Abstract
DNA hypomethylation on pericentromeric satellite regions is an early and frequent event associated with heterochromatin instability during human hepatocarcinogenesis. A DNA methyltransferase, DNMT3b, is required for methylation on pericentromeric satellite regions during mouse development. To clarify the molecular mechanism underlying DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis, we examined mutations of the DNMT3b gene and mRNA expression levels of splice variants of DNMT3b in noncancerous liver tissues showing chronic hepatitis and cirrhosis, which are considered to be precancerous conditions, and in hepatocellular carcinomas (HCCs). Mutation of the DNMT3b gene was not found in HCCs. Overexpression of DNMT3b4, a splice variant of DNMT3b lacking conserved methyltransferase motifs IX and X, significantly correlated with DNA hypomethylation on pericentromeric satellite regions in precancerous conditions and HCCs (P = 0.0001). In particular, the ratio of expression of DNMT3b4 to that of DNMT3b3, which is the major splice variant in normal liver tissues and retains conserved methyltransferase motifs I, IV, VI, IX, and X, showed significant correlation with DNA hypomethylation (P = 0.009). Transfection of human epithelial 293 cells with DNMT3b4 cDNA induced DNA demethylation on satellite 2 in pericentromeric heterochromatin DNA. These results suggest that overexpression of DNMT3b4, which may lack DNA methyltransferase activity and compete with DNMT3b3 for targeting to pericentromeric satellite regions, results in DNA hypomethylation on these regions, even in precancerous stages, and plays a critical role in human hepatocarcinogenesis by inducing chromosomal instability.
DNA methylation plays important roles in gene silencing, chromatin remodeling, and genome stability (1–4). Aberrant DNA methylation is one of the most consistent epigenetic changes in human cancers (1–4). Generally, the overall level of DNA methylation is lower in cancer cells than in normal cells (5, 6), although a number of tumor suppressor genes are silenced by DNA methylation on CpG islands around their promoter regions in cancer cells (1–4, 7, 8).
We have carefully examined alterations of DNA methylation status on pericentromeric satellite regions and CpG islands of specific genes, and expression of DNA methyltransferases and methyl-CpG-binding proteins, in noncancerous liver tissues showing chronic hepatitis and cirrhosis, which are considered to be precancerous conditions (9, 10), and in hepatocellular carcinomas (HCCs) (11–17). Among these alterations, DNA hypomethylation on pericentromeric satellite regions was detected even in precancerous conditions and appears to be one of the earliest epigenetic changes during human hepatocarcinogenesis (17). Satellite regions are located in pericentromeric heterochromatin DNA, and DNA hypomethylation on these regions is known to result in centromeric decondensation, enhancing chromosome recombinations (18, 19). In fact, frequent chromosome 1q copy gain with a pericentromeric breakpoint was reported in HCCs showing DNA hypomethylation on satellite 2 (20). DNA hypomethylation on pericentromeric satellite regions may induce chromosomal instability during hepatocarcinogenesis, even in precancerous conditions.
A newly identified DNA methyltransferase, DNMT3b (21), is specifically required for DNA methylation on pericentromeric satellite regions in embryonic stem cells and early mouse embryos (22). Germ-line mutations of the DNMT3b gene have been reported in patients with immunodeficiency/centromeric instability/facial anomalies syndrome (23, 24), a rare recessive autosomal disorder characterized by DNA hypomethylation on pericentromeric satellite regions (25). These findings encouraged us to examine genetic alterations of the DNMT3b gene in HCCs.
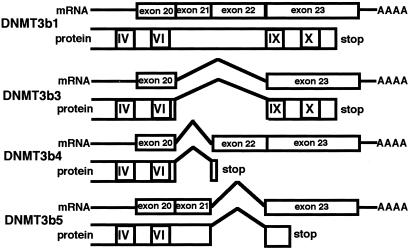
Down-regulation of DNMT3b is unlikely to underlie DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis, because we and other groups have reported increased expression of the mRNA for DNMT3b in human cancers, including HCCs (17, 26, 27). However, four splice variants of human DNMT3b in the C-terminal catalytic domain are known (ref. 26, Fig. 1). DNMT3b4 and DNMT3b5, without conserved methyltransferase motifs IX and X, probably lack DNA methyltransferase activity. So far, no studies of the expression of DNMT3b in human cancers have discriminated between the splice variants.
Figure 1.
mRNA structure and protein structure of splice variants of human DNMT3b. The four splice variants in the C-terminal catalytic domain (26) and the numbers of exons and the exon-intron boundaries (28) have been described.
To clarify the molecular mechanism underlying DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis, we examined mutations of the DNMT3b gene and expression levels of splice variants of DNMT3b in noncancerous liver tissues showing chronic hepatitis and cirrhosis and in HCCs.
Materials and Methods
Patients and Tissue Specimens.
Fifty-nine primary HCCs and the corresponding noncancerous liver tissues were obtained from surgically resected materials from 49 patients (cases H1 to H49) who were treated at the National Cancer Center Hospital, Tokyo. Twelve patients were hepatitis B virus surface antigen (HBs-Ag)-positive, 29 were anti-hepatitis C virus antibody (anti-HCV)-positive, one was both HBs-Ag and anti-HCV positive, and seven were both HBs-Ag and anti-HCV negative. Histological examination of noncancerous liver tissues from HCC patients revealed no remarkable findings, findings compatible with chronic hepatitis, and findings compatible with cirrhosis in 1, 24, and 24 tissues, respectively. For comparison, normal liver tissues showing no remarkable histological findings were also obtained from eight patients (cases C1 to C8) who were both HBs-Ag and anti-HCV negative and underwent partial hepatectomy for liver metastasis of primary colon cancer.
PCR–Single-Strand Conformation Polymorphism Analysis and Direct Sequencing.
Genomic DNA was amplified by PCR with 22 rhodamine-labeled intronic oligonucleotide primer sets encompassing all coding exons of the DNMT3b gene (GenBank accession no. AL035071 and Table 1). Primers were designed on the basis of the exon-intron boundaries as described (28). PCR products were electrophoresed on 6% polyacrylamide gel. DNA was recovered from the mobility-shifted bands and sequenced in both directions with the ABI PRISM BigDye Terminator kit and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems).
Table 1.
The primer sets for PCR–single-strand conformation polymorphism to detect mutation of the DNMT3b gene
| Target exons | Primer sets | Target exons | Primer sets |
|---|---|---|---|
| Exon 2 | 5′-TCCCTGCTTCCCTTTCACC-3′ (sense) | Exon 13 | 5′-ACCCCAGGCTTTAGCAGCT-3′ (sense) |
| 5′-TTGTCTGCAGTGACCGCTC-3′ (antisense) | 5′-GGAGTTAGAGGAGGCAAGGG-3′ (antisense) | ||
| Exon 3 | 5′-TTAGCAAGGCCGTTCCC-3′ (sense) | Exon 14 | 5′-CCCTTCTCTGGTCTCCGATT-3′ (sense) |
| 5′-CCACGTGATGAAAGCCAAAG-3′ (antisense) | 5′-GACTGCAGGAACGTAGGAGC-3′ (antisense) | ||
| Exon 4 | 5′-TGACTTGCTGATACCCTGGG-3′ (sense) | Exon 15 | 5′-CAGGAGACCAGCTCTGACAAAG-3′ (sense) |
| 5′-GAGTGTTGGCCACAAGTGCT-3′ (antisense) | 5′-TTTCCAACACCCTGTGCC-3′ (antisense) | ||
| Exon 5 | 5′-AGGCCTCCAGTCACCTAAGG-3′ (sense) | Exon 16 | 5′-GTCTTTGCCCTGTGCCTTC-3′ (sense) |
| 5′-TGCAGTGAGTGAGGCATATCTC-3′ (antisense) | 5′-CCTGGCTACCCTGTTGTGAC-3′ (antisense) | ||
| Exon 6 | 5′-TTGCTCTGGCCCAAACTATG-3′ (sense) | Exon 17 | 5′-GAACTGTTCATTTTACCATAGCAGG-3′ (sense) |
| 5′-GGTCCTCAGTCACCCTGG-3′ (antisense) | 5′-GGGAAAAAGACAGGAAGAGATG-3′ (antisense) | ||
| Exon 7 | 5′-CCTCTCCTCACTGGGATTTCTT-3′ (sense) | Exon 18 | 5′-TCTAGAAGTGGGTCCAGCTCTC-3′ (sense) |
| 5′-TTCAAAGGGAGGCAGGC-3′ (antisense) | 5′-AAGCAAGTGGCTCCTTCTCAG-3′ (antisense) | ||
| Exon 8 | 5′-AGACATGGCACCTGGGACA-3′ (sense) | Exon 19 | 5′-GAACCTGCTGGTCTCAGGGA-3′ (sense) |
| 5′-TCTTGCTTCATCCCTGCTCT-3′ (antisense) | 5′-CCTGCCCTGGCTGTCTGT-3′ (antisense) | ||
| Exon 9 | 5′-CACCCCCCCATTCATCA-3′ (sense) | Exon 20 | 5′-CTGCAAAGGTCTGGTTGACAC-3′ (sense) |
| 5′-GACTCTCCCAAGAAGTGGTCC-3′ (antisense) | 5′-CCAGGTCTTTCTAGGAAGGCTT-3′ (antisense) | ||
| Exon 10 | 5′-TGGGTGACAGAGCAAGACC-3′ (sense) | Exon 21 | 5′-TGTGCCTAGCAGAGGACCCT-3′ (sense) |
| 5′-CAGAAGAAAGTGCATAGAAAACAGG-3′ (antisense) | 5′-AGTCCCCACTTGGAGGTCAC-3′ (antisense) | ||
| Exon 11 | 5′-ACCCAGGCATAGCATGGTCT-3′ (sense) | Exon 22 | 5′-TTGAGCCTTGACTCCCCAG-3′ (sense) |
| 5′-TGATCTGCAGCGTCCTCCT-3′ (antisense) | 5′-GCCCATGTCTGCCCATTT-3′ (antisense) | ||
| Exon 12 | 5′-GGAATTGATCTGTACCCGGC-3′ (sense) | Exon 23 | 5′-GGTTGAGGCTGTCAACATCC-3′ (sense) |
| 5′-TGGGTTAAACCCTGACAGGG-3′ (antisense) | 5′-ATCACACCTCCTGGGTCCTG-3′ (antisense) |
Reverse-Transcription (RT)-PCR and Cloning Sequencing.
First-strand cDNA was prepared from total RNA with random hexadeoxynucleotide primers and SuperScript RNase H– reverse transcriptase (GIBCO/BRL). cDNA derived from human testis total RNA (CLONTECH) was used as control. Subsequent PCR with a primer set of (forward) 5′-CCT GCT GAA TTA CTC ACG CCC C-3′ and (reverse) 5′-GTC TGT GTA GTG CAC AGG AAA GCC-3′ (26) amplified all four splice variants in the C-terminal catalytic domain of the DNMT3b gene. For visual confirmation, the PCR products were separated electrophoretically on 1.5% agarose gel. The PCR products were then cloned into the pCRII vector (TA cloning kit; Invitrogen). The inserts were amplified by colony PCR and sequenced with both vector primers: M13 forward, 5′-GTA AAA CGA CGG CCA G-3′; M13 reverse, 5′-CAG GAA ACA GCT ATG AC-3′.
Splice Variant-Specific Quantitative RT-PCR.
Oligonucleotide primer sets specific for DNMT3b3 and DNMT3b4 were designed. Each primer spans the splice variant-specific exon-exon boundary: 3b3 forward, 5′-GAT GAA CAG GAT CTT TGG CTT T-3′ (exon 20/23); 3b3 reverse, 5′-GCC TGG CTG GAA CTA TTC ACA-3′ (exon 23); 3b4 forward, 5′-CGG GAT GAA CAG TTA AAG AAA GTA C-3′ (exon 20/22); 3b4 reverse, 5′-CCA AAG ATC CTT TCG AGC TC-3′ (exon 22/23). The PCRs were performed with the SYBR Green PCR Core Reagents kit (Applied Biosystems). Real-time detection of the emission intensity of SYBR Green bound to double-stranded DNAs was performed with the ABI PRISM 7700 Sequence Detection System. cDNAs derived from the HCC cell line Alexander (29) were used as the calibrator samples. Quantitative PCRs were performed in triplicate for each sample-primer set, and the mean of the three experiments was used as the relative quantification value. At the end of 40 PCR cycles, the reaction products were separated electrophoretically on 3% agarose gel to confirm that no nonspecific product was obtained at each amplification.
Transfection with DNMT3b4 cDNA.
Full-length human DNMT3b4 cDNA was prepared by RT-PCR of total RNA from the Alexander cell line. The primer set (forward, 5′-CGC GGA TCC TGG AAA GCA TGA AGG GAG ACA C-3′; reverse, 5′-GCT CTA GAA CTG TTC ATC CCG GGT AGG TTG-3′) was designed such that after a BamHI/XbaI double digest the coding sequence of DNMT3b4 could be ligated in-frame into the expression vector with a myc tag at the C terminus (pcDNA3.1-myc; Invitrogen). To confirm the fidelity of the PCR, the insert was fully sequenced. The cloned cDNA was linearized and transfected into 293 cells—human embryonic kidney cells transformed with adenovirus type 5 DNA (30)—using FuGENE 6 Transfection Reagent (Roche Diagnostics). After G418 selection of resistant cells, myc-tagged DNMT3b4-expressing clones were chosen by Western blotting. Mock-transfected 293 cells were generated by transfection with vector alone.
Western Blotting.
Total cell extracts were separated by SDS/PAGE, transferred electrophoretically onto poly(vinylidene difluoride) filters, and then incubated with anti-myc mAb 9E10 (Santa Cruz Biotechnology).
Southern Blotting.
High molecular weight DNA (5 μg) was digested for 24 h with 10 units of either MspI or HpaII, which cut at the sequence CCGG, per μg of DNA. HpaII does not cut when the internal cytosine is methylated. The DNA fragments were separated by electrophoresis, transferred to nitrocellulose membranes, and hybridized with 32P-labeled oligonucleotide probe for satellite 2 (31). Signal intensities were measured with an image analyzer (model BAS-2500; Fujifilm, Tokyo).
Statistics.
Correlations between mRNA levels for DNMT3b splice variants and DNA methylation status were analyzed with the Kruskal–Wallis test. Differences in ratios for signal intensities of Southern blotting were analyzed with the Mann–Whitney U test. Differences with P values of less than 0.05 were considered significant.
Results
No Mutation of the DNMT3b Gene in HCCs.
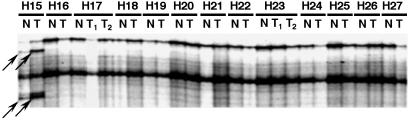
Fig. 2 shows examples of PCR–single-strand conformation polymorphism analysis of the DNMT3b gene in HCC cases. Because all of the shifted bands were detected in both the noncancerous liver tissue and the HCC of a particular case, they were considered to be polymorphisms. Sequencing revealed that the polymorphisms were located in introns 2 and 13 near the exon-intron boundaries (Table 2). No mutation of any coding exon of the DNMT3b gene was detected in 59 HCCs.
Figure 2.
Examples of PCR–single-strand conformation polymorphism analysis of the DNMT3b gene in HCC cases with the intronic primer set encompassing exon 13. N, noncancerous liver tissue; T, HCC. The shifted bands of H15N and H15T (arrows) were sequenced and revealed to be a polymorphism in intron 13 (Table 2).
Table 2.
Polymorphisms of the DNMT3b gene
| Position | Genotype | HCC patients (n = 49) | Patients with liver metastasis of primary colon cancer (n = 8) |
|---|---|---|---|
| Intron 2 (22 bp upstream of the 3′ splice site) | −/− | 38 | 5 |
| −/c | 9 | 3 | |
| c/c | 2 | 0 | |
| Intron 13 (16–18 bp downstream of the 5′ splice site) | act/act | 48 | 8 |
| act/— | 1 | 0 |
DNMT3b3 and DNMT3b4 Are Major Splice Variants in HCCs.
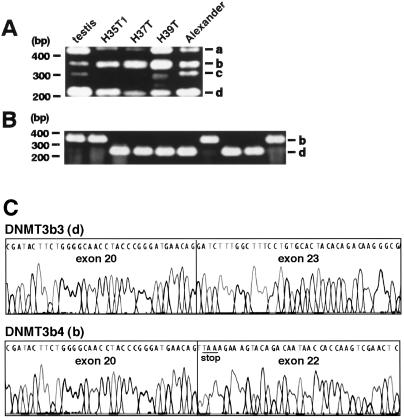
Fig. 3A shows examples of RT-PCR products obtained with a primer set that amplifies all four splice variants of DNMT3b in the C-terminal catalytic domain. cDNA derived from total RNA of human testis, in which significant expression of all four splice variants has been reported (26), was used as control. At the end of 35 cycles of PCR amplification, PCR products of about 230 bp were visible in almost all samples (band d in Fig. 3A). PCR products of about 350 bp were clearly visible in HCCs and Alexander cells, but control testis contained little of this product (band b in Fig. 3A). RT-PCR products of sample H35T1 were then cloned into the pCRII vector and the inserts were amplified by colony PCR (Fig. 3B). Almost all clones examined possessed inserts of about 230 bp (band d) or 350 bp (band b). Sequencing confirmed that the inserts of about 230 bp (band d) and 350 bp (band b) corresponded to DNMT3b3, skipping exons 21 and 22, and DNMT3b4, skipping exon 21, respectively (ref. 26, Figs. 1 and 3C).
Figure 3.
Examples of RT-PCR with a primer set that will amplify all four splice variants in the C-terminal catalytic domain of DNMT3b in HCCs (H) and Alexander cells. (A) Alexander cells were derived from human HCC (29) and showed DNA hypomethylation on pericentromeric satellite regions (data not shown). PCR products of about 420 bp (a), 350 bp (b), 300 bp (c), and 230 bp (d) will correspond to DNMT3b1, DNMT3b4, DNMT3b5, and DNMT3b3, respectively. (B) RT-PCR products of H35T1 were cloned into the pCRII vector, and the inserts were examined by colony PCR. Almost all clones examined possessed inserts of about 350 bp (b) and 230 bp (d). (C) Sequencing confirmed that the insert of about 350 bp (b) corresponded to DNMT3b4, which lacks the conserved methyltransferase motifs IX and X (Fig. 1) because of the premature stop codon (underlined) arising from skipping of exon 21.
Overexpression of DNMT3b4 Significantly Correlates with DNA Hypomethylation on Pericentromeric Satellite Regions in Precancerous Conditions and HCCs.
To accurately quantify levels of mRNA for DNMT3b3 and DNMT3b4, which were the major splice variants in HCCs (Fig. 3), splice variant-specific quantitative RT–PCR was performed in eight normal liver tissues from patients with liver metastasis of primary colon cancer and in 49 noncancerous liver tissues and 59 HCCs from 49 HCC patients.
We have previously reported the DNA methylation status on pericentromeric satellite regions in this cohort (17). In the eight normal liver tissues, satellites 2 and 3 were heavily methylated (17). We then categorized the degree of DNA hypomethylation in Southern blotting as −, +, or 2+: − indicates that the HpaII digest showed the same hybridization pattern as normal liver tissues, + indicates that smaller fragments were detected in the HpaII digest compared with normal liver tissues, and 2+ indicates that the HpaII digest showed the same hybridization pattern as the MspI digest of the same sample and normal liver tissues (17). Nine (18%) of 49 noncancerous liver tissues from HCC patients and 39 (66%) of 59 HCCs showed + or 2+ DNA hypomethylation (17).
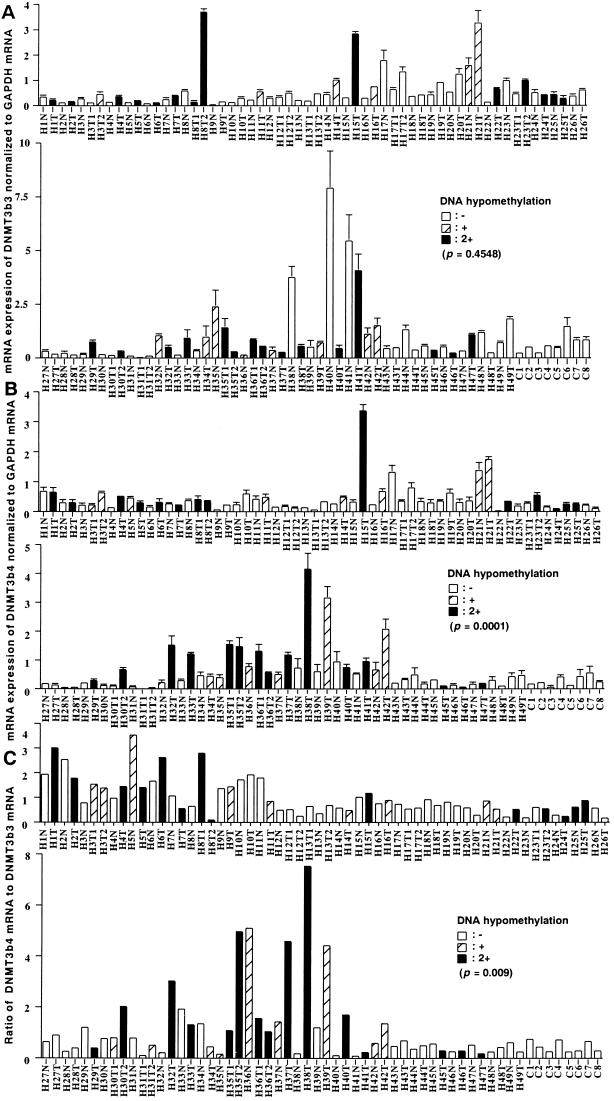
Fig. 4A shows mRNA expression levels for DNMT3b3 normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The average levels of mRNA for DNMT3b3, normalized to GAPDH mRNA, in tissue samples from HCC cases with −, +, and 2+ DNA hypomethylation were 0.72 ± 1.20, 0.87 ± 0.85, and 0.81 ± 1.01 (mean ± SD), respectively. There was no significant correlation between mRNA levels for DNMT3b3 and DNA methylation status on pericentromeric satellite regions.
Figure 4.
Splice variant-specific quantitative RT–PCR for DNMT3b in HCC cases (H) and cases with liver metastasis of primary colon cancer (C). N, noncancerous liver tissue; T, HCC. (A) mRNA levels for DNMT3b3 normalized to GAPDH mRNA. (B) mRNA levels for DNMT3b4 normalized to GAPDH mRNA. (C) The ratio of DNMT3b4 mRNA to DNMT3b3 mRNA. We have previously reported DNA methylation status on pericentromeric satellite regions for these cases (17). In normal liver tissues (C1 to C8), satellites 2 and 3 were heavily methylated (17). The samples indicated by white, hatched, and black bars show no (−), moderate (+), and strong (2+) DNA hypomethylation, respectively, as defined in ref. 17. mRNA levels for DNMT3b4 normalized to GAPDH mRNA (B, P = 0.0001, Kruskal–Wallis test) and the ratios of DNMT3b4 mRNA to DNMT3b3 mRNA (C, P = 0.009, Kruskal–Wallis test) each were significantly correlated with DNA methylation status on pericentromeric satellite regions.
Fig. 4B shows mRNA expression levels for DNMT3b4 normalized to GAPDH mRNA. The average levels of mRNA for DNMT3b4, normalized to GAPDH mRNA, in tissue samples from HCC cases with −, +, and 2+ DNA hypomethylation were 0.31 ± 0.23, 0.78 ± 0.79, and 0.83 ± 0.94, respectively. There was a significant correlation between mRNA levels for DNMT3b4 and DNA methylation status on pericentromeric satellite regions (P = 0.0001, Kruskal–Wallis test). Fig. 4C shows the ratio of DNMT3b4 mRNA to DNMT3b3 mRNA. The averages of the ratios of DNMT3b4 mRNA to DNMT3b3 mRNA in tissue samples from HCC cases with −, +, and 2+ DNA hypomethylation were 0.71 ± 0.52, 1.40 ± 1.40, and 1.65 ± 1.70, respectively. There was a significant correlation between the ratio of DNMT3b4 mRNA to DNMT3b3 mRNA and DNA methylation status on pericentromeric satellite regions (P = 0.009, Kruskal–Wallis test).
Expression levels of mRNA for DNMT3b1 and DNMT3b5 in all tissue samples were too low to be detected by this quantitative RT–PCR system.
Transfection of Human Epithelial 293 Cells with DNMT3b4 cDNA Induces DNA Demethylation on Satellite 2.
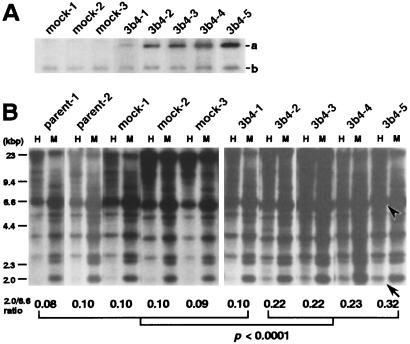
Five clones expressing myc-tagged DNMT3b4, 3b4–1 to 3b4–5, were obtained. The expression of myc-tagged DNMT3b4 was weak in 3b4–1, whereas it was particularly strong in 3b4–5 (Fig. 5A). About 50 days after transfection, DNA methylation status on satellite 2 was analyzed by Southern blotting. As shown in Fig. 5B, in 293 parent cells (individually cultured parent-1 and parent-2), mock transfectants (mock-1 to mock-3), and the DNMT3b4-transfected 3b4–1 clone, which only weakly expresses myc-tagged DNMT3b4, larger DNA fragments were detected in the HpaII digest compared with the MspI digest. On the other hand, in clones 3b4–2 to 3b4–5, which strongly express myc-tagged DNMT3b4, smaller fragments were detected in the HpaII digest compared with parent cells, mock transfectants, and clone 3b4–1. The signal intensity of the 6.6-kbp HpaII digest was strong in all parent cells, mock transfectants, and DNMT3b4 transfectants. The ratios of the signal intensity of the 2.0-kbp HpaII digest to that of the 6.6-kbp HpaII digest in clones 3b4–2 to 3b4–5 were significantly higher than those in parent cells, mock transfectants, and clone 3b4–1 (Fig. 5B, P < 0.0001, Mann–Whitney U test). Moreover, as shown in Fig. 5, there was a correlation between expression level of myc-tagged DNMT3b4 and the ratio of signal intensity of the 2.0-kbp HpaII digest to that of the 6.6-kbp HpaII digest among all DNMT3b4 transfectants. These results suggest that overexpression of DNMT3b4 induces DNA demethylation on satellite 2 in human epithelial cells.
Figure 5.
Transfection of human epithelial 293 cells with DNMT3b4 cDNA. (A) Western blotting with monoclonal anti-myc antibody. Band a, myc-tagged DNMT3b4; band b, endogenous c-myc. Among DNMT3b4 transfectants, expression of myc-tagged DNMT3b4 was weak in 3b4–1, whereas it was particularly strong in 3b4–5. About 50 days after transfection, DNA methylation status on satellite 2 was analyzed by Southern blotting (B). In parent cells (individually cultured parent-1 and parent-2), mock transfectants, and 3b4–1, larger DNA fragments were detected in the HpaII (H) digest compared with the MspI (M) digest. In clones 3b4–2 to 3b4–5, smaller fragments were detected in the H digest compared with parent-1 to parent-2, mock-1 to mock-3, and 3b4–1. The ratios of signal intensity of the 2.0-kbp H digest (arrow) to that of the 6.6-kbp H digest (arrowhead), whose intensity was strong in all parent cells, mock transfectants, and DNMT3b4 transfectants, are shown at the bottom (P < 0.0001, Mann–Whitney U test).
Discussion
We have reported DNA hypomethylation on pericentromeric satellite regions, DNA hypermethylation on CpG islands of genes such as p16, E-cadherin, and HIC-1 (hypermethylated-in-cancer), overexpression of DNA methyltransferases, and reduced expression of methyl-CpG-binding proteins during hepatocarcinogenesis (12–17). Among these findings, DNA hypomethylation on pericentromeric satellite regions was observed even in precancerous conditions and appears to be one of the earliest events during hepatocarcinogenesis. To prevent the development of HCCs in hepatitis virus carriers suffering from chronic hepatitis or cirrhosis, clarification of the molecular mechanisms underlying early events during hepatocarcinogenesis is needed. We have focused on the mechanism of DNA hypomethylation on pericentromeric satellite regions.
DNMT3b is required for de novo DNA methylation on pericentromeric satellite regions in embryonic stem cells and during mouse development (22). In adult cells, the role of DNMT3b remains unclear and it is unknown how DNA methylation on pericentromeric satellite regions is maintained. DNMT1 has so far been recognized as the “maintenance” DNA methyltransferase. Once established by DNMT3b during development, DNA methylation on pericentromeric satellite regions is likely to be maintained by DNMT1 in adult cells. In cancer cells, however, targeting of DNMT1 to substrate DNA may be disrupted by mechanisms such as dysfunction of p21WAF1 (32), which competes with DNMT1 for binding to proliferating cell nuclear antigen (33). In fact, in human cancers, overexpression of DNMT1 is significantly correlated with CpG island methylation phenotype, which is defined by frequent DNA hypermethylation on CpG islands that are not methylated in normal cells (27). If DNMT1 does not maintain DNA methylation on pericentromeric satellite regions because of disturbance of its targeting in cancer cells, DNMT3b must rescue DNA methylation on these regions. Moreover, some recent studies have proposed that all active DNA methyltransferases, DNMT1 and members of the DNMT3 family, probably possess both de novo and maintenance DNA methyltransferase activity in vivo, regardless of their preference for hemimethylated or unmethylated substrates in vitro (4, 34–36). Therefore, it is likely that DNMT3b is required to maintain DNA methylation on pericentromeric satellite regions even in somatic cells, including cancer cells. Therefore, we focused on alterations of DNMT3b during hepatocarcinogenesis.
When analyzed previously by quantitative RT-PCR with a primer set that cannot discriminate between splice variants of DNMT3b, expression of total mRNA for DNMT3b did not correlate with DNA hypomethylation on pericentromeric satellite regions in HCC cases (17). Thus, it is unlikely that reduced expression of DNMT3b simply causes DNA hypomethylation on these regions during hepatocarcinogenesis. Although germ-line mutations of the DNMT3b gene have been reported in patients with immunodeficiency/centromeric instability/facial anomalies syndrome (23, 24), in HCCs we detected no somatic mutations of the DNMT3b gene in any coding exons.
There have been a small number of studies on expression of the four splice variants in the C-terminal catalytic domain of DNMT3b in vivo. DNMT3b3 possesses the N-terminal region and conserved methyltransferase motifs I, IV, VI, IX, and X, but lacks 63 aa residues between motifs VI and IX. Because mouse DNMT3b3 did not methylate an applied unmethylated substrate in an in vitro study (37), DNA methyltransferase activity of human DNMT3b3 should also be carefully discussed. However, it has been reported that DNMT3b3 is ubiquitously expressed in normal human tissues (26). Our data also indicate that the major variant in normal liver tissues is DNMT3b3. Therefore, we cannot rule out the possibility that DNMT3b3 carries the DNMT3b activity at least in vivo, for example in human liver tissue. DNMT3b4 probably does not show methyltransferase activity because it lacks the conserved methyltransferase motifs IX and X, although it retains the N-terminal domain required for targeting to heterochromatin sites through binding to RP58 (38, 39). In vivo, it has been reported that normal human tissues, with the exception of testis, do not express significant levels of DNMT3b4 (26). Here, we confirmed the trace level of DNMT3b4 expression in normal liver tissues.
Overexpression of DNMT3b4 and elevation of the ratio of DNMT3b4 mRNA to DNMT3b3 mRNA were both significantly correlated with the degree of DNA hypomethylation on pericentromeric satellite regions in precancerous conditions and HCCs. DNMT3b4 may compete with the major variant, DNMT3b3, for targeting to pericentromeric satellite regions. This may be the reason overexpression of DNMT3b4, especially in relation to the expression of DNMT3b3, results in DNA hypomethylation on pericentromeric satellite regions in precancerous conditions and HCCs. To confirm this possibility, we introduced DNMT3b4 into human epithelial 293 cells, which express a significant level of DNMT3b3 mRNA and a trace of endogenous DNMT3b4 mRNA (data not shown). DNA demethylation on satellite 2 was observed in DNMT3b4 transfectants, depending on the expression level of myc-tagged DNMT3b4. Although the overexpression levels in DNMT3b4 transfectants may exceed the levels of DNMT3b4 in tissue samples from HCC patients, the results of the transfection studies support the possibility that overexpression of DNMT3b4 induces DNA demethylation on pericentromeric satellite regions in human epithelial cells. On the other hand, a few exceptional tissue samples from HCC patients without overexpression of DNMT3b4 or an elevated ratio of DNMT3b4 mRNA to DNMT3b3 mRNA also showed DNA hypomethylation on pericentromeric satellite regions. Therefore, overexpression of DNMT3b4 should not be considered as the only mechanism for DNA hypomethylation on pericentromeric satellite regions.
We and other groups have shown that chromosomal instability, e.g., allelic imbalance on chromosomes 1 and 16, accumulates even in noncancerous liver tissues showing chronic hepatitis and cirrhosis, and is more frequent in HCCs (15, 16, 40). Satellite 2 is the major sequence of the heterochromatin region adjacent to the centromeres of chromosomes 1 and 16. In fact, frequent 1q copy gain with a breakpoint in heterochromatin DNA was reported in HCCs with DNA hypomethylation on satellite 2 (20). Overexpression of DNMT3b4 may lead to chromosomal instability through induction of DNA hypomethylation on pericentromeric satellite regions, even in the precancerous stages of HCCs. On the other hand, it was recently shown that CpG islands and genes are relatively commonly located in heterochromatin regions (41). The growth rate of DNMT3b4 transfectants became about double that of mock transfectants soon after introduction of DNMT3b4, when allelic imbalance may not yet have accumulated (unpublished data). This growth change may be caused by alterations in the expression of genes in regions in which DNA methylation status is affected by DNMT3b activity. In fact, the expression levels of several genes that potentially participate in signal transduction pathways and/or cell growth were altered in the DNMT3b4 transfectants (unpublished data).
We conclude, therefore, that overexpression of DNMT3b4 induces DNA demethylation on pericentromeric satellite regions even in precancerous stages and may play critical roles in the development of HCC through chromosomal instability and aberrant expression of cancer-related genes. It would be of further interest to investigate whether aberrant expression of DNMT3b splice variants is of general significance during carcinogenesis, even in organs other than the liver.
Acknowledgments
This study was supported by a Grant-in-Aid for the Second Term Comprehensive 10-Year Strategy for Cancer Control and a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor, and Welfare of Japan. Y.S. is a recipient of a Research Resident Fellowship from the Foundation for Promotion of Cancer Research in Japan.
Abbreviations
- DNMT3b
DNA methyltransferase 3b
- HCC
hepatocellular carcinoma
- RT
reverse transcription
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 2.Jones P A, Takai D. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 3.Baylin S B, Esteller M, Rountree M R, Bachman K E, Schuebel K, Herman J G. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 4.Robertson K D. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 5.Gama-Sosa M A, Slagel V A, Trewyn R W, Oxenhandler R, Kuo K C, Gehrke C W, Ehrlich M. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vachtenheim J, Horakova I, Novotna H. Cancer Res. 1994;54:1145–1148. [PubMed] [Google Scholar]
- 7.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Proc Natl Acad Sci USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eguchi K, Kanai Y, Kobayashi K, Hirohashi S. Cancer Res. 1997;57:4913–4915. [PubMed] [Google Scholar]
- 9.Tsuda H, Hirohashi S, Shimosato Y, Terada M, Hasegawa H. Gastroenterology. 1988;95:1664–1666. doi: 10.1016/s0016-5085(88)80093-3. [DOI] [PubMed] [Google Scholar]
- 10.Yasui H, Hino O, Ohtake K, Machinami R, Kitagawa T. Cancer Res. 1992;52:6810–6814. [PubMed] [Google Scholar]
- 11.Kanai Y, Ushijima S, Tsuda H, Sakamoto M, Sugimura T, Hirohashi S. Jpn J Cancer Res. 1996;87:1210–1217. doi: 10.1111/j.1349-7006.1996.tb03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai Y, Ushijima S, Hui A M, Ochiai A, Tsuda H, Sakamoto M, Hirohashi S. Int J Cancer. 1997;71:355–359. doi: 10.1002/(sici)1097-0215(19970502)71:3<355::aid-ijc8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, Hui A M, Kanai Y, Sakamoto M, Hirohashi S. Jpn J Cancer Res. 1997;88:1165–1170. doi: 10.1111/j.1349-7006.1997.tb00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanai Y, Hui A M, Sun L, Ushijima S, Sakamoto M, Tsuda H, Hirohashi S. Hepatology. 1999;29:703–709. doi: 10.1002/hep.510290338. [DOI] [PubMed] [Google Scholar]
- 15.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Hepatology. 2000;32:970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 16.Kanai Y, Ushijima S, Tsuda H, Sakamoto M, Hirohashi S. Cancer Lett. 2000;148:73–80. doi: 10.1016/s0304-3835(99)00316-x. [DOI] [PubMed] [Google Scholar]
- 17.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 18.Kokalj-Vokac N, Almeida A, Viegas-Pequignot E, Jeanpierre M, Malfoy B, Dutrillaux B. Cytogenet Cell Genet. 1993;63:11–15. doi: 10.1159/000133492. [DOI] [PubMed] [Google Scholar]
- 19.Jeanpierre M, Turleau C, Aurias A, Prieur M, Ledeist F, Fischer A, Viegas-Pequignot E. Hum Mol Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 20.Wong N, Lam W C, Lai P B, Pang E, Lau W Y, Johnson P J. Am J Pathol. 2001;159:465–471. doi: 10.1016/S0002-9440(10)61718-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okano M, Xie S, Li E. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 22.Okano M, Bell D W, Haber D A, Li E. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 23.Hansen R S, Wijmenga C, Luo P, Stanek A M, Canfield T K, Weemaes C M, Gartler S M. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu G L, Bestor T H, Bourc'his D, Hsieh C L, Tommerup N, Bugge M, Hulten M, Qu X, Russo J J, Viegas-Pequignot E. Nature (London) 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 25.Maraschio P, Zuffardi O, Dalla Fior T, Tiepolo L. J Med Genet. 1988;25:173–180. doi: 10.1136/jmg.25.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson K D, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales F A, Jones P A. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S. Int J Cancer. 2001;91:205–212. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Xie S, Wang Z, Okano M, Nogami M, Li Y, He W W, Okumura K, Li E. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 29.Alexander J J, Bey E M, Geddes E W, Lecatsas G. S Afr Med J. 1976;50:2124–2128. [PubMed] [Google Scholar]
- 30.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 31.Tagarro I, Fernandez-Peralta A M, Gonzalez-Aguilera J J. Hum Genet. 1994;93:383–388. doi: 10.1007/BF00201662. [DOI] [PubMed] [Google Scholar]
- 32.Baylin S B. Science. 1997;277:1948–1949. doi: 10.1126/science.277.5334.1948. [DOI] [PubMed] [Google Scholar]
- 33.Chuang L S, Ian H I, Koh T W, Ng H H, Xu G, Li B F. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 34.Vertino P M, Yen R W, Gao J, Baylin S B. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee I, Jair K W, Yen R W, Lengauer C, Herman J G, Kinzler K W, Vogelstein B, Baylin S B, Schuebel K E. Nature (London) 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 36.Liang G, Chan M F, Tomigahara Y, Tsai Y C, Gonzales F A, Li E, Laird P W, Jones P A. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aoki A, Suetake I, Miyagawa J, Fujio T, Chijiwa T, Sasaki H, Tajima S. Nucleic Acids Res. 2001;29:3506–3512. doi: 10.1093/nar/29.17.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachman K E, Rountree M R, Baylin S B. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 39.Fuks F, Burgers W A, Godin N, Kasai M, Kouzarides T. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishimoto Y, Shiota G, Wada K, Kitano M, Nakamoto K, Kamisaki Y, Suou T, Itoh T, Kawasaki H. J Cancer Res Clin Oncol. 1996;122:585–589. doi: 10.1007/BF01221189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barry A E, Howman E V, Cancilla M R, Saffery R, Choo K H. Hum Mol Genet. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]