Abstract
The EGF receptor partitions into lipid rafts made using a detergent-free method but is extracted from the low density fraction by Triton X-100. By screening several detergents, we identified Brij 98 as a detergent in which the EGF receptor is retained in the detergentresistant membrane fraction. To identify the difference in lipid composition between those rafts that harbored the EGF receptor (detergent-free and Brij 98-resistant) and those that did not (Triton X-100-resistant), we used multi-dimensional ESI/MS to perform a lipidomics study on these three raft preparations. While all three raft preparations were similarly enriched in cholesterol, the EGF receptor-containing rafts contained more PE and less SPM than did the non-EGF receptorcontaining Triton X-100 rafts. As a result, the detergent-free and Brij 98-resistant rafts exhibited a balance of inner and outer leaflet lipids while the Triton X-100 rafts contained a preponderance of outer leaflet lipids. Furthermore, in all raft preparations, the outer leaflet raft phospholipid species were significantly different from those of the bulk membrane whereas the inner leaflet raft lipids were quite similar to those found in bulk membrane. These findings indicate that the EGF receptor is retained only in rafts that exhibit a lipid distribution compatible with a bilayer structure and that the selection of phospholipids for inclusion into rafts occurs mainly on the outer leaflet lipids.
Lipid rafts are small, low-density, plasma membrane domains that contain high levels of cholesterol and sphingolipids (1-3). Tight interactions between the sterol and the sphingolipids result in the formation of a domain that is resistant to solubilization in detergents (4- 6). This property is often used to separate lipid rafts from bulk plasma membrane fractions (1).
GPI-anchored proteins (1,7-9) and dually acylated proteins (10-12) selectively partition into lipid rafts by virtue of the interaction of their hydrophobic anchors with raft domains. Transmembrane proteins such as flotillin have also been shown to be enriched in lipid rafts as compared to bulk plasma membrane (13). Of special interest has been the finding that many molecules involved in cell signaling are enriched in lipid rafts. This includes proteins such as receptor and non-receptor tyrosine kinases, serpentine receptors, and heterotrimeric and low molecular weight G proteins (for review see (14,15). As a result of the selective localization of signaling molecules in lipid rafts, these domains are thought to serve as organizational platforms for the process of signal transduction.
Recent studies have suggested that lipid rafts represent a heterogeneous collection of domains showing differences in both protein and lipid composition. For example, Madore et al. (16) showed that in lipid raft preparations, the GPI-anchored prion protein could be selectively immunoprecipitated away from a second GPIanchored protein, Thy-1, suggesting that the two GPI-anchored proteins existed in physically separate domains. Gomez-Mouton et al. (17) used immunofluorescence to demonstrate that the raft proteins, urokinase plasminogen activator receptor and CD44 and the raft lipids, GM1 and GM3, distribute asymmetrically in cells. Urokinase plasminogen activator receptor and GM3 localized to the leading edge of the migrating T cells whereas CD44 and GM1 were found at the trailing edge of the cells. Since all four components were isolated in the same lipid raft fraction, these findings suggest that rafts with distinct protein and lipid compositions co-exist within cells and show differences in spatial localization.
Differential sensitivity of proteins to extraction by various detergents has provided additional evidence for heterogeneity among lipid rafts (18-20). The classic method for the preparation of lipid rafts involves the extraction of cells in 1% Triton X-100 followed by separation of the low density raft membranes in a sucrose gradient (1). The use of other detergents to extract membranes has demonstrated that even among a single class of raft proteins, there is variability in their resistance to detergent extraction. For example, GPI-anchored Thy-1 was shown to be associated with low density membrane domains when cells were extracted with 0.5% Triton X-100 or 0.5% Brij 96. However, another GPI-anchored protein, NCAM-120, was completely solubilized by both detergents (16). Thus, these two similarly-anchored proteins must exist in domains of different composition that are differentially sensitive to detergent extraction. Schuck et al. (21) reported that rafts made using different detergents did indeed contain different complements of proteins and were variably enriched in cholesterol and sphingolipids as compared to total cell membranes.
The EGF receptor, a type I transmembrane protein with tyrosine kinase activity (22), has been shown to be enriched in lipid rafts (23,24). Localization of the EGF receptor to rafts appears to modulate both its ligand binding and tyrosine kinase activity since the disruption of lipid rafts by acute cholesterol depletion leads to an enhancement of both these activities (24-27). Unlike traditional raft proteins, the EGF receptor is solubilized by treatment with 1% Triton X-100 (28) but is enriched in lipid rafts that are prepared using a detergent-free protocol (23,24). This suggests that the rafts into which the EGF receptor partitions may be different from classical Triton X-100-resistant rafts.
In the present work, we screened a variety of detergents to determine which supported the retention of the EGF receptor in a low density, detergent-resistant fraction. Among the detergents tested, only Brij 98 produced a distinct, EGF receptor-containing raft fraction. Subsequently, multi-dimensional ESI/MS was used to quantitate the differences in lipid composition of rafts that contained the EGF receptor (Brij 98-resistant membranes and detergent-free raft preparations (29)) and those that did not retain the EGF receptor (Triton X-100-resistant membranes). The results of this lipidomics analysis demonstrate that while all rafts are similarly enriched in cholesterol, the EGF receptor-containing rafts possess a balance of inner and outer leaflet lipids whereas non-EGF receptor-containing rafts contain principally outer leaflet lipids. In addition, the data demonstrate that phospholipids in the outer leaflet of rafts undergo a significant selection for inclusion into rafts whereas inner leaflet lipids show relatively little selection compared to bulk membrane.
Experimental Procedures
Materials
Triton X-100, Tween 20 and Brij 98 were obtained from Sigma Chemical Co. (St. Louis, MO). Octylglucoside was purchased from Calbiochem. Brij 96 was from Fluka. The polyclonal anti-EGF receptor antibody and polyclonal anti-Gq antibody were from Santa Cruz. The monoclonal anti-transferrin receptor antibody was obtained from Zymed. The monoclonal antibodies against flotillin-1 and annexin II and the polyclonal antibody against caveolin-1 were purchased from Transduction Laboratories. The polyclonal anti-ß-COP antibody was from Sigma and the polyclonal anticalnexin antibody was from Stressgen. The monoclonal antibody against the Na+/K+-ATPase ß-subunit was from Biomol. The monoclonal anti-prohibitin antibody was from Neomarkers. Horse radish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG and chemiluminescence reagents were from Amersham. Effectene transfection reagent was obtained from Qiagen. OptiPrep was purchased from Granier BioOne. Percoll was obtained from Sigma Chemical Co. All of the lipid internal standards were purchased from Avanti Polar Lipids. All the solvents used for sample preparation and for mass spectrometric analyses were obtained from Burdick and Jackson.
Cells and tissue culture
Chinese hamster ovary cells (CHO cells) were maintained in Ham’s F12 medium containing 10% fetal calf serum in 5% CO2. Cells were transfected with wild type human EGF receptor in pcDNA3.1(-) (Invitrogen) using Effectene according to the manufacturer’s instructions. Transfected cells were passaged in Ham’s F12 medium containing 10% fetal calf serum and colonies stably expressing the EGF receptor were selected by addition of 400 μg/ml G418 to the growth medium. Isolated clones were maintained in Ham’s F12 medium containing 10% fetal calf serum plus 200 μg/ml G418.
Preparation of detergent-resistant lipid rafts
One confluent D150 plate of cells was washed three times in phosphate-buffered saline and drained well. To the plate was added one ml of MES-buffered saline (50 mM MES, pH 6.5, 150 mM NaCl) containing Triton X-100, Brij 98, Brij 96, Tween 20 or octylglucoside. All detergents were used at a concentration of 1%, except octylglucoside which was used at a concentration of 2%. The detergent to protein ratio was 10:1 for the four detergents used at 1% and was 20:1 for octylglucoside. Cells were scraped into the detergent-containing buffer and mechanically disrupted by passage through a 3” × 22g needle 20 times. The lysate was mixed with an equal volume of 80% sucrose in MES-buffered saline. The material was placed in the bottom of a 12-ml ultracentrifuge tube and a 10 ml linear 5% to 30% sucrose gradient in MES-buffered saline was poured on top. The gradients were centrifuged for 3 h at 175,000 x g and then fractionated into 12 one-ml fractions. Fractions 3 through 5 of the sucrose gradients were used for the MS analysis of lipids.
Preparation of non-detergent lipid rafts
The method of Macdonald and Pike (30) was used for the preparation of non-detergent lipid rafts. Briefly, four confluent D150 plates of cells were scraped into base buffer (250 mM sucrose, 20 mM Tris-HCl, pH 7.8) to which had been added 1 mM CaCl2 and 1 mM MgCl2. Cells were pelleted and then lysed in one ml base buffer with calcium and magnesium by passage through a 3” x 22g needle 20 times and a post-nuclear supernatant obtained by low speed centrifugation. The post-nuclear supernatant was made 25% in Opti-Prep by the addition of an equal volume of 50% Opti-Prep in base buffer. Rafts were isolated by centrifugation in a 0 to 20% Opti-Prep gradient in base buffer. Fractions 1 and 2 were pooled for MS lipid analysis.
Western Blotting
For analysis, 100 μl of each fraction from a gradient was separated by SDS polyacrylamide gel electrophoresis. Gels were transferred electrophoretically to nitrocellulose which was blocked by incubation with 10% non-fat powdered milk. The nitrocellulose strips were incubated for 2 hr at room temperature with primary antibody, washed and then incubated with the appropriate horse radish peroxidaseconjugated secondary antibody. After washing, antibodies were detected by chemiluminescence.
Preparation of lipid extracts and mass spectrometric analysis of lipids
Lipids were extracted by the Bligh and dyer procedure with modification as previously described (31–33). Briefly, to each lipid raft sample (approximately 100 μg of protein), internal standards including 14:0-14:0 PS (40 nmol/mg protein), 15:0-15:0 PtdGro (9 nmol/mg protein), 15:0-15:0 PE (57 nmol/mg protein), and 14:1-14:1 PC (45.0 nmol/mg protein) were added. Lipids from each sample were extracted against 2 ml of 50 mM LiCl twice, back extracted against 2 ml of 10 mM LiCl twice, filtered with a 0.2-μm PFTE syringe filter, and finally stored in 200 μl of 1:1 (v/v) chloroform/methanol. Each lipid solution was further diluted approximately 20-fold just prior to infusion and lipid analysis.
Multi-dimensional ESI/MS analyses were performed utilizing a triple-quadrupole mass spectrometer (ThermoFinnigan TSQ Quantum Ultra, San Jose, CA) equipped with an electrospray ion source as described previously (31,32). Typically, a 1-min period of signal averaging in the profile mode was employed for each MS spectrum and a 1 to 2 min period of signal averaging for each MS/MS spectrum. Identification and quantitation of each individual molecular species were performed in a multidimensional mass spectrometric array format as described previously (31–34)
Protein and cholesterol assays
Proteins were determined using the precipitation Lowry method described by Peterson (35). Cholesterol was determined using the Wako CII Cholesterol Assay kit.
Results
EGF Receptors in Detergent-Resistant Membranes
Five different detergents were screened for their ability to generate EGF receptor-containing lipid rafts. These included: Triton X-100, Tween 20, Brij 98, Brij 96 and octylglucoside. The former four detergents were used at 1% while the latter detergent was used at 2%. Solubilization was aided by passage of the detergent lysates through a 22 g needle. After cell solubilization and centrifugation through a 5% to 30% sucrose gradient as described in Experimental Procedures, the gradients were fractionated and analyzed by Western blotting for the distribution of a variety of plasma membrane proteins. The distribution of marker proteins in a detergent-free raft preparation was included for comparison. The results are shown in Figure 1.
Figure 1. Density gradient analysis of lipid rafts prepared using different detergents.
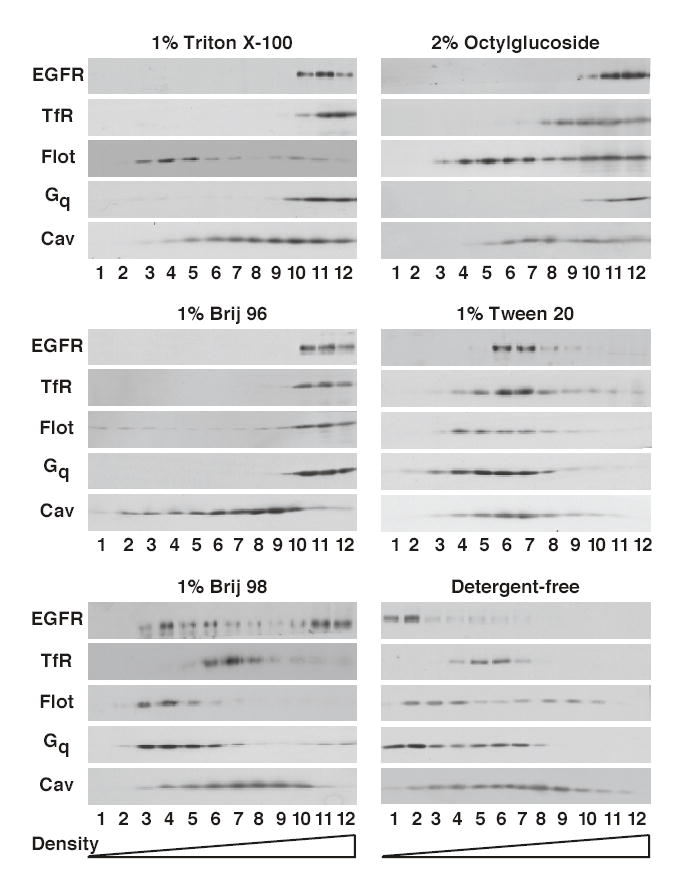
Detergentresistant membranes or detergent-free lipid rafts were prepared from CHO cells as described in Materials and Methods. Extracts were separated by density gradient centrifugation and the gradients fractionated into 12 fractions. An equal volume of each fraction was analyzed by SDS polyacrylamide gel electrophoresis followed by Western blotting with the indicated antibody.
In the detergent-free raft preparation, the EGF receptor was recovered in the three lightest fractions of the gradient along with other raft proteins such as flotillin and the dually acylated Gq protein. These fractions were distinct from those that contained the plasma membrane marker protein, the transferrin receptor, indicating that rafts had been separated from bulk plasma membrane. Caveolin was broadly distributed in this gradient possibly due to the interaction of caveolae with cytoskeletal elements.
Extraction of cells with 1% Triton X-100 resulted in the complete solubilization of the EGF receptor, the transferrin receptor and the heterotrimeric Gq protein, as evidenced by the recovery of these proteins in the high density portion of the gradient. By contrast the raft protein, flotillin, floated into the low density region of the gradient, identifying the location of the lipid raft fraction in this gradient. As in the detergent-free preparation, caveolin was recovered throughout the gradient. Membranes solubilized with 2% octylglucoside showed a pattern of marker protein distribution similar to that observed for Triton X-100-solubilized membranes in which the EGF receptor was excluded from the low density fraction marked by flotillin.
Unexpectedly, solubilization of CHO cells with 1% Brij 96 led to the recovery of almost all proteins, including the raft marker flotillin, in the high density, non-raft fractions of the gradient. The lone exception to this rule was caveolin, which was partially recovered in the upper fractions of the gradient. At the other end of the spectrum, treatment of cells with 1% Tween 20 resulted in the recovery of all marker proteins in the middle third of the gradient, with little distinction in the distribution of the different proteins. This indicates that even at a high concentration and when used with mechanical agitation, Tween 20 does not differentially solubilize raft and non-raft membranes and thus does not permit isolation of a distinct low density raft fraction.
Among the detergents tested, only Brij 98 appeared to generate a distinct low density, detergent-resistant fraction that contained the EGF receptor as well as known raft proteins. Approximately one-third to one-half of the EGF receptor was recovered in the low density region of the gradient (fractions 3–5) that also contained flotillin and Gq. The plasma membrane marker, transferrin receptor, was found in the middle of the gradient at a position distinct from that of the lipid raft proteins. Caveolin again distributed broadly throughout the gradient.
Characterization of Brij 98-Resistant Membranes
Additional studies were undertaken to determine whether the low density membrane fraction obtained by solubilization of cells in Brij 98 effectively separated plasma membrane raft proteins from proteins present on intracellular membranes. In addition to the distribution of the EGF receptor, flotillin, Gq, the transferrin receptor and caveolin, Figure 2 (left) shows the distribution of Na+-K+-ATPase, an intrinsic plasma membrane protein, annexin-2 an extrinsic plasma membrane protein, calnexin, a marker for endoplasmic reticulum, ß-COP, a marker for Golgi, and prohibitin, a mitochondrial membrane protein. As can be seen from the figure, all of the intracellular membrane marker proteins were recovered in the high density portion of the gradient, well-separated from the raft fractions that contained the EGF receptor, flotillin, and Gq. Thus, solubilization of membranes with Brij 98 results in the production of a low density fraction, devoid of markers for intracellular membranes and non-raft plasma membrane proteins, but which contains the EGF receptor and other known raft proteins.
Figure 2. Characterization of detergent-resistant membranes prepared using 1% Brij 98.
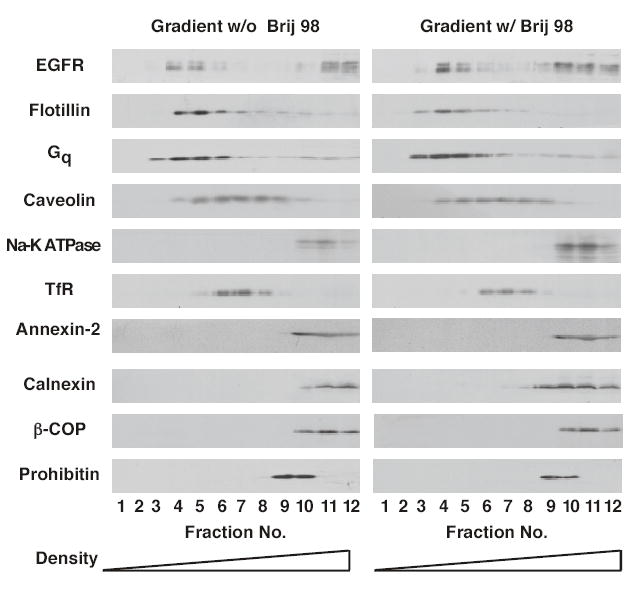
CHO cells were solubilized with 1% Brij 98 and the extracts analyzed by sucrose density gradient centrifugation as described in Materials and Method. Gradients were fractionated and equal volumes of each fraction were separated by SDS polyacrylamide gel electrophoresis. Gels were transferred to Immobilon and subjected to Western blotting using the indicated antibody. Left gradient, sucrose solutions contained no Brij 98. Right gradient, sucrose solutions contained 0.5% Brij 98.
The data on the left of Figure 2 were generated under conditions in which there was no detergent present in the sucrose gradient. However, as shown in the right side of Figure 2, inclusion of 0.5% Brij 98 in the gradient fractions did not alter the distribution of any of the proteins. These data indicate that the distribution of proteins observed using this procedure is not the result of a “reconstitution” of membrane domains associated with removal of detergent during gradient centrifugation.
Mass Spectrometric Analysis of Raft Lipid Composition
To begin to identify the differences between raft preparations that retained the EGF receptor (detergent-free preparations and Brij 98-resistant membranes) and those that did not (Triton X-100- resistant membranes), lipid rafts of each type were prepared and subjected to analysis using multidimensional ESI/MS. Table 1 compares the lipid composition by class of each of the three raft preparations as well as membranes from the PNS fraction. The values for the abundance of each individual species are presented in Supplemental Tables 1, 2 and 3.
Table 1.
Lipid Content of Rafts and PNS Membranes
|
PNS |
Detergent-free Rafts |
Brij 98 Rafts |
Triton X-100 Rafts |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | |
| Ethanolamine glycerophospholipids | 127.17 | 45.11 | 49.95 | 384.24 | 36.33 | 41.25 | 1394.70 | 99.38 | 41.46 | 700.30 | 59.28 | 28.20 |
| Choline glycerophospholipids | 62.24 | 14.59 | 24.44 | 158.64 | 17.52 | 17.03 | 332.73 | 87.35 | 9.89 | 285.30 | 64.65 | 11.50 |
| Sphingomyelin | 35.17 | 4.96 | 13.81 | 257.99 | 44.56 | 27.70 | 1057.54 | 143.69 | 31.44 | 1142.18 | 202.56 | 46.04 |
| PhosphatidyIserine | 14.37 | 3.75 | 5.65 | 84.35 | 10.53 | 9.06 | 428.90 | 56.09 | 12.75 | 272.70 | 6.36 | 10.99 |
| Phosphatidylinositol | 11.02 | 2.14 | 4.33 | 20.05 | 2.67 | 2.15 | 48.06 | 7.53 | 1.43 | 42.74 | 4.33 | 1.72 |
| Phosphatidic acid | 2.41 | 1.17 | 0.95 | 5.73 | 1.10 | 0.62 | 72.99 | 7.17 | 2.17 | 28.84 | 5.40 | 1.16 |
| Phosphatidylglycerol | 2.22 | 1.23 | 0.87 | 20.52 | 3.11 | 2.20 | 29.10 | 2.89 | 0.87 | 8.55 | 1.06 | 0.34 |
| Total Phospholipid | 254.60 | 25.30 | 100.00 | 931.52 | 53.60 | 100.00 | 3364.02 | 399.80 | 100.00 | 2480.61 | 331.90 | 100.00 |
| Cholesterol | 67.80 | 2.35 | 884 | 79 | 2688 | 320 | 2215 | 103 | ||||
| mol% Cholesterol | 21.03 | 48.69 | 44.41 | 47.17 | ||||||||
PNS membranes and lipid rafts were isolated from CHO cells as outlined in Experimental Procedures. Lipids were extracted from samples containing 100 μg of membrane protein and were analyzed for phospholipid content by multi-dimensional ESI/MS. Results represent the mean +/− SD of three separate experiments.
Supplemental Table 1.
Choline-Containing Phospholipids
|
PNS |
Detergent-free |
Brij 98 |
Triton X-100 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m/z | Assignment | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid |
| Choline glycerophospholipids | |||||||||||||
| 738.5 | D16:1-16:0 | 3.84 | 1.55 | 1.51 | 21.29 | 2.54 | 2.29 | 23.26 | 4.12 | 0.69 | 27.47 | 6.28 | 1.11 |
| 740.5 | D16:0-16:0 | 1.54 | 0.35 | 0.60 | 5.78 | 0.69 | 0.62 | 39.72 | 19.76 | 1.18 | 22.68 | 2.81 | 0.91 |
| 752.5 | A16:0-18:1 | 1.62 | 0.65 | 0.64 | 6.34 | 0.43 | 0.68 | 9.23 | 3.79 | 0.27 | 9.02 | 2.67 | 0.36 |
| 764.5 | D16:0-18:2 | 5.53 | 1.19 | 2.17 | 16.41 | 4.73 | 1.76 | 18.01 | 7.93 | 0.54 | 17.24 | 4.17 | 0.69 |
| 766.5 | D16:0-18:1 | 19.51 | 4.70 | 7.66 | 59.76 | 5.19 | 6.42 | 130.75 | 33.57 | 3.89 | 109.57 | 35.37 | 4.42 |
| 788.6 | D16:0-20:4 | 5.06 | 2.06 | 1.99 | 1.41 | 0.25 | 0.15 | 13.28 | 2.52 | 0.39 | 13.74 | 3.01 | 0.55 |
| 790.6 | D18:1-18:2 | 3.13 | 0.65 | 1.23 | 2.57 | 0.77 | 0.28 | 8.50 | 4.01 | 0.25 | 6.91 | 3.42 | 0.28 |
| 792.6 | D18:0-18:2/D18:1-18:1 | 10.31 | 2.62 | 4.05 | 2.99 | 1.39 | 0.32 | 38.62 | 5.17 | 1.15 | 32.01 | 7.15 | 1.29 |
| 794.6 | D18:0-18:1 | 3.08 | 0.91 | 1.21 | 5.18 | 0.85 | 0.56 | 26.92 | 4.39 | 0.80 | 23.73 | 4.01 | 0.96 |
| 800.6 | P18:0-20:4 | 0.75 | 0.32 | 0.30 | 3.93 | 1.74 | 0.42 | 4.13 | 0.40 | 0.12 | 10.93 | 1.46 | 0.44 |
| 812.6 | D16:0-22:6 | 1.30 | 0.55 | 0.51 | 20.21 | 3.25 | 2.17 | 2.24 | 0.02 | 0.07 | 1.35 | 0.82 | 0.05 |
| 814.6 | D18:1-20:4 | 2.87 | 0.48 | 1.13 | 6.27 | 2.40 | 0.67 | 4.76 | 0.40 | 0.14 | 4.41 | 1.87 | 0.18 |
| 816.6 | D18:0-20:4 | 2.50 | 0.27 | 0.98 | 1.69 | 0.54 | 0.18 | 11.11 | 0.13 | 0.33 | 5.16 | 1.92 | 0.21 |
| 836.6 | D18:2-22:6 | 0.49 | 0.30 | 0.19 | 3.01 | 0.80 | 0.32 | 1.12 | - | 0.03 | 0.05 | 0.07 | 0.00 |
| 838.6 | D18:1-22:6 | 0.71 | 0.31 | 0.28 | 2.66 | 1.53 | 0.29 | 1.09 | 1.54 | 0.03 | 1.07 | 0.04 | |
| Total | 62.24 | 14.59 | 24.44 | 158.64 | 17.52 | 17.03 | 332.73 | 87.35 | 9.89 | 285.34 | 64.65 | 11.50 | |
| Sphingomyelin | |||||||||||||
| 765.5 | N20:0 | 11.36 | 8.73 | 4.46 | 6.09 | 0.53 | 0.65 | 145.45 | 32.22 | 4.32 | 121.80 | 24.83 | 4.91 |
| 707.5 | N16:1 | 1.27 | 0.35 | 0.50 | 12.88 | 1.15 | 1.38 | 71.26 | 7.10 | 2.12 | 47.60 | 7.81 | 1.92 |
| 709.5 | N16:0 | 17.87 | 6.50 | 7.02 | 209.07 | 41.49 | 22.44 | 703.74 | 91.17 | 20.92 | 840.36 | 217.64 | 33.88 |
| 735.5 | N18:1 | 0.26 | 0.41 | 0.10 | 0.99 | 0.50 | 0.11 | 2.51 | 0.37 | 0.07 | 2.51 | 1.47 | 0.10 |
| 737.5 | N18:0 | 0.69 | 0.43 | 0.27 | 3.50 | 0.34 | 0.38 | 7.16 | 0.19 | 0.21 | 10.76 | 4.29 | 0.43 |
| 817.5 | N24:2 | 0.96 | 0.50 | 0.38 | 3.69 | 0.45 | 0.40 | 22.38 | 4.62 | 0.67 | 1802 | 1.56 | 0.73 |
| 819.5 | N24:1 | 2.24 | 0.60 | 0.88 | 17.15 | 0.98 | 1.84 | 84.18 | 7.71 | 2.50 | 80.10 | 21.08 | 3.23 |
| 821.5 | N24:0 | 0.51 | 0.37 | 0.20 | 4.63 | 1.94 | 0.50 | 20.87 | 1.43 | 0.62 | 21.04 | 7.68 | 0.85 |
| Total | 35.17 | 4.96 | 13.81 | 257.99 | 44.56 | 27.70 | 1057.54 | 143.69 | 31.44 | 1142.18 | 202.56 | 46.04 | |
Lipids were extracted from membranes and analyzed for choline-containing phospholipid content using multi-dimensional ESI/MS as described in Experimental Procedures. The first number in each pair refers to the number of carbon atoms in the fatty acid chain. The number after the colon refers to the number of double bonds. The two fatty acyl chain designations are separated by a hyphen. If isobaric molecular species are present in an ion peak, all the major isobaric species are given separated by a back slash. The prefix D indicates a diacylglycerol compound. The prefix P indicates a plasmenyl compound. The prefix A indicates a plasmanyl compound. For spingomyelin, only a single fatty acid chain is designated because the other is the 18:1 aliphatic chain included in the sphingosine backbone. The prefix N indicates that the fatty acid is in an amide linkage with sphingosine. Results represent the mean +/− SD of three experiments.
Supplemental Table 2.
Ethanolamine-containing Glycerophospholipids
|
PNS |
Detergent free |
Brij 98 |
Triton X 100 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m/z | Assignment | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid |
| 658.5 | D14:1-16:1 | 1.43 | 0.67 | 0.56 | 2.38 | 1.70 | 0.26 | 0.74 | 0.06 | 0.02 | 1.52 | 0.01 | 0.06 |
| 672.5 | P16:0-16:1 | 2.10 | 1.04 | 0.82 | 3.41 | 0.59 | 0.37 | 4.35 | 0.25 | 0.13 | 5.29 | 0.26 | 0.21 |
| 686.5 | D14:0-18:2/D16:1-16:1 | 0.99 | 1.03 | 0.39 | 2.67 | 1.20 | 0.29 | 2.85 | 0.18 | 0.08 | 2.10 | 0.17 | 0.08 |
| 688.5 | D14:0-18:1/D16:0-16:1 | 0.88 | 0.62 | 0.34 | 3.02 | 1.52 | 0.32 | 2.65 | 0.04 | 0.08 | 2.02 | 0.10 | 0.08 |
| 698.5 | P16:0-18:2/P18:1-16:1 | 1.45 | 0.76 | 0.57 | 7.22 | 1.64 | 0.77 | 28.47 | 3.37 | 0.85 | 26.61 | 3.37 | 1.07 |
| 700.5 | P18:1-16:0/P16:0-18:1 | 4.83 | 1.87 | 1.90 | 28.63 | 7.52 | 3.07 | 109.88 | 4.86 | 3.27 | 88.59 | 11.82 | 3.57 |
| 702.5 | P18:0-16:0 | 3.41 | 1.84 | 1.34 | 0.55 | 0.67 | 0.06 | 0.00 | 1.88 | 2.66 | 0.08 | ||
| 712.5 | D16:1-18:2 | 1.35 | 0.93 | 0.53 | 2.01 | 0.52 | 0.22 | 3.60 | 0.18 | 0.11 | 2.64 | 0.37 | 0.11 |
| 714.5 | P16:0-19:1/D16:0-18:2/D16:1-18:1 | 1.76 | 0.89 | 0.69 | 8.27 | 1.45 | 0.89 | 19.28 | 0.80 | 0.57 | 11.28 | 1.62 | 0.45 |
| 716.5 | D16:0-18:1 | 3.05 | 1.24 | 1.20 | 11.18 | 2.87 | 1.20 | 31.66 | 3.40 | 0.94 | 31.68 | 7.54 | 1.28 |
| 720.5 | P16:1 -20:4 | 10.63 | 1.72 | 4.17 | 9.56 | 1.04 | 1.03 | 7.73 | 0.43 | 0.23 | 5.28 | 0.99 | 0.21 |
| 722.5 | P14:0-22:4/P16:0-20:4 | 4.04 | 1.78 | 1.59 | 29.04 | 14.00 | 3.12 | 143.02 | 4.31 | 4.25 | 47.95 | 17.01 | 1.93 |
| 724.5 | P16:0-20:3/P18:1-18:2 | 0.99 | 0.58 | 0.39 | 3.09 | 0.71 | 0.33 | 5.36 | 0.11 | 0.16 | 6.47 | 2.00 | 0.26 |
| 726.5 | P16:0-20: 2/P18:0-18:2/P18:1-18:1 | 2.45 | 1.01 | 0.96 | 13.14 | 4.91 | 1.41 | 45.40 | 1.94 | 1.35 | 20.78 | 2.08 | 0.84 |
| 728.5 | D17:1-18:1/P16:0-20:1/P18:0-18:1 | 3.75 | 1.62 | 1.47 | 22.08 | 2.60 | 2.37 | 70.35 | 1.41 | 2.09 | 57.18 | 9.54 | 2.31 |
| 730.5 | A18:0-18:1 | 1.29 | 0.68 | 0.51 | 3.34 | 2.20 | 0.36 | 4.31 | 1.91 | 0.13 | 1.33 | 0.38 | 0.05 |
| 732.5 | D14:1-22:6 | 2.08 | 0.74 | 0.82 | 5.40 | 3.73 | 0.58 | 2.05 | 0.45 | 0.06 | 1.04 | 0.37 | 0.04 |
| 734.5 | D16:2-20:4/D14:1-22:5 | 1.01 | 0.47 | 0.40 | 1.78 | 0.56 | 0.19 | 8.42 | 1.24 | 0.25 | _ | _ | 0.00 |
| 736.5 | D16:1-20:4 | 1.28 | 0.69 | 0.50 | 2.93 | 0.50 | 0.31 | 12.31 | 0.69 | 0.37 | 5.44 | 0.06 | 0.22 |
| 740.5 | D16:0-20:3/D18:1-18:2 | 2.94 | 1.10 | 1.15 | 4.15 | 0.28 | 0.45 | 12.22 | 1.39 | 0.36 | 8.55 | 1.00 | 0.34 |
| 742.5 | D18:0-18:2/D18:1-18:1 | 4.00 | 1.52 | 1.57 | 20.03 | 1.51 | 2.15 | 76.27 | 6.77 | 2.27 | 30.46 | 4.75 | 1.23 |
| 744.5 | D18:0-18:1 | 4.75 | 1.45 | 1.86 | 14.30 | 4.86 | 1.54 | 53.67 | 0.40 | 1.60 | 37.49 | 2.36 | 1.51 |
| 746.5 | D18:0-18:0/P16:0-22:6 | 2.22 | 1.07 | 0.87 | 6.59 | 5.10 | 0.71 | 48.02 | 4.14 | 1.43 | 10.88 | 0.63 | 0.44 |
| 748.5 | P18: 1-20:4/P16:0-22:5 | 8.95 | 1.37 | 3.52 | 16.92 | 11.91 | 1.82 | 92.06 | 13.63 | 2.74 | 23.29 | 2.96 | 0.94 |
| 750.5 | P18:1 -20:3/P18:0-20:4/P16:0-22:4 | 5.07 | 1.67 | 1.99 | 22.50 | 9.67 | 2.42 | 127.83 | 14.29 | 3.80 | 50.25 | 7.30 | 2.03 |
| 752.5 | P18:1 -20: 2/P18:0-20:3/P16:0-22:3 | 1.25 | 0.50 | 0.49 | 2.89 | 0.16 | 0.31 | 5.69 | 1.22 | 0.17 | 6.94 | 0.60 | 0.28 |
| 754.5 | P18:1-20:1/P18:1-20:2/P16:0-22:2 | 1.59 | 0.69 | 0.62 | 5.99 | 0.54 | 0.64 | 14.11 | 0.59 | 0.42 | 7.00 | 0.27 | 0.28 |
| 756.5 | P18:1 -20:0/P18:0-20:1/P16:0-22:1 | 1.67 | 0.81 | 0.65 | 7.44 | 0.60 | 0.80 | 17.55 | 1.44 | 0.52 | 13.42 | 0.28 | 0.54 |
| 758.5 | P18:0-20:0 | 2.43 | 1.05 | 0.95 | 9.09 | 2.93 | 0.98 | 10.19 | 0.20 | 0.30 | 10.17 | 0.44 | 0.41 |
| 760.5 | D16:1-22:6 | 1.72 | 0.26 | 0.67 | 3.59 | 1.08 | 0.39 | 11.64 | 2.97 | 0.35 | 5.89 | 1.41 | 0.24 |
| 762.5 | D16:0-22:6 | 1.38 | 0.71 | 0.54 | 2.27 | 0.28 | 0.24 | 7.70 | 1.12 | 0.23 | 3.09 | 0.47 | 0.12 |
| 764.5 | D18:1-20:4 | 2.80 | 0.81 | 1.10 | 5.20 | 0.72 | 0.56 | 20.46 | 3.85 | 0.61 | 10.12 | 0.59 | 0.41 |
| 766.5 | D16:0-22:4/D18:0-20:4 | 4.34 | 1.12 | 1.70 | 9.36 | 1.21 | 1.00 | 49.00 | 6.68 | 1.46 | 18.27 | 2.81 | 0.74 |
| 768.5 | D18:2-20:1/D18:1-20:27 | 1.44 | 0.81 | 0.57 | 3.61 | 1.29 | 0.39 | 7.65 | 0.51 | 0.23 | 3.96 | 0.25 | 0.16 |
| 770.5 | D18:1-20:1/D18:0-20:2 | 1.84 | 0.85 | 0.72 | 5.81 | 1.99 | 0.62 | 9.23 | 0.22 | 0.27 | 2.27 | 0.54 | 0.09 |
| 772.5 | D18:0-20:1/P18:1 -22:6 | 2.01 | 0.92 | 0.79 | 6.53 | 1.12 | 0.70 | 24.42 | 1.85 | 0.73 | 4.31 | 0.30 | 0.17 |
| 774.5 | P18: 1-22:5/P18:0-22:6 | 3.62 | 1.01 | 1.42 | 11.68 | 1.93 | 1.25 | 54.54 | 6.01 | 1.62 | 20.35 | 2.54 | 0.82 |
| 776.5 | P18:0-22:5/P18-1 -22-4 | 2.35 | 0.83 | 0.92 | 5.40 | 2.63 | 0.58 | 37 go | 7.64 | 1.12 | 10.62 | 0.93 | 0.43 |
| 778.5 | P18:0-22:4/P18:1 -22:3 | 3.23 | 1.32 | 1.27 | 5.71 | 1.81 | 0.61 | 33.28 | 4.96 | 0.99 | 9.76 | 0.54 | 0.39 |
| 780.5 | P18:0-22:3 | 1.96 | 0.62 | 0.77 | 6.48 | 1.23 | 0.70 | 14.88 | 0.98 | 0.44 | 8.97 | 0.49 | 0.36 |
| 782.5 | P18:0-22:2 | 1.60 | 0.40 | 0.63 | 4.72 | 1.24 | 0.51 | 11.77 | 0.09 | 0.35 | 8.53 | 0.83 | 0.34 |
| 786.5 | D18:2-22:6 | 1.70 | 0.76 | 0.67 | 6.33 | 1.29 | 0.68 | 15.72 | 0.86 | 0.47 | 9.74 | 0.08 | 0.39 |
| 788.5 | D18:1-22:6 | 2.13 | 0.13 | 0.84 | 7.28 | 0.51 | 0.78 | 41.63 | 3.36 | 1.24 | 21.45 | 1.08 | 0.86 |
| 790.5 | D18:1-22:5/D18:0-22:6 | 2.17 | 0.54 | 0.85 | 5.26 | 2.39 | 0.57 | 10.74 | 1.03 | 0.32 | 4.46 | 1.01 | 0.18 |
| 792.5 | D18-1-22-4/D18:0-22:5 | 2.50 | 0.88 | 0.98 | 6.54 | 0.21 | 0.70 | 22.50 | 3.32 | 0.67 | 9.24 | 1.13 | 0.37 |
| 794.5 | D18:1-22:3/D18:0-22:4 | 3.56 | 0.39 | 1.40 | 12.13 | 0.06 | 1.30 | 45.23 | 7.76 | 1.34 | 20.51 | 2.73 | 0.83 |
| 796.5 | D18:0-22:3 | 1.30 | 0.62 | 0.51 | 3.41 | 1.51 | 0.37 | 2.71 | 0.32 | 0.08 | 2.20 | 0.04 | 0.09 |
| Total | 127.17 | 45.11 | 49.95 | 384.24 | 36.33 | 41.25 | 1.394.68 | 99.38 | 41.46 | 700.30 | 59.28 | 28.23 | |
| Plasmalogens | 73.31 | 236.94 | 925.74 | 455.78 | |||||||||
| % Plasmalogens | 57.64 | 61.66 | 66.37 | 65.08 | |||||||||
Lipids were extracted from membranes and analyzed for ethanolamine glycerophospholipid content using multi-dimensional ESI/MS, as described in Experimental Procedures. Results represent the mean +/− SD of three separate experiments. The first number in each pair refers to the number of carbon atoms in the fatty acyl chain. The number after the colon refers to the number of double bonds. The two fatty acyl chain designations are separated by a hyphen. If isobaric molecular species are present in an ion peak, all the major isobaric species are given separated by a back slash. The prefix D indicates a diacyl compound. The prefix P indicates a plasmenyl compound. The prefix A indicates a plasmanyl compound. Results represent the mean +/− SD of three separate experiments.
Supplemental Table 3.
Acidic Phospholipids
|
PNS |
Detergent-free |
Brij 98 |
Triton X-00 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m/z | Assignment | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid | Average (nmol/mg protein) | S.D. | mol% Phospholipid |
| Phosphatidic Acid | |||||||||||||
| 673.5 | 16:0-18:1 | 0.38 | 0.19 | 0.15 | 1.04 | 0.26 | 0.11 | 18.44 | 4.12 | 0.55 | 7.54 | 1.36 | 0.30 |
| 699.5 | 18:1-18:1 | 0.85 | 0.60 | 0.33 | 0.93 | 0.15 | 0.10 | 17.95 | 1.77 | 0.53 | 4.77 | 0.88 | 0.19 |
| 701.5 | 18:0-18:1 | 1.18 | 0.67 | 0.46 | 3.77 | 0.83 | 0.40 | 36.61 | 1.28 | 1.09 | 16.53 | 3.15 | 0.67 |
| Total PtdH | 2.41 | 1.17 | 0.95 | 5.73 | 1.10 | 0.62 | 72.99 | 7.17 | 2.17 | 28.84 | 5.40 | 1.16 | |
| Phosphatidylglycerol | |||||||||||||
| 745.5 | 16:0-18:2 | 0.23 | 0.07 | 0.09 | 2.11 | 0.71 | 0.23 | 1.24 | 0.14 | 0.04 | 0.40 | 0.13 | 0.02 |
| 747.6 | 16:0-18:1 | 0.77 | 0.15 | 0.30 | 3.60 | 0.38 | 0.39 | 13.36 | 0.93 | 0.40 | 3.59 | 0.80 | 0.14 |
| 749.5 | 16:0-18:0 | 0.13 | 0.13 | 0.05 | 0.16 | 0.08 | 0.02 | 10.19 | 1.08 | 0.30 | 1.57 | 0.51 | 0.06 |
| 771.5 | 18:1-18:2 | 0.17 | 0.18 | 0.07 | 1.65 | 0.53 | 0.18 | 0.49 | 0.85 | 0.01 | 0.46 | 0.20 | 0.02 |
| 773.5 | 18:1-18:1 | 0.75 | 0.60 | 0.29 | 11.54 | 1.73 | 1.24 | 3.53 | 2.45 | 0.10 | 2.53 | 0.32 | 0.10 |
| 775.6 | 18:0-18:2 | 0.16 | 0.28 | 0.06 | 1.46 | 0.50 | 0.16 | 0.34 | 0.59 | 0.01 | 0.00 | 0.00 | 0.00 |
| Total PtdGro | 2.22 | 1.23 | 0.87 | 20.52 | 3.11 | 2.20 | 29.15 | 2.89 | 0.87 | 8.55 | 1.06 | 0.34 | |
| Phosphatidylserine | |||||||||||||
| 73.5 | 16-0-16-1 | 009 | 0.14 | 0.04 | 0.62 | 0.17 | 0.07 | 2.36 | 0.93 | 0.07 | 0.71 | 0.37 | 0.03 |
| 746.5 | P16:0-18:0 | 0.01 | 0.02 | 0.00 | 0.68 | 0.12 | 0.07 | 2.36 | 0.32 | 0.07 | 1.96 | 0.38 | 0.08 |
| 758.5 | 16:0-18:2 | 0.19 | 0.15 | 0.08 | 0.63 | 0.09 | 0.07 | 3.53 | 0.88 | 0.10 | 1.64 | 0.59 | 0.07 |
| 760.5 | 16:0-18:1 | 0.80 | 0.25 | 0.31 | 11.21 | 1.57 | 1.20 | 49.77 | 14.57 | 1.48 | 36.80 | 8.20 | 1.48 |
| 770.8 | P18:1-18:1 | 0.59 | 1.01 | 0.23 | 0.10 | 0.05 | 0.01 | 0.46 | 0.09 | 0.01 | 0.34 | 0.06 | 0.01 |
| 772.8 | P18:0-18:1 | 0.40 | 0.32 | 0.16 | 0.58 | 0.10 | 0.06 | 2.84 | 0.24 | 0.08 | 1.70 | 0.35 | 0.07 |
| 774.8 | P18:0-18:0 | 0.33 | 0.08 | 0.13 | 3.02 | 0.49 | 0.32 | 12.27 | 1.67 | 0.36 | 9.28 | 0.66 | 0.37 |
| 784.5 | 18:1-18:2 | 0.10 | 0.09 | 0.04 | 0.36 | 0.22 | 0.04 | 2.90 | 0.76 | 0.09 | 1.48 | 0.05 | 0.06 |
| 786.5 | 18:1-18:1 | 1.18 | 0.48 | 0.46 | 9.75 | 0.97 | 1.05 | 44.55 | 6.97 | 1.32 | 17.73 | 3.76 | 0.71 |
| 788.5 | 18:0-18:1 | 5.04 | 1.06 | 1.98 | 38.21 | 2.59 | 4.10 | 172.22 | 19.36 | 5.12 | 143.77 | 6.90 | 5.80 |
| 802.5 | P18:0-20:0 | 0.36 | 0.32 | 0.14 | 2.20 | 1.55 | 0.24 | 3.52 | 0.58 | 0.10 | 3.58 | 0.54 | 0.14 |
| 810.5 | 18:0-20:4 | 0.70 | 0.72 | 0.28 | 0.76 | 0.49 | 0.08 | 11.92 | 2.79 | 0.35 | 3.19 | 1.03 | 0.13 |
| 812.5 | 18:0-20:3 | 0.69 | 0.20 | 0.27 | 1.69 | 1.26 | 0.18 | 19.42 | 2.42 | 0.58 | 7.57 | 2.74 | 0.31 |
| 814.5 | 18:0-20:2 | 0.19 | 0.17 | 0.07 | 1.30 | 0.16 | 0.14 | 5.34 | 0.44 | 0.16 | 2.57 | 0.13 | 0.10 |
| 816.5 | 18:1-20:0 | 0.27 | 0.12 | 0.11 | 2.23 | 0.43 | 0.24 | 7.94 | 0.95 | 0.24 | 8.58 | 0.05 | 0.35 |
| 834.5 | 18:0-22:6 | 0.60 | 0.33 | 0.24 | 1.99 | 1.68 | 0.21 | 33.69 | 8.44 | 1.00 | 11.83 | 3.70 | 0.48 |
| 836.5 | 18:0-22:5 | 1.02 | 0.24 | 0.40 | 2.10 | 1.58 | 0.23 | 30.53 | 4.50 | 0.91 | 8.01 | 0.85 | 0.32 |
| 838.5 | 18:0-22:4 | 0.46 | 0.62 | 0.18 | 2.79 | 2.63 | 0.30 | 4.52 | 0.39 | 0.13 | 2.30 | 0.56 | 0.09 |
| 844.8 | 22:0-18:1 | 0.84 | 1.14 | 0.33 | 1.59 | 0.40 | 0.17 | 7.33 | 0.02 | 0.22 | 5.60 | 0.66 | 0.23 |
| 858.9 | A24:0-18:1 | 0.03 | 0.02 | 0.01 | 0.46 | 0.17 | 0.05 | 1.06 | 0.09 | 0.03 | 0.42 | 0.10 | 0.02 |
| 870.9 | 24:1-18:1 | 0.28 | 0.25 | 0.11 | 0.76 | 0.21 | 0.08 | 4.23 | 0.33 | 0.13 | 1.17 | 0.16 | 0.05 |
| 872.9 | 24:0-18:1 | 0.20 | 0.03 | 0.08 | 1.31 | 0.04 | 0.14 | 6.16 | 1.07 | 0.18 | 2.48 | 0.08 | 0.10 |
| Total PtdSer | 14.37 | 3.75 | 5.65 | 84.35 | 10.53 | 9.06 | 428.89 | 56.09 | 12.75 | 272.67 | 6.36 | 10.99 | |
| Phosphatidylinositol | |||||||||||||
| 833.6 | 16:0-18:2 | 0.41 | 0.14 | 0.16 | 0.45 | 0.06 | 0.05 | 3.24 | 0.01 | 0.10 | 1.74 | 0.23 | 0.07 |
| 835.6 | 16:0-18:1 | 0.90 | 0.15 | 0.35 | 2.56 | 0.33 | 0.27 | 6.13 | 1.19 | 0.18 | 6.02 | 1.86 | 0.24 |
| 857.6 | 16:0-20:4 | 0.91 | 0.18 | 0.36 | 1.15 | 0.39 | 0.12 | 2.91 | 0.44 | 0.09 | 0.73 | 1.26 | 0.03 |
| 859.6 | 18:1-18:2 | 0.41 | 0.16 | 0.16 | 0.50 | 0.05 | 0.05 | 1.13 | 0.27 | 0.03 | 4.32 | 0.33 | 0.17 |
| 861.6 | 18:1-18:1 | 0.97 | 0.18 | 0.38 | 2.51 | 0.19 | 0.27 | 4.79 | 1.25 | 0.14 | 3.13 | 0.79 | 0.13 |
| 863.6 | 18:0-18:1 | 0.71 | 0.18 | 0.28 | 1.81 | 0.24 | 0.19 | 4.66 | 1.04 | 0.14 | 4.20 | 0.88 | 0.17 |
| 883.6 | 18:1-20:4 | 0.67 | 0.11 | 0.26 | 0.68 | 0.27 | 0.07 | 1.64 | 0.63 | 0.05 | 1.22 | 0.15 | 0.05 |
| 885.6 | 18:0-20:4 | 4.44 | 0.47 | 1.74 | 8.39 | 1.40 | 0.90 | 20.03 | 2.47 | 0.60 | 16.61 | 2.33 | 0.67 |
| 887.6 | 18:0-20:3 | 0.40 | 0.19 | 0.16 | 0.77 | 0.11 | 0.08 | 1.24 | 0.47 | 0.04 | 1.64 | 0.90 | 0.07 |
| 889.6 | 18:0-20:2 | 0.27 | 0.25 | 0.10 | 0.31 | 0.10 | 0.03 | 0.28 | 0.04 | 0.01 | 0.74 | 0.02 | 0.03 |
| 909.6 | 18:0-22:6 | 0.26 | 0.12 | 0.10 | 0.22 | 0.02 | 0.02 | 0.50 | 0.07 | 0.01 | 0.50 | 0.09 | 0.02 |
| 911.6 | 18:0-22:5 | 0.31 | 0.12 | 0.12 | 0.30 | 0.08 | 0.03 | 0.62 | 0.08 | 0.02 | 0.93 | 0.04 | 0.04 |
| 913.6 | 18:0-22:4 | 0.37 | 0.18 | 0.15 | 0.42 | 0.12 | 0.05 | 0.92 | 0.19 | 0.03 | 0.98 | 0.17 | 0.04 |
| Total Ptdlns | 11.02 | 2.14 | 4.33 | 20.05 | 2.67 | 2.15 | 48.06 | 7.53 | 1.43 | 42.74 | 4.33 | 1.72 | |
| Total Anionic Lipids | 30.03 | 3.39 | 11.79 | 130.66 | 14.16 | 14.03 | 579.08 | 69.42 | 17.21 | 352.79 | 17.15 | 14.22 | |
Lipids were extracted from membranes isolated from CHO cells. Extracts were analyzed for anionic phospholipid content using multi-dimensional ESI/MS, as described in Experimental Procedures. Results represent the mean +/− SD of two separate experiments. The first number in each pair refers to the number of carbon atoms in the fatty acyl chain. The number after the colon refers to the number of double bonds. The two fatty acyl chain designations are separated by a hyphen. The prefix P indicates a plasmenyl compound. The prefix A indicates a plasmanyl compound. All other species are diacyl compounds. Results represent the mean +/− SD of three experiments.
All three raft fractions had a substantially higher mol% of cholesterol and sphingomyelin than did the PNS membranes. By contrast, the mol% of PC and PtdIns in the raft preparations were ~2-fold lower than those seen in the PNS. As observed previously (36), PS was enriched approximately two-fold in the raft preparations as compared to the PNS. These findings demonstrate that all three lipid raft preparations have a composition that is distinct from that of the starting membranes and exhibit the enrichment in cholesterol and sphingomyelin that is characteristic of these domains.
While many of the general characteristics of the lipid raft fractions were shared among all three preparations, the phospholipid composition of the two raft preparations that retained EGF receptors were similar to each other but differed significantly from the lipid composition of the Triton X-100-resistant rafts that excluded EGF receptors (Table I). For example, the detergentfree and Brij 98 raft preparations contained ~40 mol% PE whereas the Triton X-100 rafts contained about one-third less of this lipid. Conversely, the detergent-free and Brij 98-resistant membranes contained ~30 mol% SPM while the Triton X-100-resistant rafts contained ~46 mol% SPM, a 50% increase compared to the receptor-containing rafts. Thus, the EGF receptor-containing rafts had a phospholipid composition that was distinct from that of the non-EGF receptor containing Triton X-100 rafts.
PE and the acidic phospholipids tend to be found mainly in the inner leaflet of the membrane whereas PC and SPM are most often found in the outer leaflet of membranes. The data in Table 1 indicate that the PNS and the EGF receptorcontaining lipid raft fractions have slightly more of these inner leaflet-preferring lipids than of the outer leaflet-preferring lipids whereas the situation is reversed in the non-EGF receptorcontaining Triton X-100 rafts. To determine whether there were any additional general differences between inner and outer leaflet lipids in these membrane preparations, the chain length and saturation of the fatty acyl groups were compared in these subsets of lipids.
Figure 3 shows the distribution of inner and outer leaflet phospholipids with respect to the chain length of the fatty acyl groups. For purposes of this analysis PC and SPM were considered to be outer leaflet lipids whereas PE and the anionic phospholipids were considered to be inner leaflet lipids. As can be seen from the figure, there appears to be relatively little selection of inner leaflet lipids based on chain length. The fraction of inner leaflet lipids containing C16, C18, C22 and C24 fatty acyl side chains was similar in the PNS and all three lipid raft preparations. Only for the lipids containing C20 fatty acyl groups were there any differences, with the EGF receptor-containing rafts containing modestly fewer such lipids and the EGF receptorexcluding Triton rafts showing significantly fewer such lipids than the PNS. By contrast, there are significant differences in the chain length of outer leaflet lipids in the PNS as compared to the three raft preparations. In particular, the rafts appear to select for phospholipids containing C16 and C24 fatty acyl groups and to select against phospholipids containing C18, C20 and C22 fatty acyl groups.
Figure 3.
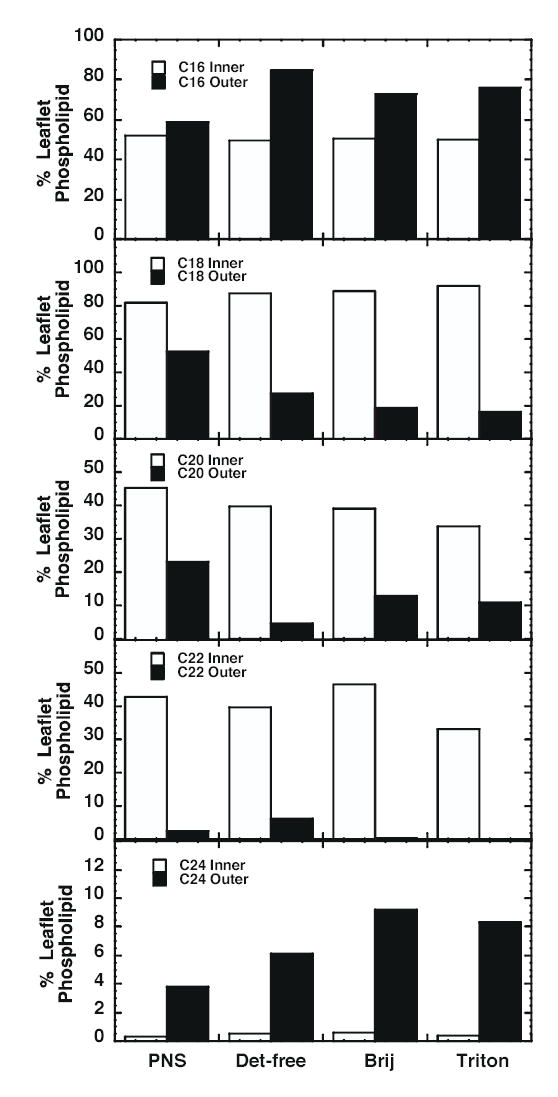
Comparison of fatty acyl chain length in inner and outer leaflet lipids. Inner leaflet lipids were defined as PE, PS, PtdIno, PtdH and PtdGro. Outer leaflet lipids included PC and SPM. The values were calculated as the nmol/mg protein of all species of inner or outer leaflet phospholipid containing at least one chain of a given length divided by the total nmol/mg protein of inner or outer leaflet lipids. The value was multiplied by 100 to obtain a percent of total. Because each phospholipid species contains 2 fatty acyl groups, the inner leaflet lipids total ~200%. SPM has only one fatty acid group (in addition to the C18 backbone of sphingosine) and thus the outer leaflet lipids show variable totals depending on the mol% of SPM in the membrane.
Figure 4 compares the saturation of fatty acyl groups in inner and outer leaflet lipids. For purposes of this analysis, a phospholipid was deemed saturated if it contained no more than one double bond between the two fatty acyl chains. As can be seen from the figure, inner leaflet lipids (PE and acidic phospholipids) were significantly less saturated than outer leaflet lipids (PC and SPM). This is largely due to the highly saturated nature of SPM. For both inner and outer leaflet lipids, however, raft lipids showed a higher degree of saturation than those in the PNS. Among the raft preparations, the EGF receptor-excluding Triton X-100 rafts were more saturated than either of the EGF receptor containing rafts. Overall, the EGF receptor-containing detergent-free and Brij 98 rafts exhibited greater similarity to each other in terms of lipid saturation than they did to the non-EGF receptor-containing Triton X-100 rafts.
Figure 4. Fractional saturation of fatty acyl chains in inner and outer leaflet lipids.
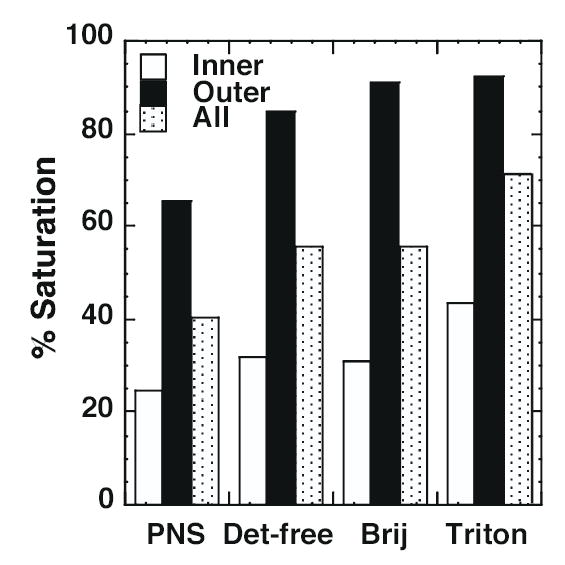
A saturated phospholipid was defined as one in which there was ≤ 1 double bond between the two fatty acyl chains in the lipid. The total nmol/mg protein of saturated species was divided by the total nmol/mg protein of that class of phospholipid and multiplied by 100 to obtain the % saturation. Results are from the averaged data sets for each class of phospholipids.
The individual classes of phospholipids were then examined for differences between PNS and the various raft preparations. Figure 5 presents a scatter plot of the mole fraction (relative abundance) of the individual molecular species of PC obtained by multi-dimensional ESI/MS analysis in positive-ion mode in the presence of LiOH. The data are calculated as the nmol/mg protein of a particular species divided by the total amount of PC present in the preparation. This allows a determination of whether a particular species is selectively enriched or depleted in a membrane preparation, regardless of the overall mol% of that class of phospholipid in a given preparation. Species are organized into groups that show enrichment in two or more of the raft preparations relative to the PNS, depletion in the rafts relative to the PNS, or no consistent change relative to PNS.
Figure 5. Mole fraction of PC species in PNS and lipid raft preparations.
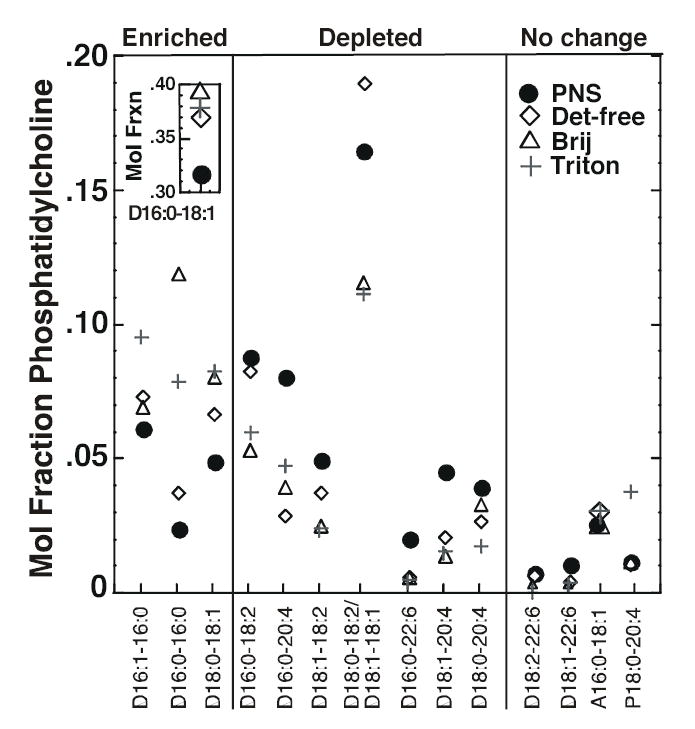
The mole fraction (relative abundance) of each species was calculated by dividing the actual abundance of that species by the total amount of PC present in that particular membrane preparation. Each symbol represents the mole fraction of the species indicated on the X-axis in the indicated membrane preparation. The first number in each pair on the X-axis refers to the number of carbon atoms in the fatty acyl chain. The number after the colon refers to the number of double bonds. The two fatty acyl chain designations are separated by a hyphen. The prefix P indicates a plasmenyl compound. The prefix A indicates a plasmanyl compound. All other species are diacyl compounds. A species was designated as “enriched” if at least two of the three raft preparations showed a greater mole fraction of that species as compared to the PNS. Data represent the average of three experiments. The absolute abundance data are given in Supplemental Table 1.
Among PC species, the most prominent was 16:0-18:1 phosphatidylcholine (inset) which accounted for ~40% of the total phospholipid species in all three raft preparations. This represents an enrichment relative to the PNS in which this represented only ~30% of the total PC. Other PC species that were enriched relative to the PNS included 16:0-16:0, 16:1-16:0 and 18:0-18:1. In general, the PC species that were depleted in the raft preparations were those that contained polyunsaturated chains such as 20:4 and 22:6. These data indicate that PC species containing more saturated fatty acyl groups were enriched in all three raft preparations while species containing polyunsaturated fatty acyl groups were relatively depleted.
For all three raft preparations, the major SPM species was N16:0 SPM (Table 2). However, this species accounted for ~50% of the total SPM in PNS but ~70-80% of the total SPM in the three raft preparations. Interestingly, the N20:0 species of SPM was nearly as abundant (32%) as the N16:0 species in the PNS membranes but was significantly less represented in the raft preparations. In the detergent-free rafts, it accounted for only ~2% of the total SPM whereas this species represented ~10% of the total SPM in the Brij 98- and Triton X-100-resistant rafts. These findings suggest that there is some selectivity with respect to which SPM species partition into what type of lipid raft. Detergent-free rafts have a clear preference for SPM species with shorter fatty acyl groups. The fact that this preference is not as sharp in Brij 98 and Triton X-100 rafts suggests that these detergents may extract out the shorter chain SPM species giving a somewhat skewed composition relative to the original membranes.
Table 2.
Relative Abundance of Sphingomyelin Species in Lipid Rafts and PNS
|
PNS |
Detergent-free |
Brij 98 |
Triton X-100 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| m/z | Assignment | Average (nmol/mg protein) | % Total Sphingomyelin | Average (nmol/mg protein) | % Total Sphingomyelin | Average (nmol/mg protein) | % Total Sphingomyelin | Average (nmol/mg protein) | % Total Sphingomyelin |
| 707.5 | N16:1 | 1.27 | 3.61 | 12.88 | 4.99 | 71.26 | 6.74 | 47.60 | 4.17 |
| 709.5 | N16:0 | 17.87 | 50.82 | 209.07 | 81.04 | 703.74 | 66.54 | 840.36 | 73.58 |
| 735.5 | N18:1 | 0.26 | 0.75 | 0.99 | 0.38 | 2.51 | 0.24 | 2.51 | 0.22 |
| 737.5 | N18:0 | 0.69 | 1.95 | 3.50 | 1.36 | 7.16 | 0.68 | 10.76 | 0.94 |
| 765.5 | N20:0 | 11.36 | 32.31 | 6.09 | 2.36 | 145.45 | 13.75 | 121.80 | 10.66 |
| 817.5 | N24:2 | 0.96 | 2.73 | 3.69 | 1.43 | 22.38 | 2.12 | 18.02 | 1.58 |
| 819.5 | N24:1 | 2.24 | 6.36 | 17.15 | 6.65 | 84.18 | 7.96 | 80.10 | 7.01 |
| 821.5 | N24:0 | 0.51 | 1.46 | 4.63 | 1.79 | 20.87 | 1.97 | 21.04 | 1.84 |
| Total | 35.17 | 100.00 | 257.99 | 100.00 | 1057.54 | 100.00 | 1142.18 | 100.00 | |
The fractional abundance of each species was calculated by dividing the nmol/mg protein of that species by the total nmol/mg protein of Sphingomyelin present in that particular membrane preparation and multiplying by 100. Data represent the average of three experiments. The absolute abundance data are given in Supplemental Table I.
Figure 6 shows the mole fraction (relative abundance) of phosphatidylethanolamine species present in each of the membrane preparations. These data were obtained using negative-ion multi-dimensional ESI/MS. Unlike PC and SPM, there was not a single major species of phosphatidylethanolamine but rather a collection of many species that represented 6-12% of the total. As noted previously, the Triton X-100 rafts contained substantially less PE than the EGF receptor-containing rafts. Nonetheless, all three raft preparations showed similar patterns of enrichment or depletion of specific species as compared to the PNS membranes. For example, all raft preparations tended to be depleted in phosphatidylethanolamine species that contained a 14 carbon fatty acyl group. Such PE species were not highly represented within this class of phospholipids nor were they present in any other phospholipids in these cells. Nonetheless, as a group, they appear to be excluded from lipid rafts. All raft preparations were also relatively depleted in species that contained polyunsaturated fatty acyl groups as compared to PNS, consistent with a preference for more saturated acyl groups. By contrast, the raft preparations were enriched in ethanolamine plasmalogens relative to the PNS (Figure 7), and this enrichment was apparent in the overall composition of these preparations as well (Supplemental Table 3).
Figure 6. Mole fraction of phosphatidylethanolamine species in PNS and lipid raft preparations.
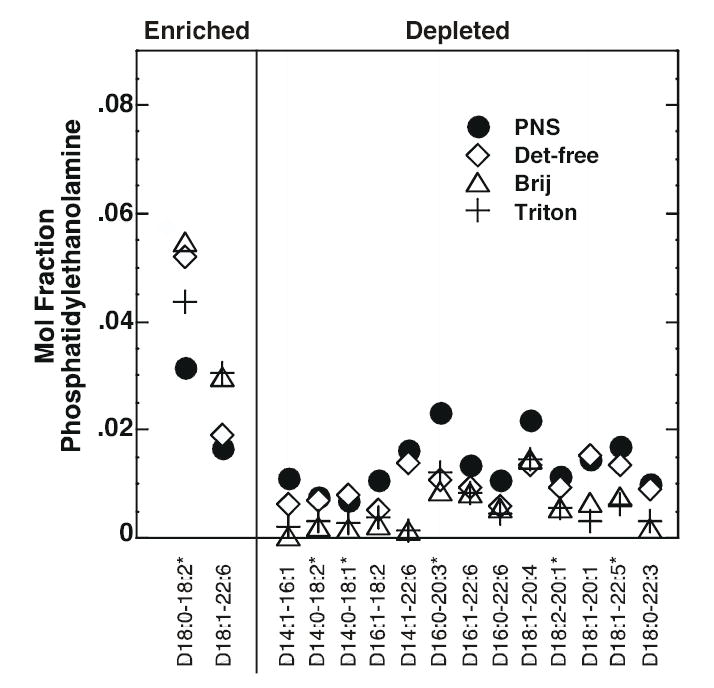
The mole fraction (relative abundance) of each species was calculated by dividing the actual abundance of that species by the total amount of PE present in that particular membrane preparation. Each symbol represents the mole fraction of the species indicated on the X-axis in the indicated membrane preparation. The first number in each pair on the X-axis refers to the number of carbon atoms in the fatty acyl chain. The number after the colon refers to the number of double bonds. The two fatty acyl chain designations are separated by a hyphen. All species are diacyl compounds. Species marked with an asterisk are those for which there are other isobaric species. A species was designated as “enriched” if at least two of the three raft preparations showed a greater relative abundance of that species as compared to the PNS. Data represent the average of three experiments. The absolute abundance data are given in Supplemental Table 2.
Figure 7. Mole fraction of ethanolamine plasmalogens in PNS and lipid raft preparations.
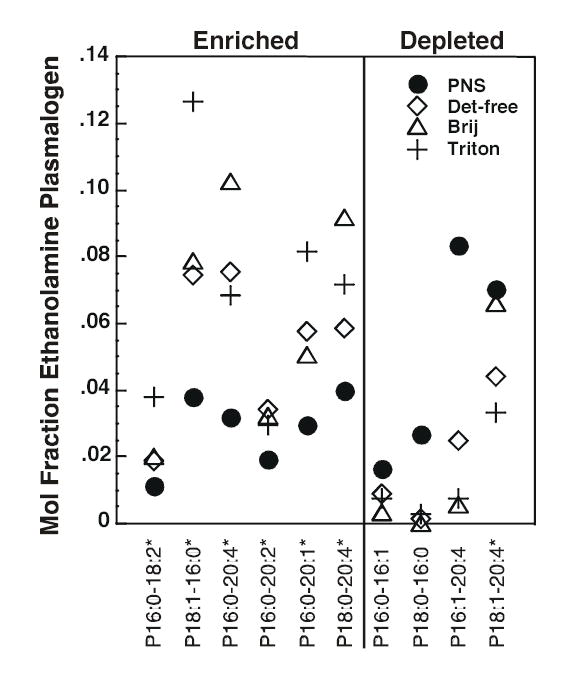
The mole fraction (relative abundance) of each species was calculated by dividing the actual abundance of that species by the total amount of PE present in that particular membrane preparation. Each symbol represents the fractional abundance of the species indicated on the X-axis in the indicated membrane preparation. The first number in each pair on the X-axis refers to the number of carbon atoms in the fatty acyl chain. The number after the colon refers to the number of double bonds. The two fatty acyl chain designations are separated by a hyphen. The prefix P indicates a plasmenyl compound. Species marked with an asterisk are those for which there are other isobaric species. A species was designated as “enriched” if at least two of the three raft preparations showed a greater relative abundance of that species as compared to the PNS. Data represent the average of three experiments. The absolute abundance data are given in Supplemental Table II.
Anionic phospholipids were quantitated by multi-dimensional ESI/MS in negative-ion mode without the addition of LiOH. Figure 8 shows the mole fraction (relative abundance) of the various species of PS. There was a single major species of PS, 18:0-18:1, in all membrane preparations and this species was more abundant in rafts as compared to the PNS. Similarly, the 16:0–18:1 species was markedly enriched in all raft preparations. Together, the increased absolute abundance of these two species accounted for the majority of the increase in PS observed in the three raft preparations. Overall, the lipid rafts tended to be enriched in saturated PS species and depleted in species containing polyunsaturated fatty acyl groups.
Figure 8. Mole fraction of PS species in PNS and lipid raft preparations.
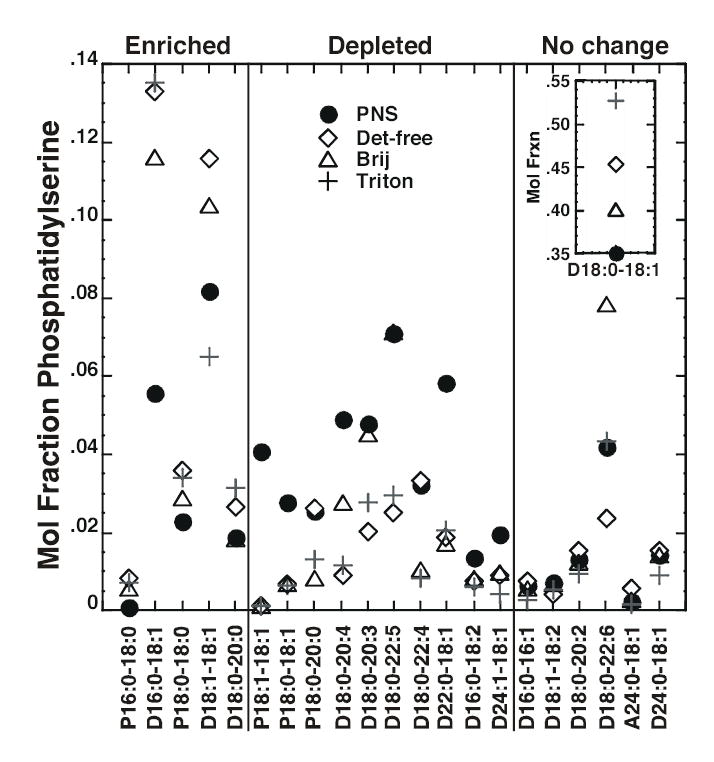
The mole fraction (relative abundance) of each species was calculated by dividing the actual abundance of that species by the total amount of PS present in that particular membrane preparation. Each symbol represents the fractional abundance of the species indicated on the X-axis in the indicated membrane preparation. The first number in each pair on the X-axis refers to the number of carbon atoms in the fatty acyl chain. The number after the colon refers to the number of double bonds. The two fatty acyl chain designations are separated by a hyphen. The prefix P indicates a plasmenyl compound. The prefix A indicates a plasmanyl compound. All other species are diacyl compounds. A species was designated as “enriched” if at least two of the three raft preparations showed a greater relative abundance of that species as compared to the PNS. Data represent the average of three experiments. The absolute abundance data are given in Supplemental Table 3.
Although as a class, PtdIns was depleted in the lipid raft preparations, no species were significantly enriched or depleted in this class (see Supplemental Table 3). This suggests that outside of the selection based on head group, there was no selective partitioning of specific PtdIns species into or out of lipid rafts.
Discussion
The traditional method for the preparation of lipid rafts involves solubilization of cells in Triton X-100, followed by isolation of a low buoyant density fraction by density gradient centrifugation. Many different variations on this method have been used to isolate lipid rafts, in particular, changes in the concentration and type of detergent used for extraction. These changes lead to the inclusion or exclusion of a variety of different proteins and lipids in the resulting rafts (16,17,21). While it is clear that there are differences between the rafts prepared by these various procedures, little is known about how such rafts differ from each other and why some proteins are retained in the detergent-resistant domains while others are not.
In this study, we focused on the behavior of the EGF receptor, a transmembrane protein known to be present in detergent-free preparations of lipid rafts (23,37). A screen of five different detergents demonstrated that under most conditions, the EGF receptor is not isolated in the detergent-resistant fraction. Triton X-100 and octylglucoside both produced rafts that contained the raft marker flotillin and some caveolin but they lacked EGF receptors and Gq. Brij 96 appeared to have a greater tendency than either Triton X-100 or octylglucoside to disrupt lipid rafts, since even flotillin was excluded from the low density fractions prepared using this detergent. In addition, only a small portion of caveolin was found in the low density region of the gradient. This differs from previous reports that suggested that Brij 96 was a less stringent solubilizer of cell membranes than Triton X-100 (21). The difference may be due to the fact that in the earlier experiments, 0.5% Brij 96 and 1% Triton X-100 were compared, while in the current experiments, both detergents were used at a final concentration of 1%. In addition, the protocols for solubilization were different in the two studies. These results make it clear that methodology plays a key role in the outcome of any detergent solubilization experiment.
Although the EGF receptor was not retained in most detergent-resistant membrane fractions, our studies indicated that solubilization of cells in 1% Brij 98 resulted in the generation of a distinct low density membrane fraction that contained the EGF receptor and other raft markers but was devoid of non-raft, plasma membrane or intracellular membrane proteins. Of interest is the observation that the transferrin receptor was recovered in a portion of the gradient that was of intermediate density, at a position distinct from that occupied by other plasma membrane proteins, such as the Na+-K+-ATPase. The transferrin receptor, a non-raft protein, is known to be palmitoylated (38), and may therefore be solubilized in a more lipid-rich, lower density complex than non-acylated proteins. That Brij 98 solubilization can distinguish this class of proteins from others in the membrane may be useful in studies of acylated proteins.
We next addressed the question of why the EGF receptor was included in some detergentresistant membrane fractions but not in others. Analyses of the lipid composition of the two raft preparations that retained the EGF receptor and one (Triton X-100 resistant rafts) that did not was used to determine whether there was a correlation between lipid content and retention of the EGF receptor.
The lipid analyses indicated many general similarities among the three raft preparations. For example, all three preparations were enriched in cholesterol, SPM and saturated acyl side chains as compared to PNS membranes. In addition, all were enriched in PS and ethanolamine plasmalogens relative to PNS. Thus, all three preparations exhibited characteristics consistent with the known properties of lipid rafts.
While many general characteristics were similar among the lipid rafts examined, our findings indicate that there are clear cut differences in the lipid composition of rafts that retain the EGF receptor and those that do not. The major difference observed was in the relative abundance of the major phospholipids, PE and SPM. In both detergent-free and Brij 98 EGF receptor-containing rafts, PE accounts for ~40 mol% of the total phospholipid while in EGF receptor-excluding Triton X-100 rafts, PE represents only ~28% of the total phospholipid. Conversely, SPM represents ~30 mol% in the EGF receptor-containing rafts but 47 mol% in the Triton X-100 rafts.
Because of these differences in the abundance of these major phospholipid species, there is a difference in the relative levels of inner and outer leaflet lipids in the three raft preparations. PE is an inner leaflet-preferring lipid while PC and SPM are outer leafletpreferring lipids. Typically, the ratio of PE/(PC+SPM) is near unity in any given membrane. And this is the case for both of the EGF receptor-containing raft preparations (0.92 and 1.0 for detergent-free and Brij 98 rafts, respectively). However, this ratio is only 0.47 in the receptor-excluding, Triton X-100 rafts. These data indicate that the Triton X-100-resistant rafts are relatively depleted of inner leaflet lipids.
Looking at the inner and outer leaflet lipids as groups, the outer leaflet lipids appear to undergo a more stringent selection for inclusion into lipid rafts than do the inner leaflet lipids. In terms of head group and fatty acyl chain length and saturation, raft outer leaflet lipids are distinctly different from those of the PNS whereas inner leaflet lipids differ only marginally from those found in the PNS membranes. An exception to this rule is PS which is selected for inclusion in lipid rafts, representing 9% of the inner leaflet lipids in PNS but 16%, 22% and 26% of the inner leaflet lipids in detergent-free, Brij 98, and Triton X-100 rafts, respectively. Similarly, PtdIns appears to be specifically excluded from rafts, representing 7% of the inner leaflet lipids in PNS but only 4%, 2.5% and 4% in detergent-free, Brij 98, and Triton X-100 rafts, respectively. The observation that all lipid rafts, no matter how they were made, exhibit this leaflet-dependent difference in lipid selectivity suggests that it is an intrinsic feature of lipid rafts, not one that is introduced by methodological differences in preparation. These findings imply that lipid rafts are largely outer leaflet structures with substantially less rigorously selected inner leaflet lipids.
These data also provide insight into the compositional differences of rafts made by extracting with different detergents. The data in Figure 3 suggest that extraction with Brij 98 or Triton X-100 results in a similar degree of selection for phospholipids containing fatty acyl groups of particular lengths. In this regard, the two detergent raft preparations are more similar to each other than either is to the detergent-free raft preparation. In addition, both detergents appear to preferentially exclude the N16:0 sphingomyelin species from rafts since they contain 5-fold less of this lipid than does the detergent-free raft preparation. However, Triton X-100 selectively extracted inner leaflet lipids but Brij 98-resistant rafts had a normal balance of inner and outer leaflet lipids. Thus, the ability to deplete inner leaflet lipids is not a general feature of all detergents but rather depends on the properties of the individual detergents.
Several studies have suggested that Triton X-100 induces the formation of lipid domains in ternary mixtures of SPM, PC and cholesterol (39,40). Triton X-100 is membrane-disordering, but through unfavorable interactions with SPM, it drives the separation of SPM and cholesterol into a liquid ordered phase, distinct from the liquid disordered phase that contains most of the PC. When applied to our data, these findings suggest that outer leaflet phospholipids are likely to be positively selected for retention in lipid rafts rather than selectively extracted from these domains. While this suggests that the detergent-resistant domains isolated here may not accurately reflect the domains that exist within the cell, it should be noted that all raft preparations showed a similar pattern of selection of outer leaflet lipids. Thus, while detergent extraction may enhance or promote the formation of domains of specific lipid content, it builds on a foundation that is already apparent in rafts made using detergent-free methods.
Rafts appear to exist on both the outer and inner leaflets of the membrane, with outer leaflet rafts harboring GPI-anchored proteins and inner leaflet rafts containing acylated proteins. Outer leaflet rafts are stabilized by the interaction of sphingomyelin and cholesterol. By comparison, inner leaflet rafts are significantly less stable due to the lack of sphingomyelin in this leaflet (41) and hence the absence of its stabilizing interaction with cholesterol. Our data suggest that inner leaflet rafts are preferentially disrupted by treatment with Triton X-100, giving rise to membrane preparations with a preponderance of outer leaflet lipids. By contrast, inner leaflet rafts are retained in both detergent-free and Brij 98-resistant raft preparations. The hypothesis that Brij 98, but not Triton X-100, solubilization results in the maintenance of inner leaflet rafts is supported by the observation that Gq, which is targeted to the cytoplasmic face of the membrane via protein acylation, is retained in Brij 98-resistant membranes but is lost from Triton X-100 resistant membrane fractions.
Together, these data provide a picture of the type of raft into which the EGF receptor partitions. The observation that the EGF receptor is present only in rafts, detergent-resistant or detergent-free, that contain significant levels of both inner and outer leaflet lipids suggests that for this transmembrane protein, the presence of a bilayer structure reminiscent of the original membrane is required for the retention of the receptor in the lipid raft. Furthermore, the lower degree of saturation suggests that these bilayer rafts are likely to be less ordered and hence, more fluid, than other types of rafts. This may be important for enabling the types of conformational changes than must occur in the receptor when it dimerizes and transduces its signal through the membrane. Indeed, Evans and Needham (42) showed that incorporation of a transbilayer peptide into PC/cholesterol mixtures, reduced the compressibility modulus of the resulting bilayers, enhancing their elasticity. Thus, the transmembrane EGF receptor may play a role in defining the properties of the rafts into which it partitions.
Recent studies have suggested that outer leaflet and inner leaflet rafts are only loosely associated under steady state conditions (43). However, co-localization of inner leaflet rafts containing H-ras with outer leaflet rafts is observed when the outer leaflet rafts are aggregated with antibodies directed against a GPI-anchored protein (43). Transmembrane domain proteins such as the EGF receptor that appear to interact with both outer and inner leaflet rafts, may enhance the coupling of rafts in the two leaflets. This could promote the co-localization of signaling molecules present in the different leaflets, thereby enhancing the efficiency of downstream signaling upon receptor activation.
In summary, our studies demonstrate that rafts made using different methodologies exhibit significant differences in lipid composition. Despite these differences, all raft preparations show a more stringent selection for specific characteristics in outer leaflet as compared to inner leaflet lipid species. These findings suggest that raft biogenesis may be driven by the formation of an outer leaflet structure, with inner leaflet rafts forming in response to an additional organizing element. The fact that the EGF receptor is only able to partition into rafts that exhibit a bilayer-like composition raises the possibility that this receptor as well as other transmembrane raft proteins may participate in the organization of rafts on the inner leaflet of the membrane.
Footnotes
This work was supported by NIH grants RO1 GM064491 (LJP), PO1 HL57278 (RWG, XH) and RO1 HL41250 (RWG).
Abbreviations used are: CHO cells, Chinese hamster ovary cells; EGF, epidermal growth factor; ESI, electrospray ionization; GPI, glycosylphosphatidylinositol; MS, mass spectrometry; MS/MS, tandem mass spectrometry; PNS, post-nuclear supernatant; PC, choline glycerophospholipids; PE, ethanolamine glycerophospholipids; PS, serine glycerophospholipids; PtdGro, phosphatidylglycerol; PtdH, phosphatidic acid; PtdIns, phosphatidylinositol; SPM, sphingomyelin.
References
- 1.Brown DA, Rose JK. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 2.Fiedler K, Kobayashi T, Kurzchalia TV, Simons K. Biochem. 1993:6365–6373. doi: 10.1021/bi00076a009. [DOI] [PubMed] [Google Scholar]
- 3.Prinetti A, Chigorno V, Tettamanti G, Sonnino S. J Biol Chem. 2000;275:11658–11665. doi: 10.1074/jbc.275.16.11658. [DOI] [PubMed] [Google Scholar]
- 4.Brown DA, London E. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 5.Edidin M. Ann Rev Biophy Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 6.Shogomori H, Brown DA. Biol Chem. 2003;384:1259–1263. doi: 10.1515/BC.2003.139. [DOI] [PubMed] [Google Scholar]
- 7.Zurzolo C, van't Hof W, van Meer G, Rodriguez-Boulan E. EMBO J. 1994;13:42–53. doi: 10.1002/j.1460-2075.1994.tb06233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milhiet PE, Giocondi MC, Baghdadi O, Ronzon F, Roux B, Le Grimellec C. EMBO Reports. 2002;3:485–490. doi: 10.1093/embo-reports/kvf096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saslowsky DE, Larence J, Ren X, Brown DA, Henderson RM, Edwardson JM. J Biol Chem. 2002;277:26966–26970. doi: 10.1074/jbc.M204669200. [DOI] [PubMed] [Google Scholar]
- 10.Moffett S, Brown DA, Linder ME. J Biol Chem. 2000;275:2191–2198. doi: 10.1074/jbc.275.3.2191. [DOI] [PubMed] [Google Scholar]
- 11.Melkonian Ka, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. J Biol Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- 12.Zacharias DA, Violin JD, Newton AC, Tsien RY. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 13.Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. J Biol Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 14.Smart EJ, Graf GA, McNiven MA, Sessa WC, Engelman JA, Scherer PE, Okamoto T, Lisanti MP. Mol Cell Biol. 1999;19:7289–7304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pike LJ. J Lipid Res. 2003;44:655–667. doi: 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Madore N, Smith KL, Graham CH, Jen A, Brady K, Hall S, Morris R. EMBO J. 1999;18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Mouton C, Abad JL, Mira E, Lacalle RA, Gallardo E, Jimenez-Baranda S, Illa I, Bernad A, Manes S, Martinez-A C. Proc Natl Acad Sci USA. 2001;98:9642–9647. doi: 10.1073/pnas.171160298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roper K, Corbeil D, Huttner WB. Nature Cell Biol. 2000;2:582–592. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 19.Slimane TA, Trugnan G, van Izendoorn SCD, Hoekstra D. Mol Biol Cell. 2003;14:611–624. doi: 10.1091/mbc.E02-08-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilangumaran S, Arni S, van Echten- Deckert G, Borisch B, Hoessli DC. Mol Biol Cell. 1999;10:891–905. doi: 10.1091/mbc.10.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Proc Natl Acad Sci USA. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullrich A, Coussens L, Hayflick JS, Dull TJ, Gray A, Tam AW, Lee J, Yarden Y, Libermann TA, Schlessinger J, Downward J, Mayes ELV, Whittle N, Waterfield ND, Seeburg PH. Nature. 1984;309:418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 23.Mineo C, James GL, Smart EJ, Anderson RGW. J Biol Chem. 1996;271:11930–11935. doi: 10.1074/jbc.271.20.11930. [DOI] [PubMed] [Google Scholar]
- 24.Pike LJ, Casey L. Biochem. 2002;41:10315–10322. doi: 10.1021/bi025943i. [DOI] [PubMed] [Google Scholar]
- 25.Furuchi T, Anderson RGW. J Biol Chem. 1998;273:21099–21104. doi: 10.1074/jbc.273.33.21099. [DOI] [PubMed] [Google Scholar]
- 26.Ringerike T, Glystad FD, Levy FO, Madshus IH, Stang E. J Cell Sci. 2002;115:1331–1340. doi: 10.1242/jcs.115.6.1331. [DOI] [PubMed] [Google Scholar]
- 27.Roepstorff K, Thomsen P, Sandvig K, van Deurs B. J Biol Chem. 2002;277:18954–18960. doi: 10.1074/jbc.M201422200. [DOI] [PubMed] [Google Scholar]
- 28.Pike LJ, Casey L. J Biol Chem. 1996;271:26453–26456. doi: 10.1074/jbc.271.43.26453. [DOI] [PubMed] [Google Scholar]
- 29.Smart EJ, Ying YS, Mineo C, Anderson RGW. Proc Natl Acad Sci USA. 1995;92:10104–10108. doi: 10.1073/pnas.92.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macdonald, J. L., and Pike, L. J. (2005) J. Lipid Res.46, in press [DOI] [PubMed]
- 31.Han X, Yang J, Cheng H, Ye H, Gross RW. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Han X, Cheng H, Mancuso DJ, Gross RW. Biochem. 2004;43:15584–15594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- 33.Han X, Gross RW. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 34.Han, X., and Gross, R. W. (2005) Expert Rev. ProteomicsIn press [DOI] [PubMed]
- 35.Peterson GL. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 36.Pike LJ, Han X, Chung KN, Gross R. Biochem. 2002;41:2075–2088. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
- 37.Pike LJ, Miller JM. J Biol Chem. 1998;273:22298–22304. doi: 10.1074/jbc.273.35.22298. [DOI] [PubMed] [Google Scholar]
- 38.Omary MB, Trowbridge IS. J Biol Chem. 1981;256:4715–4718. [PubMed] [Google Scholar]
- 39.Heerklotz H. Biophys J. 2002;83:2693–2701. doi: 10.1016/S0006-3495(02)75278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heerklotz H, Szadkowska H, Anderson T, Seelig J. J Mol Biol. 2003;329:793–799. doi: 10.1016/s0022-2836(03)00504-7. [DOI] [PubMed] [Google Scholar]
- 41.Niu SL, Litman BJ. Biophys J. 2002;83:3408–3415. doi: 10.1016/S0006-3495(02)75340-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans E, Needhan D. Journal of Physical Chemistry. 1987;91:4219–4228. [Google Scholar]
- 43.Prior IA, Muncke C, Parton RG, Hancock JF. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]


