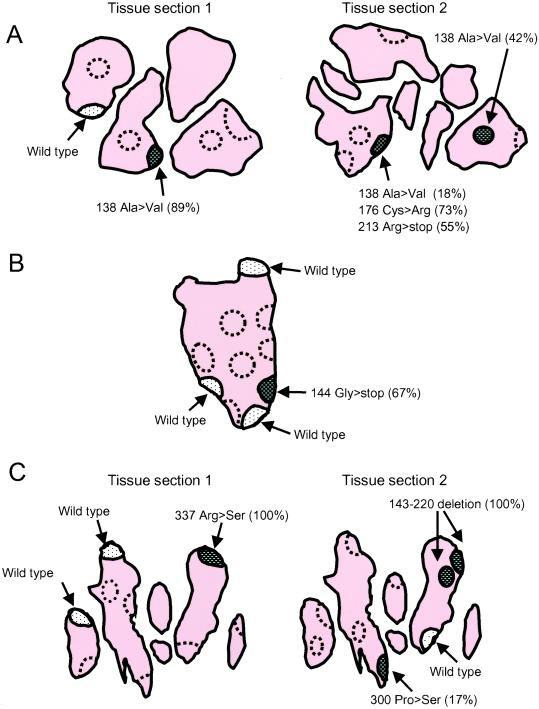
The majority of microdissected regions contained a relatively low abundance of individual p53 mutations (≤15% of the total cDNA pool). However, several microdissected regions contained a high percentage of an individual mutation, suggesting oligoclonal expansion. In some cases, an individual mutation could be identified in separate regions within a single section or even from two sections cut from the same tissue block. Fig. 4A (tissue section 1) shows an example from patient RA1 in which six regions (three lining regions and three sublining regions) were analyzed. p53 mRNA was detected in two of three lining regions, whereas none of the three sublining regions contained detectable p53 mRNA (noted by dashed lines). Surprisingly, 89% of the p53 subclones contained codon 138 Ala (GCC) to Val (GTC) mutation in one lining region (hatched area). In contrast, wild-type p53 predominated in the other region (dotted area). To confirm the results, a nearby section from the same tissue block was also examined (Fig. 4A, tissue section 2). The p53 codon 138 Ala to Val mutation was detected in the adjacent regions in a serial section. Additional mutations were also present in the same region (codon 176 Cys>Arg, codon 213 Arg>stop).
Figure 4.
Localization of p53 mutations in RA synovial tissues. Several schematic drawings of frozen tissue sections are shown to illustrate the location of p53 expression and mutations. The outer edge of the section indicates lining regions, and interior is sublining. Note that folds in the tissue result in the appearance of several discrete pieces of tissue from a single block. The regions shown by dashed lines contained no detectable p53 mRNA by a nested PCR. The predominant p53 sequences are show for each p53 mRNA-positive region. (A) Patient RA1. Six regions (three lining regions and three sublining regions) were microdissected in the first section (tissue section 1). p53 mRNA was detected in two of three lining regions, whereas none of the three sublining regions contained detectable p53 mRNA. In one lining region (hatched area), 89% of the p53 subclones contained codon 138 Ala (GCC) to Val (GTC) missense mutation. Many of the mutant subclones contained more than one mutation, suggesting sequential accumulation of mutations by various cells. In contrast, most of the p53 subclones contained wild-type p53 in the other region (dotted area). In an adjacent section from the same tissue block (tissue section 2), the codon 138 Ala to Val mutation was also detected. Additional mutations were also abundant in the same region (codon 176 Cys>Arg, codon 213 Arg>stop). (B) Patient RA2. Ten regions (seven lining regions and three sublining regions) were microdissected. p53 mRNA was detected in four of seven lining regions; however, none of the three sublining regions contained detectable p53 mRNA. In one lining region (hatched area), 67% of p53 subclones contained the codon 144 Gly (CAG) to stop codon (TAG) mutation, whereas the remaining three lining regions (dotted area) were primarily wild-type p53. (C) Patient RA8. Nine regions (eight lining and one sublining) were microdissected. p53 mRNA was detected in three of eight lining regions, but not in the sublining. In one lining region (hatched area), 100% of p53 subclones contained the codon 337 Arg (CGC) to Ser (AGC) mutation, whereas the remaining two lining regions (dotted area) were primarily wild-type p53.
A second example of a tissue with clustered p53 mutations is shown in Fig. 4B (patient RA2). Ten regions (seven lining and three sublining) were microdissected. p53 mRNA was detected in four of seven lining regions. However, none of the three sublining locations contained detectable p53 mRNA. In one lining region (hatched area), 67% of p53 subclones contained the codon 144 Gly (CAG) to stop codon (TAG) mutation, whereas the remaining three lining regions (dotted area) were primarily wild-type p53.
Fig. 4C shows the third example of a tissue with a cluster of p53 mutations (patient RA8). Nine regions (eight lining and one sublining) were microdissected (tissue section 1). p53 mRNA was detected in three of eight lining regions, but not in the sublining. In one lining region (hatched area), 100% of p53 subclones contained the codon 337 Arg (CGC) to Ser (AGC) mutation, whereas the remaining two lining regions were primarily wild-type p53. As shown in Fig. 4C, a second microdissected section from this patient also demonstrated an island containing a codon 143–220 deletion in 100% of subclones.
IL-6 mRNA Expression in Microdissected RA Synovial Tissues.
After evaluating the frequency and location of p53 mutations in microdissected synovial tissues, we determined whether the presence of mutations correlated with cytokine expression. IL-6 was selected because of its relative abundance in RA synovium and the fact that it is transcriptionally regulated by p53 (27). Regions with high or low mutations rates (>15% and ≤15% of the cDNA pool, respectively) in the p53 cDNA were assayed for relative IL-6 expression with real-time quantitative reverse transcriptase–PCR. Samples were available for analysis from six patients, with 15 high mutation and 34 low mutation regions. As shown in Fig. 5, the regions with high rates of p53 mutations contained significantly greater amounts of IL-6 mRNA compared with the low p53 mutation samples (P < 0.02). Therefore, the sites within RA synovium containing the greatest number of mutations also had the highest levels of IL-6 gene expression.
Figure 5.
IL-6 mRNA expression in microdissected RA synovial tissues. Microdissected samples previously evaluated for p53 mutations were subsequently examined for IL-6 gene expression by real-time PCR. IL-6 mRNA expression was normalized to GAPDH. Regions with high p53 mutation rates (>15%) contained significantly greater amounts of IL-6 mRNA (P < 0.02).
Discussion
Environmental exposure to mutagens and production of endogenous genotoxic molecules can damage genomic DNA. Over time, this process can overwhelm repair mechanisms and cause permanent alterations in susceptible genes. In the skin, for instance, chronic exposure to UV light can induce base substitutions in key regulatory genes that ultimately lead to actinic keratoses (28). Mutations in genes such as p53 have been documented in non-neoplastic diseases like ulcerative colitis (5, 29, 30). More recently, DNA damage and somatic mutations have also been implicated in longstanding RA (7, 19–21). However, this mutagenesis does not appear to cause malignancy; instead it enhances the aggressive nature of the disease (24, 25).
The purpose of the current study was not to determine the uniqueness of this phenomenon to RA; rather we sought to define the location and functional significance of mutations within the rheumatoid synovium of patients with severe destructive arthritis—to date the only rheumatic disease where somatic mutations have been identified. Ideally it would be of interest to evaluate p53 mutations in other forms of joint disease; however, the only common chronic arthritis with comparable disease duration and destruction requiring joint replacement surgery is osteoarthritis. Because no mutations have been found in osteoarthritis, microdissection could not provide additional information. We have considered analyzing joint tissues from other matched non-RA inflammatory arthropathies. Unfortunately, destructive arthritis leading to joint replacement is rare in these patients and synovial tissues of comparable disease duration and destruction are unavailable. Evaluation of a surrogate marker for oxidative DNA damage, i.e., total p53 expression, showed very low p53 protein levels in one form of chronic non-RA arthritis (i.e., reactive arthritis), which makes the likelihood of demonstrating mutations very low (31).
In any case, the goal of this study was to determine the distribution of mutations and their potential functional sequelae in chronic destructive RA. Additional studies examining synovial biopsies from early RA to determine when in the course of disease the mutations occur and how they compare with matched arthroscopic specimens from early, nondestructive reactive arthritis are required. For the time being, therefore, the present study is limited to the primary goal as stated above. The nonmutant microdissected regions from each patient serve as an additional internal control for the mutant sites. Most of the regions evaluated contained an abundance of wild-type p53 genes, whereas the mutant islands contain a high percentage of a particular transition mutation. This study, along with the use of high-fidelity polymerase and experiments demonstrating accurate control amplification of template, addresses the question of PCR fidelity and regional variation of p53 structure and function in RA joint tissues.
Although many p53 mutations have been described in cDNA and genomic DNA from RA synovial samples, their anatomic location and the lineage of the mutant cells have been difficult to determine. Oligoclonal expansion of cells have been observed in RA synovium, but the relatively low abundance of individual mutations suggests that only a small percentage of clusters or “islands” of cells are affected. This situation is analogous to sun-exposed skin, where mutant p53 cells are limited to discreet clusters (32). These mutations do not necessarily cause neoplasia, but can ultimately lead to precancerous lesions like actinic keratoses. Similarly, longstanding inflammation in ulcerative colitis is associated with oligoclonal expansion of mutant cells (5, 29, 30). Although limited clonal expansion might not be pathogenic in sun-exposed skin, increased production of soluble mediators by a relatively small number of synovial cells could have important consequences.
To address these questions, we microdissected frozen sections of RA synovial tissue and analyzed the sequence of p53 genes in the cDNA pool. Initial experiments were performed by using laser capture microscopy, but the relatively small amount of material obtained and the difficulty maintaining RNA integrity limited the utility of this technique. Instead, intimal lining and sublining regions were manually dissected guided by light microscopy. Although these regions were larger than those obtained by laser capture techniques, the low yield of p53 cDNA in the smaller samples precluded extensive analysis. Also, because the number of mutant cells likely represents a small percentage of the total in each region, analysis of cDNA enabled us to focus on cells with high p53 gene expression within a very heterogeneous population. This finding, along with the fact that mutations are distributed across many exons, made an exon by exon evaluation of genomic DNA impractical.
Microdissection analysis showed that the p53 gene is expressed mainly in the synovial intimal lining. Previously, using immunohistochemistry to localize p53 protein, both sublining and intimal lining cells were identified as the primary location (31, 33). Technical limitations, including the effectiveness of antigen retrieval methods and the specificities of different antibodies, probably account for the variability. By focusing on gene expression, we found the intimal lining to be the major location for p53 gene expression. The site of mutant cells within the synovium was also determined; most were in the intimal lining. Individual p53 mutations were present in relatively low abundance and typically comprised <10% of the p53 cDNA pool in a particular location. Some of the subclones also contained more than one mutation. Based on the location of mutations and the presence of similar mutations in cultured fibroblast-like synoviocytes (7, 19–21), it is likely that the mutant genes are harbored in type B fibrobroblast-like synoviocytes within the lining. We cannot exclude that type A lining cells (macrophage-like synoviocytes) also contain base changes but sublining T cells do not have an abundance of p53 abnormalities based on micrcodissection data.
Missense transition mutations were observed in the microdissected tissue sections, similar to previous reports on whole synovium (7, 21). This finding is consistent with the hypothesis that the DNA damage results from local oxidative exposure (7, 34, 35). A notable exception in one patient was a highly abundant long deletion. Long deletions of p53 gene are uncommon, but have been reported in several malignant tissues (36–38). Such mutations could result from defective mismatch repair mechanisms in synoviocytes, as has recently been correlated with the presence of synovial microsatellite instability in RA.§ Moreover, the intimal lining has an extraordinarily high metabolic activity and is likely a major source of genotoxic molecules, such as reactive oxygen and reactive nitrogen, in the synovium (39). Local expression of inducible NO synthase along with direct exposure to the toxic synovial fluid environment probably contributes to the DNA damage (39). In addition, synovial fibroblasts that reside in the intimal lining probably do not recirculate to a significant degree, whereas lymphocytes can emigrate from the joint. Under these circumstances, the number of sublining cells that are permanently anchored would be less than in the lining where they can accumulate over time. This possibility is supported by the observation that T cells bearing mutations in the hprt gene are present in both the joints and blood of patients with RA (40).
Perhaps the most intriguing finding of the microdissection studies is that p53 mutations exist in islands within synovial tissue. Three of the eight patients had an abundance of specific mutations within a particular region. Although this result could be caused by individual cells with extremely high gene expression, it is more likely that these mutations afforded a growth advantage and permitted limited monoclonal expansion. In another patient, we even identified the same mutation in analogous regions of an adjacent tissue section. All mutations forming clusters were missense mutations, and the codons in which these mutations reside are characteristic hotspots in transformed tissues (22, 41–45). The presence of mutant p53 islands is consistent with studies in other tissues exposed to genotoxic stimuli, including the sun-exposed skin and colonic epithelium in inflammatory bowel disease (30, 32).
To determine whether the p53 mutations in RA synovial tissues have functional sequelae, we evaluated IL-6 mRNA expression in the microdissected regions. IL-6 is a key proinflammatory cytokine that is abundant in RA synovium and synovial tissue (46, 47) and its transcription is suppressed by wild-type p53 (27). We have shown that some p53 mutations from RA patients interfere with the ability of wild-type p53 to inhibit IL-6 gene expression (23). Real-time PCR analysis of microdissected synovium indicated that those regions that had abundant p53 mutations expressed significantly greater amounts of IL-6 mRNA compared with regions containing predominantly wild-type p53. The mechanism of IL-6 production relates to either a direct effect of p53 on IL-6 transcription or secondary to other changes in the inflammatory milieu. This key observation provides an in vivo correlation between the abundance of p53 mutations and the expression of genes that participate in the pathogenesis of RA.
The IL-6 data are consistent with our recent studies on the role of p53 in murine collagen-induced arthritis (CIA). Although p53 is considered mainly a key intracellular protein that regulates cell proliferation and survival, we proposed that it also serves important anti-inflammatory functions (48). When susceptible strains of mice that lack p53 are immunized with type II collagen they get a more aggressive, inflammatory, and destructive CIA than wild-type mice (48). Of particular interest, p53 knockout mice with CIA have significantly greater synovial IL-6 expression, which is not observed in nonarthritic joints (where p53 is not expressed) or in serum of arthritic mice. Therefore, p53 serves a local homeostatic function by suppressing cytokine production and assisting in the resolution of inflammation. The implications for RA are apparent because defective p53 expression could potentially exacerbate synovitis even if only a relatively low percent of cells is affected.
In summary, the presence of p53 mutations in RA joint tissues prompted us to evaluate regional variations of p53 expression and mutation abundance. The synovial intimal lining was the primary site of p53 gene expression and the location of most mutations. Regions with an abundance of p53 mutations correlated with increased local expression of IL-6. These findings support the hypothesis that genotoxic stimuli in the inflamed RA synovium can lead to localized monoclonal expansion of pathogenic cells. Furthermore, we suspect that this is a general property of chronically inflamed tissues and could explain destruction seen in other chronic inflammatory diseases.
Acknowledgments
This work was supported in part by National Institutes of Health Grants AR45347 and AR44850.
Abbreviations
- RA
rheumatoid arthritis
- GAPDH
glyceraldehyde-3-phosophodehydrogenase
- Ct
cycle threshold
Footnotes
Lee, S.-H., Chang, D. K., Boland, C. R., Han, Z. & Firestein, G. S. (2001) Arthritis Rheum. 44, S213 (abstr.).
References
- 1.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 4.Levine A J, Momand J, Finlay C A. Nature (London) 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 5.Hussain S P, Amstad P, Raja K, Ambs S, Nagashima M, Bennett W P, Shields P G, Ham A J, Swenberg J A, Marrogi A J, Harris C C. Cancer Res. 2000;60:3333–3337. [PubMed] [Google Scholar]
- 6.Jonason A S, Kunala S, Price G J, Restifo R J, Spinelli H M, Persing J A, Leffell D J, Tarone R E, Brash D E. Proc Natl Acad Sci USA. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firestein G S, Echeverri F, Yeo M, Zvaifler N J, Green D R. Proc Natl Acad Sci USA. 1997;94:10895–10900. doi: 10.1073/pnas.94.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firestein G S. In: Textbook of Rheumatology. Ruddy S, Harris E D, Sledge C B, Budd R C, Sergent J S, editors. Philadelphia: Saunders; 2001. pp. 921–966. [Google Scholar]
- 9.Gay S, Gay R E, Koopman W J. Ann Rheum Dis. 1993;52, Suppl. 1:S39–S47. doi: 10.1136/ard.52.suppl_1.s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller-Ladner U, Kriegsmann J, Franklin B N, Matsumoto S, Geiler T, Gay R E, Gay S. Am J Pathol. 1996;149:1607–1615. [PMC free article] [PubMed] [Google Scholar]
- 11.Firestein G S. Arthritis Rheum. 1996;39:1781–1790. doi: 10.1002/art.1780391103. [DOI] [PubMed] [Google Scholar]
- 12.Müller-Ladner U, Kriegsmann J, Gay R E, Koopman W J, Gay S, Chatham W W. Arthritis Rheum. 1995;38:1328–1332. doi: 10.1002/art.1780380922. [DOI] [PubMed] [Google Scholar]
- 13.Mulherin D, Fitzgerald O, Bresnihan B. Br J Rheumatol. 1996;35:1263–1268. doi: 10.1093/rheumatology/35.12.1263. [DOI] [PubMed] [Google Scholar]
- 14.Joosten L A, Helsen M M, Saxne T, van De Loo F A, Heinegard D, van Den Berg W B. J Immunol. 1999;163:5049–5055. [PubMed] [Google Scholar]
- 15.Lafyatis R, Remmers E F, Roberts A B, Yocum D E, Sporn M B, Wilder R L. J Clin Invest. 1989;83:1267–1276. doi: 10.1172/JCI114011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamanishi Y, Firestein G S. Rheum Dis Clin N Am. 2001;27:355–371. doi: 10.1016/s0889-857x(05)70206-4. [DOI] [PubMed] [Google Scholar]
- 17.Tak P P, Zvaifler N J, Green D R, Firestein G S. Immunol Today. 2000;21:78–82. doi: 10.1016/s0167-5699(99)01552-2. [DOI] [PubMed] [Google Scholar]
- 18.Firestein G S, Nguyen K, Aupperle K R, Yeo M, Boyle D L, Zvaifler N J. Am J Pathol. 1996;149:2143–2151. [PMC free article] [PubMed] [Google Scholar]
- 19.Reme T, Travaglio A, Gueydon E, Adla L, Jorgensen C, Sany J. Clin Exp Immunol. 1998;111:353–358. doi: 10.1046/j.1365-2249.1998.00508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kullmann F, Judex M, Neudecker I, Lechner S, Justen H P, Green D R, Wessinghage D, Firestein G S, Gay S, Scholmerich J, Muller-Ladner U. Arthritis Rheum. 1999;42:1594–1600. doi: 10.1002/1529-0131(199908)42:8<1594::AID-ANR5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 21.Inazuka M, Tahira T, Horiuchi T, Harashima S, Sawabe T, Kondo M, Miyahara H, Hayashi K. Rheumatology. 2000;39:262–266. doi: 10.1093/rheumatology/39.3.262. [DOI] [PubMed] [Google Scholar]
- 22.Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, Harris C C, Montesano R. Nucleic Acids Res. 1998;26:205–213. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Z, Boyle D L, Shi Y, Green D R, Firestein G S. Arthritis Rheum. 1999;42:1088–1092. doi: 10.1002/1529-0131(199906)42:6<1088::AID-ANR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 24.Aupperle K R, Boyle D L, Hendrix M, Seftor E A, Zvaifler N J, Barbosa M, Firestein G S. Am J Pathol. 1998;152:1091–1098. [PMC free article] [PubMed] [Google Scholar]
- 25.Pap T, Aupperle K R, Gay S, Firestein G S, Gay R E. Arthritis Rheum. 2001;44:676–681. doi: 10.1002/1529-0131(200103)44:3<676::AID-ANR117>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 26.Arnett F C, Edworthy S M, Bloch D A, McShane D J, Fries J F, Cooper N S, Healey L A, Kaplan S R, Liang M H, Luthra H S, et al. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 27.Santhanam U, Ray A, Sehgal P B. Proc Natl Acad Sci USA. 1991;88:7605–7609. doi: 10.1073/pnas.88.17.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomura T, Nakajima H, Hongyo T, Taniguchi E, Fukuda K, Li L Y, Kurooka M, Sutoh K, Hande P M, Kawaguchi T, et al. Cancer Res. 1997;57:2081–2084. [PubMed] [Google Scholar]
- 29.Yin J, Harpaz N, Tong Y, Huang Y, Laurin J, Greenwald B D, Hontanosas M, Newkirk C, Meltzer S J. Gastroenterology. 1993;104:1633–1639. doi: 10.1016/0016-5085(93)90639-t. [DOI] [PubMed] [Google Scholar]
- 30.Brentnall T A, Crispin D A, Rabinovitch P S, Haggitt R C, Rubin C E, Stevens A C, Burmer G C. Gastroenterology. 1994;107:369–378. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 31.Tak P P, Smeets T J, Boyle D L, Kraan M C, Shi Y, Zhuang S, Zvaifler N J, Breedveld F C, Firestein G S. Arthritis Rheum. 1999;42:948–953. doi: 10.1002/1529-0131(199905)42:5<948::AID-ANR13>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Ling G, Persson A, Berne B, Uhlen M, Lundeberg J, Ponten F. Am J Pathol. 2001;159:1247–1253. doi: 10.1016/S0002-9440(10)62511-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou C T, Yang J S, Lee M R. J Pathol. 2001;193:110–116. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH746>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 34.Wink D A, Kasprzak K S, Maragos C M, Elespuru R K, Misra M, Dunams T M, Cebula T A, Koch W H, Andrews A W, Allen J S, et al. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen T, Brunson D, Crespi C L, Penman B W, Wishnok J S, Tannenbaum S R. Proc Natl Acad Sci USA. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burns J E, Baird M C, Clark L J, Burns P A, Edington K, Chapman C, Mitchell R, Robertson G, Soutar D, Parkinson E K. Br J Cancer. 1993;67:1274–1284. doi: 10.1038/bjc.1993.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plummer S J, Santibanez-Koref M, Kurosaki T, Liao S, Noble B, Fain P R, Anton-Culver H, Casey G. Oncogene. 1994;9:3273–3280. [PubMed] [Google Scholar]
- 38.Nakai H, Kaneko H, Horiike S, Ariyama Y, Misawa S, Kashima K, Ishizaki K. Br J Haematol. 1994;87:839–842. doi: 10.1111/j.1365-2141.1994.tb06747.x. [DOI] [PubMed] [Google Scholar]
- 39.Hilliquin P, Borderie D, Hernvann A, Menkes C J, Ekindjian O G. Arthritis Rheum. 1997;40:1512–1517. doi: 10.1002/art.1780400820. [DOI] [PubMed] [Google Scholar]
- 40.Cannons J L, Karsh J, Birnboim H C, Goldstein R. Arthritis Rheum. 1998;41:1772–1782. doi: 10.1002/1529-0131(199810)41:10<1772::AID-ART9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 41.Willems P M, Kuypers A W, Meijerink J P, Holdrinet R S, Mensink E J. Leukemia. 1993;7:986–991. [PubMed] [Google Scholar]
- 42.Sedlacek Z, Kodet R, Seemanova E, Vodvarka P, Wilgenbus P, Mares J, Poustka A, Goetz P. Br J Cancer. 1998;77:1034–1039. doi: 10.1038/bjc.1998.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuyer M, Henzen-Logmans S C, van der Burg M E, Fieret E J, Klijn J G, Foekens J A, Berns E M. Int J Cancer. 1998;76:299–303. doi: 10.1002/(sici)1097-0215(19980504)76:3<299::aid-ijc2>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 44.Lomax M E, Barnes D M, Gilchrist R, Picksley S M, Varley J M, Camplejohn R S. Oncogene. 1997;14:1869–1874. doi: 10.1038/sj.onc.1201133. [DOI] [PubMed] [Google Scholar]
- 45.DiGiammarino E L, Lee A S, Cadwell C, Zhang W, Bothner B, Ribeiro R C, Zambetti G, Kriwacki R W. Nat Struct Biol. 2002;9:12–16. doi: 10.1038/nsb730. [DOI] [PubMed] [Google Scholar]
- 46.Houssiau F A, Devogelaer J P, Van Damme J, de Deuxchaisnes C N, Van Snick J. Arthritis Rheum. 1988;31:784–788. doi: 10.1002/art.1780310614. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto H, Yamamura M, Morita Y, Harada S, Makino H, Ota Z. Arthritis Rheum. 1997;40:1096–1105. doi: 10.1002/art.1780400614. [DOI] [PubMed] [Google Scholar]
- 48.Yamanishi Y, Boyle D L, Pinkoski M J, Mahboubi A, Lin T, Han Z, Zvaifler N J, Green D R, Firestein G S. Am J Pathol. 2002;160:123–130. doi: 10.1016/S0002-9440(10)64356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]