Abstract
The latent membrane protein (LMP1) of Epstein–Barr virus (EBV) is expressed in EBV-associated nasopharyngeal carcinoma, which is notoriously metastatic. Although it is established that LMP1 represses E-cadherin expression and enhances the invasive ability of carcinoma cells, the mechanism underlying this repression remains to be elucidated. In this study, we demonstrate that LMP1 induces the expression and activity of the DNA methyltransferases 1, 3a, and 3b, using real-time reverse transcription–PCR and enzyme activity assay. This results in hypermethylation of the E-cadherin promoter and down-regulation of E-cadherin gene expression, as revealed by methylation-specific PCR, real-time reverse transcription–PCR and Western blotting data. The DNA methyltransferase inhibitor, 5′-Aza-2′dC, restores E-cadherin promoter activity and protein expression in LMP1-expressing cells, which in turn blocks cell migration ability, as demonstrated by the Transwell cell migration assay. Our findings suggest that LMP1 down-regulates E-cadherin gene expression and induces cell migration activity by using cellular DNA methylation machinery.
E-cadherin is a Ca2+-dependent adhesion molecule that mediates cell–cell contact and is important for tissue morphogenesis, cell polarity, and tumor invasiveness (1). E-cadherin expression is frequently suppressed or reduced in carcinoma tissues of the breast and liver, and many carcinoma cell lines derived from colon, stomach, and prostate (2). In human cancers such as nasopharyngeal carcinoma (NPC), loss of membranous E-cadherin expression correlates significantly with tumor intracranial invasion (3), advanced disease stage, and lymph node metastasis (4). E-cadherin acts as an invasion-suppressor gene (5–7) and, consequently, knowledge of the molecular mechanism that controls its expression or function is of primary importance in understanding the process of tumor invasion.
The ubiquitous human herpesvirus, Epstein–Barr virus (EBV), is closely associated with many human malignancies, including NPC (8), Burkitt's lymphoma, T cell lymphoma, gastric carcinoma (9), and invasive breast cancer (10). NPC is a human squamous cell cancer prevalent in southeastern China and Taiwan. This cancer comprises ≈40% of the head and neck cancers and is notorious for its highly metastasic nature (11). In NPC, EBV infection is predominantly latent and viral gene expression is restricted, similar to other EBV-associated malignant tumors. One of the viral genes, latent membrane protein 1 (LMP1), is expressed in ≈70% of NPC. This protein has the ability to transform rodent cells (12) and render cell growth in soft agar (13). Recent studies showed that LMP1 mimics the CD40 signal pathway, which is important for B-lymphocyte transformation (14). LMP1 transgenic mice exhibit hyperplasia of the epidermis (15). Human epithelial cells expressing LMP1 display significant higher invasive capacity, correlating with decreased E-cadherin expression (16). In addition, LMP1 expression in Madin–Darby canine kidney (MDCK) cells (used as a model in cell migration studies) induces Ets-1 expression and invasive growth (17,18).
LMP1 is a 63-kDa integral membrane protein comprising a short N-terminal domain, six transmembrane domains and a 200-aa C-terminal domain. The C-terminal domain can be subdivided into two major activating regions, CTAR1 and CTAR2. CTAR1 associates with tumor necrosis factor receptor-associated proteins (TRAFs), whereas CTAR2 interacts with tumor necrosis factor receptor-associated death domain protein (TRADD), which mainly mediates nuclear factor κB activity (19) and induces expression of the epidermal growth factor receptor (20). Although several studies indicate that LMP1 is involved in multiple cellular functions, the mechanism used by the protein in mediating cell migration activity is still unclear.
In this study, we provide evidence for LMP1-mediated repression of E-cadherin. We demonstrate that LMP1 induces DNA methyltransferase expression and activity, which in turn results in hypermethylation of the E-cadherin promoter and repression of the E-cadherin protein.
Materials and Methods
Cell Culture.
MDCK cells (ATCC no. CCL-34), MDA-MB-468 cells (ATCC no. HTB 132), and MCF-7 cells (ATCC no. HTB22) were obtained from the American Type Culture Collection. The NPC076 cell line was kindly provided by C. T. Lin (National Taiwan University, Taiwan). LMP1-expressing cell clones were established by cotransfection with LMP1 expression plasmid pT7E (21) and pSV2-neo (Promega), followed by selection with G418. Cells were maintained in 500 μg/ml G418. All cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin.
Plasmid Construction.
The E-cadherin promoter, pEcad−1008/+49, was generated by PCR amplification (35 cycles of denaturation at 96°C for 1 min, annealing at 60°C for 1 min and extention at 72°C for 2 min) by using genomic DNA of normal skin cells and primers EcadF (5′-TTTGGTACCAATTAGGCCGCTCGAGCGAGAGTGCAG-3′) and EcadR (5′-GCTGAGCTCTGAACTGACTTCCGCAAGCTCACAGG-3′). PCR products were digested with KpnI and SacI and cloned into KpnI/SacI-treated pGL2-Basic vector (Promega). The pEcad−34/+49 construct was generated by digestion of pEcad−1008/+49 with BglI and KpnI to remove the sequence from −1008 to −34, followed by self-ligation. The pEcad−164/+49 plasmid was generated by digestion of pEcad−1008/+49 with BstEII and treatment of Klenow, digestion with SacI, followed by ligation overnight at RT to SmaI/SacI-digested pGL2Basic vector. The pEcad−74/+49 construct was generated by PCR, using Ecad-74F (5′-AACCCTCAGCCAATCAGCGGTACGGGGGGCGGTGCTCCG-3′) and EcadR as primers and pEcad−1008/+49 as the template. The −83/+49 fragment was similarly generated, except that a primer Ecad-83F (5′-CGGCAGGTGAACCCTCAGCC-3′) instead of Ecad-74F was used. The PCR product was digested with KpnI and SacI, and subsequently ligated to KpnI/SacI-treated pGL2-basic vector.
The cytomegalovirus (CMV)-LMP1 expression vector was generated by ligation of a PCR product amplified with primers LMP1+43 (5′-GACAAGCTTATGGAACGCGACCTTG-3′)/LMP1+1472 (5′-ATCCCATGGTTAGTCATAGTAGCTTAGCTG-3′) with pT7E DNA (21) as template, to HindIII/BamHI-treated pCMV2-FLAG vector (Kodak).
DNA Transfection and Luciferase Reporter Assay.
MDCK (1 × 105) cells were transfected with 0.5 μg pCMV-LMP1, 0.5 μg pEcad constructs, and 0.1 μg pSV2-gal (Promega). MCF-7 (3.5 × 105) cells were transfected with 1 μg pCMV-LMP1, 1 μg pEcad constructs and 0.2 μg pSV2-gal. Transfections were performed by using lipofectamine (GIBCO/BRL), according to manufacturer's instructions. After 48 h, cells were harvested and lysed in lysis buffer (Promega). Lysates were collected and used to determine luciferase activity in the Autolumat LB953 luminometer (Berthold, Germany). The β-galactosidase activity per transfection, determined with a spectrophotometer at OD420, was used as the internal control.
Transwell Cell Migration Assay.
Cell migration ability was evaluated by using the chemotaxis chamber (Neuroprobe, Cabin John, MD). Cells (1 × 104) in 50 μl of culture medium were applied to the upper chamber of the device, and 30 μl of medium containing 10 μg/ml collagen type IV was added to the lower chamber. A polycarbonate membrane with a pore size of 8 μm was placed in between the two chambers. After 6 h of incubation at 37°C, the membrane was fixed in methanol for 10 min and stained in 20:2 water/Giemsa solution for 1 h. Migrated cells on the membrane were counted under a microscope.
DNA Methyltransferase Activity Assay.
DNA methyltransferase activity was determined by using an experimental protocol generously provided by K.-M. Jair and K. E. Schiebel (The Johns Hopkins Oncology Center, Johns Hopkins University School of Medicine, Baltimore) (22). Briefly, the cell lysate was incubated with 0.5 μg poly d (I-C) and 5 μCi adenosyl-methionine S-[methyl-H3] (Amersham Pharmacia; 1 Ci = 37 GBq) to a final volume of 15 μl at 37°C for 2 h. The reaction was terminated with 40 μl of stop solution (1% SDS/2 mM EDTA/5% Butanol/125 mM NaCl/250 μg/ml salmon sperm DNA). The reaction product was purified by spin-column (Biomax, Rockville, MD), precipitated with ethanol, and resuspended in 30 μl of 0.3 M NaOH. The solution was spotted on GF/C filter disk (Whatman), washed with 3 ml 5% trichloroacetic acid and 3 ml 70% ethanol, dried at 80°C, and evaluated in a scintillation counter.
Methylation-Specific PCR.
Methylation-specific PCR was carried out as described (23). Briefly, genomic DNA (1 μg) was denatured and modified by treatment with 30 μl of 10 mM hydroquinone (Sigma) and 520 μl of 3 M sodium bisulfite (Sigma), pH 5.0, at 50°C for 16 h. Modified DNA was purified by using the Wizard DNA purification kit (Promega) and either analyzed immediately or stored at −20°C until use. For methylation-specific PCR analyses, 100 ng of modified DNA in a final volume of 50 μl was incubated with 1× PCR buffer (1.64 mM NH4SO4/6.4 mM MgCl2/100 mM 2-mercaptoethanol), 200 nM dNTP, and 10 pmol primers for E-cadherin (23). The PCR conditions were described previously (23). The PCR products were separated on an 8% acrylamide gel.
Real-Time Reverse Transcription (RT)-PCR.
Total RNA of the recombinant adenovirus rAdLMP1 or rAdLacZ-infected MCF-7 (1 × 106) cells was isolated with the TRIzol reagent (GIBCO/BRL). The mRNA (1 μg), which was purified by using the oligo(dT) column, was used for synthesis of first-strand cDNA with an oligo(dT) primer and Taqman Reverse transcription kit (Applied Biosystems). Primers used for the detection of DNA methyltransferases and the internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are described in the literature (24). The primers, PCNA F1 (5′-CTCGGCATATCCTGCAAATT-3′) and R1 (5′-GCGCTAGTATTTGAAGCACCA-3′) that generate a 150-bp product were used to detect the expression level of the proliferating cell nuclear antigen (PCNA). PCR with the primers for E-cadherin mRNA detection, hEcad-F (5′-TGAAGGTGACAGAGCCTCTGGAT-3′) and hEcad-R (5-TGGGTGAATTCGGGCTTGTT-3′), resulted in a 200-bp product. Quantitative RT-PCR was performed according to manufacturer's instructions on a LightCycler instrument (Roche Diagnostics) with FastStart DNA Master SYBR Green I (Roche Diagnostics), which fluoresces on binding to double-stranded DNA. Results were normalized to GAPDH or PCNA data.
Immunostaining and Western Blotting.
For immunostaining analyses, cells grown to confluence on cover-slips were fixed with methanol for 10 min at −20°C, blocked with BSA for 1 h, and incubated with individual antibodies for 1 h. The antibodies used in these experiments were α-E-cadherin antibody (Transduction Laboratories, Lexington, KY), α-γ-catenin, α-tubulin (Santa Cruz Biotechnology), or anti-LMP1 antibody S12 (25). Next, cells were washed and incubated in FITC-conjugated secondary antibody and 4′,6-diamidin-2-phenylindol-dihydrochloride (DAPI) (Roche Diagnostics) for 1 h before examination under a microscope (AxioplanII, Zeiss). Western blotting was performed as described (26). Proteins of interest were detected by using the enhanced chemiluminescence system (Amersham Pharmacia), following the manufacturers' instructions. The α-DNA methyltransferase 1(DNMT1) was purchased from Imgenex (San Diego), and the PCNA antibody PC10 was obtained from Santa Cruz Biotechnology.
Recombinant Adenovirus Construction.
Recombinant adenovirus containing the LMP1 gene (rAdLMP1) was generated by using the adenovirus construction system provided by B. Vogelstein and K. W. Kinzler (Howard Hughes Medical Institute Research Lab and Molecular Genetics Lab, The Johns Hopkins Oncology Center, Baltimore). The LMP1 gene was obtained by digestion of pT7E (21) with EcoRI. The 9.4-kb fragment was treated with Klenow and subsequently HindIII, before ligation to the EcoRV/HindIII-treated pShuttle vector at room temperature overnight. The rAdLacZ construct was kindly provided by M. Kawabata and K. Miyazono (The Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo).
Results
LMP1 Represses E-cadherin Gene Expression.
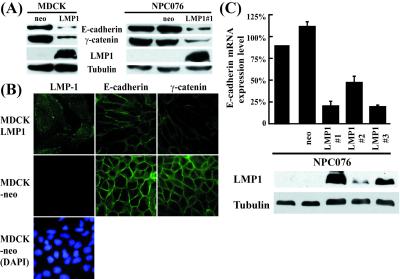
LMP1 represses E-cadherin and triggers the invasive potential of cells (16, 17). To elucidate the potential mechanism, we first determined whether these effects of LMP1 are detected at both protein and mRNA levels. As illustrated in Fig. 1, E-cadherin protein was significantly reduced in stably transfected NPC076–LMP1 cells, but no such reduction was detected in parental NPC076 and NPC076-neo control cell clones, analogous to results obtained with MDCK cells. Interestingly, γ-catenin in the E-cadherin-associated complex was simultaneously reduced in LMP1-expressing cells. Notably, LMP1-expressing cells displayed extended cytoplasm morphology, in contrast to cobblestone-like MDCK-neo cells or parental MDCK cells (data not shown) (Fig. 1B). Equivalent results were observed with NPC076–LMP1 cells (data not shown). To further determine whether LMP1 affects the E-cadherin gene at the transcription level, E-cadherin mRNA was quantitated in NPC076 and stably transfected NPC076-NLMP cells by using real-time RT-PCR. The levels of E-cadherin mRNA in two cell clones stably expressing LMP1 (clone 1 and 3) were ≈20% that detected in NPC076 cells (see Fig. 1C for details). Clone 2, expressing lower levels of LMP1, displayed ≈50% inhibition of E-cadherin gene transcription. These results suggest that LMP1 represses E-cadherin protein through down-regulation of gene transcription.
Figure 1.
Repression of E-cadherin gene expression by LMP1. (A) Western blotting of E-cadherin and its associated complex in cells stably expressing LMP1. Cell extracts (40 μg) of LMP1-expressing cells (MDCKLMP, NPC076LMP), neo-control cell clones (MDCK-neo, NPC076-neo) and parental NPC076 were separated on a SDS/8% PAGE gel, followed by electroblotting onto nitrocellulose membrane. The membrane was incubated with individual antibodies for LMP1, E-cadherin, and γ-catenin, as described in Materials and Methods. Tubulin detected by α-tubulin antibody was used to normalize the amount of total protein in each preparation. (B) Immunostaining studies. Expression of E-cadherin and γ-catenin of the E-cadherin-associated complex in LMP1-expressing MDCK cells was examined by immunfluorescence staining with antibodies specific for these proteins, as described in A. Anti-mouse-FITC antibody was used as the secondary antibody. DAPI staining of MDCK-neo cells revealed the nuclei of the cells. (C) Quantitative analysis of E-cadherin transcripts. Poly(A)+ RNA purified from parental NPC076, NPC076-neo, and NPC076-LMP1 cells was subjected to real time RT-PCR for measuring E-cadherin transcripts, as described in Materials and Methods. Western blotting data on LMP1 and tubulin for individual cell clones are additionally shown. The data represent an average of three independent experiments. The vertical bar indicates SD.
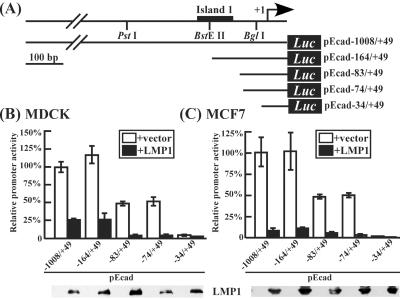
The E-cadherin Promoter (−164 to +49) Contains an LMP1-Mediated Responsive Sequence.
Because MCF-7 and MDCK cells are widely used to study the regulation of E-cadherin promoter activity, these cells were used to identify the promoter region regulated by LMP1. As shown in Fig. 2B, cotransfection of FLAG-tagged LMP1 expressing vector with pEcad−1008/+49 into MDCK and MCF-7 cells resulted in ≈75% and ≈90% reduction of promoter activity, respectively. Further deletion to −164 (pEcad−164/+49) led to similar results. Activity of the promoter constructs, pEcad−83/+49 and pEcad−74/+49 (possessing ≈50% pEcad−1008/+49 activity) was further repressed to ≈10% by LMP1. Finally, deletion to −34 resulted in complete loss of activity. These results suggest that LMP1 represses E-cadherin expression by down-regulating promoter activity, and that the proximal sequence (−164 to −34) of the E-cadherin promoter was chosen for the following study.
Figure 2.
Identification of the LMP1-responsive region of the E-cadherin promoter. (A) Schematic representation of human E-cadherin promoter-containing reporter construct and its deletion mutants. The +1 position represents the transcriptional initiation site and restriction enzyme sites used to generate deletion mutants are indicated. (B and C) LMP1 down-regulation of E-cadherin promoter activity in MDCK and MCF-7 cells. The pE-cad constructs were individually cotransfected with pCMV-LMP1 or pCMV2-FLAG. Luciferase activity was normalized by the β-galactosidase activity of pSV2-gal per transfection. Western blotting analysis of the LMP1 protein is shown at the bottom of the figure. The data are an average of at least six independent experiments, and SD is indicated by the vertical bar.
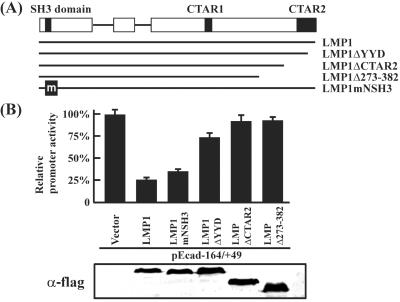
CTAR-2 Is Essential for LMP1-Mediated Down-Regulation of the E-cadherin Gene.
CTAR1 and CTAR2 of LMP1 are two critical regions responsible for several LMP1-mediated functions. Mutation or deletion of these regions often abolishes LMP1-mediated signaling. We used this criterion to confirm LMP1-mediated down-regulation of the E-cadherin gene. FLAG-tagged LMP1 and its mutants (LMP1ΔYYD, LMP1ΔCTAR2, LMP1Δ273-382, and LMP1mNSH3) were cotransfected with pEcad−164/+49 (Fig. 3A) into MDCK cells, and effects were measured by analyzing luciferase activity. Fig. 3B shows that deletion of CTAR2 alone completely abolished LMP1 ability to down-regulate the E-cadherin gene. Deletion of the last three amino acids, YYD, of CTAR2 restored activity to ≈75%. The N terminus contains a Src homology 3 (SH3)-like domain. Mutation at this site had little effect on LMP1-mediated E-cadherin regulation. These results confirm the involvement of LMP1 in E-cadherin repression and indicate that the CTAR2 region of the protein plays a critical role in this regulation process.
Figure 3.
Identification of the LMP1 domain critical for E-cadherin promoter regulation. (A) Schematic representation of LMP1 and its mutant constructs. The Src homology 3 (SH3)-like domain, CTAR1 and CTAR2 are indicated. (B) Regulation of pEcad−164/+49 by LMP1 and its mutants. Individual LMP1-expressing constructs were cotransfected with the pEcad−164/+49, and relative luciferase activity was determined and normalized as described in Materials and Methods. The percentage of promoter activity in each transfection was determined with respect to the vector control value, taken as 100%. The LMP1 protein in each transfection was determined by Western blotting analysis using α-flag antibody, M5 (Sigma).
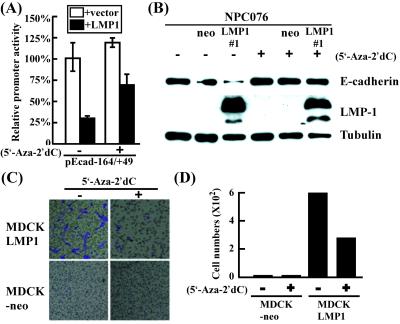
LMP1-Mediated Repression of E-cadherin Is a Methylation-Dependent Activity.
Methylation of the CpG islands within the E-cadherin promoter region inactivates gene expression (27). To analyze whether methylation plays a role in LMP1-mediated E-cadherin repression, we examined whether the DNA methyltransferase inhibitor, 5′-Aza-2′dC (5 μM) restores E-cadherin promoter activity and protein expression. As shown in Fig. 4A, cotransfection of FLAG-tagged LMP1 expressing vector with pEcad−164/+49 into MDCK cells resulted in reduction (≈25%) of the promoter activity, which can be restored (≈75%) by treatment with the inhibitor. Similarly, E-cadherin protein level was examined in treated and untreated NPC076, NPC076-neo, and NPC076-LMP1 cells. Our data show that E-cadherin protein expression in NPC076-LMP1 cells was restored after drug treatment for 5 days (Fig. 4B). In contrast, no significant changes were observed before and after drug treatment in either the parental NPC076 or NPC076-neo cells. Similar results were observed with MDCK-LMP1 cells (data not shown). These results suggest that methylation plays a crucial role in LMP1-mediated E-cadherin repression.
Figure 4.
LMP1-mediated repression of E-cadherin and cell migration ability are restored by 5′-Aza-2′dC. (A) Restoration of E-cadherin promoter activity by 5′-Aza-2′dC. NPC cells were cotransfected individually with pEcad−164/+49 with pCMV-LMP1 or pCMV2-FLAG in the presence or absence of 5′-Aza-2′dC. Luciferase activity was normalized by the β-gal activity of pSV2-gal per transfection. The data are an average of five independent experiments, and SD is indicated by the vertical bar. (B) De-repression of E-cadherin by 5′-Aza-2′dC. NPC076-LMP1 cells were treated with 5 μM 5′-Aza-2′dC for 5 days. Cells were harvested, and E-cadherin expression was analyzed by Western blotting using α-E-cadherin antibody. LMP1 expression was detected with an LMP1-specific monoclonal antibody, S12. Tubulin (identified by the α-tubulin antibody) was used as the internal control. (C and D) Inhibition of LMP1-mediated cell migration by 5′-Aza-2′dC. MDCK-LMP1 and MDCK-neo cells were treated with 5 μM 5′-Aza-2′dC for 5 days or left untreated before subjection to the Transwell migration assay. Cells migrating to the other side of the membrane were stained with Giemsa and counted under the microscope. Data shown in D represent an average of 10 wells. The experiment was repeated three times, with reproducible results. The vertical bar in the figure indicates SD.
E-cadherin repression is associated with enhanced cell migration ability. We therefore examined whether inhibition of methylation alters the cell migration activity of LMP1-expressing cells. MDCK-LMP1 cells were either left untreated or treated with 5′-Aza-2′dC and subjected to the Transwell migration assay. Of 1 × 104 MDCK-LMP1 cells, >600 untreated cells migrated to the other side of the membrane, compared with only ≈250 cells from drug-treated cultures (Fig. 4 C and D), indicating that the migration ability of LMP1-expressing cells is inhibited by 5′-Aza-2′dC treatment.
LMP1 Activates DNA Methyltransferase.
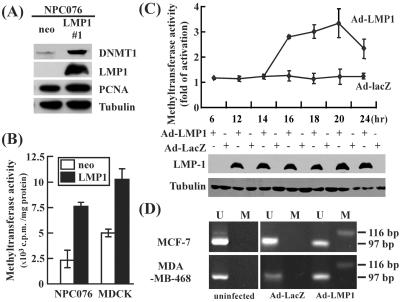
If E-cadherin promoter methylation is indeed mediated by LMP1, it is possible that this protein enhances DNA methyltransferase expression. This would result in the elevation of DNA methyltransferase enzyme activity and hypermethylation of the E-cadherin promoter. As shown in Fig. 5A, DNMT1 protein level was significantly elevated as examined in NPC076-LMP1 stable cell clone, as compared with relatively unchanged level of a cell proliferation marker PCNA (both normalized to tubulin). Accordingly, methyltransferase activity was clearly elevated in both NPC076-LMP1 (≈3-fold) and in MDCK-LMP1 (≈2-fold) cells, compared with that in control cells (Fig. 5B). The kinetics of activation of DNA methyltransferase activity was also analyzed in MCF-7 cells infected with rAdLMP1 or the control virus, rAdLacZ. Enzyme activity was elevated ≈3-fold at 16 h after infection, and reached maximum levels (≈3.5-fold) at the 20-h time point, followed by a slight drop at 24 h (Fig. 5C). Control cells infected with the rAdLacZ virus did not display elevated enzyme activity.
Figure 5.
LMP1 activates DNMT. (A) Western blot analysis of DNA methyltransferase 1 (DNMT1) in NPC076-LMP1 and NPC076-neo cells. Western blot analysis was performed as described in Fig. 1A. The protein level of DNMT1, PCNA, and LMP1 were detected by α-DNMT1 antibody 60B1220, α-PCNA antibody PC10, and α-LMP1 antibody S12, individually. Tubulin detected by α-tubulin antibody was used to normalize the amount of total protein in each preparation. (B) DNA methyltransferase activity assay in LMP1-expressing stable cell clones. DNA methyltransferase activity was measured in NPC076-LMP1 and MDCK-LMP1 or neo-control cells. (C) Kinetics of DNA methyltransferase activity in rAdLMP1-infected MCF-7 cells. Cells were infected with either rAdLMP1 or rAdLacZ at a multiplicity of infection of 100. Methyltransferase activity was determined as described in Materials and Methods. The data are an average of at least three independent experiments. The vertical bar represents standard deviation. Increase in C was determined by dividing the activity of cells infected with rAdLMP1 with that of cells infected with rAdLacZ. LMP1 and tubulin expression, shown at the bottom, were analyzed by Western blotting. (D) Methylation-specific PCR of E-cadherin promoter. MDA-MB-468 and MCF-7 cells were infected with rAdLMP1 or control virus, rAdLacZ, at multiplicity of infection of 100. Methylation-PCR analysis was carried out as described in Materials and Methods. U, unmethylated; M, methylated. The 116-bp PCR product represents the methylated state of island 1 within the E-cadherin promoter sequence, whereas the 97-bp product represents the unmethylated state. Uninfected cells were used as the unmethylated controls.
The E-cadherin promoter displayed a pattern of hypermethylation, as demonstrated by the methylation status of the E-cadherin promoter (the specific CpG island 1) in MCF-7 and MDA-MB-468 cells after infection with the rAdLMP1 virus. Genomic DNAs extracted from cells were treated with sodium bisulfite, converting deoxycytosine but not 5-methylcytosine residues into uracil through deamination. When methylation-specific primers of the E-cadherin promoter for PCR amplification were used (23), comparable high frequencies of 5-methylcytosine residues were found in rAdLMP1-infected MCF-7 and MDA-MB-468 cells, representing by the presence of 116-bp PCR products (Fig. 5D). Such hypermethylation was not detected in rAdLacZ-infected MCF-7 and MDA-MB-468 cells or uninfected control cells (Fig. 5D). Thus, our results suggest that LMP1 induces DNA methyltransferase activity and hypermethylation of the E-cadherin promoter.
LMP1 Triggers Expression of DNA Methyltransferases.
DNA methyltransferases DNMT1, DNMT3a, and DNMT3b, control the methylation status in cells. To examine whether LMP1-mediated DNA methylation is executed by these known DNA methyltransferases, dnmt gene expression was examined in MCF-7 cells infected with rAdLMP1 or rAdLacZ. The results shown in Table 1 indicate that LMP1 induces expression of all three DNMTs in LMP1-expressing NPC cell clones and rAdLMP1-infected MCF-7 cells, which correlates with elevated methyltransferase activity and significantly reduced E-cadherin transcripts. It is worth noting that the expression levels of dnmt genes were also elevated as normalized to that of a cellular proliferation marker PCNA, indicating that LMP1-mediated activation of dnmt gene expression is independent of the rate of cell proliferation. Our data collectively suggest that LMP1 induces expression of DNA methyltransferases, DNMT1, DNMT3a, and DNMT3b.
Table 1.
Relative gene expression in LMP1-expressing cells
| Cells | Genes (fold activation ± SD)/GAPDH
|
Genes (fold activation ± SD)/PCNA
|
|||||
|---|---|---|---|---|---|---|---|
| DNMT1 | DNMT3a | DNMT3b | E-cadherin | DNMT1 | DNMT3a | DNMT3b | |
| Stable clones | |||||||
| NPC-neo | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| NPC076LMP1 1 | 2.4 ± 0.25 | 2.5 ± 0.5 | 2.0 ± 0.6 | 0.25 ± 0.05 | 3.6 ± 0.62 | 6.2 ± 0.71 | 8.5 ± 2.1 |
| NPC076LMP1 2 | 1.5 ± 0.4 | 1.6 ± 0.15 | 1.4 ± 0.2 | 0.50 ± 0.12 | 3.0 ± 0.61 | 4.6 ± 0.07 | 7.6 ± 1.6 |
| NPC076LMP1 3 | 3 | 2 ± 0.3 | 1.5 ± 0.1 | 0.16 ± 0.03 | 3.7 ± 0.56 | 4.6 ± 0.21 | 6.2 ± 2.5 |
| Recombinant adenovirus infection | |||||||
| MCF7/AdLacZ/6 h | 1 | 1 | 1 | 1 | – | – | – |
| MCF7/AdLacZ/20 h | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| MCF7/AdLMP1/6 h | 1 | 1 | 1 | 1 | – | – | – |
| MCF7/AdLMP1/20 h | 1.9 ± 0.4 | 2.2 ± 0.6 | 2.4 ± 0.2 | 0.3 | 1.4 ± 0.2 | 1.4 ± 0.1 | 1.7 ± 0.5 |
DNMT and E-cadherin gene transcripts in LMP1-expressing NPC cells or the NPC-neo cells, and in the MCF-7 cells infected with rAdLMP1 or rAdLacZ (at a multiplicity of infection of 100 individually) were determined by the real-time RT-PCR. Normalization was done by using the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or PCNA. The value obtained from the NPC076-neo cells or MCF-7 cells harvested at 6 h after infection (similar to that at 0 h) is designated respectively as “1.” The data are the average of three independent experiments.
Discussion
LMP1 is an EBV-encoded oncoprotein that transforms rodent cells and enhances cell migration activity. The latter correlates with repression of E-cadherin expression. This study is a step toward elucidating the mechanism of LMP1-mediated E-cadherin repression. We demonstrate that LMP1 induces DNA methyltransferases, resulting in hypermethylation of the proximal E-cadherin promoter. Conversely, the DNA methylation inhibitor, 5′-Aza-2′dC, restores expression of E-cadherin in LMP1-expressing cells and inhibits LMP1-induced cell migration activity.
Loss or alteration of E-cadherin occurs frequently in many different epithelial cancers (28). It is generally believed that repression of E-cadherin is achieved by hypermethylation of the E-cadherin promoter, down-regulation of E-cadherin promoter by cellular transcription repressors such as Snail, Sip1, and Ets-1 (29–31), and mutations in the ORF of the E-cadherin gene (28). Data from this study indicate that LMP1 mainly induces hypermethylation of the E-cadherin promoter, which is independent to that mediated by transcription repressors, such as Snail or Ets-1 (unpublished data). Elevated expression of DNA methyltransferases is observed in some cancer cells, compared with their normal counterparts (32). DNMT3a and DNMT3b appear to be active only de novo, and do not consistently maintain DNA methylation. On the other hand, DNMT1 is highly conserved in a wide range of species, from sea urchin to human, and is required to maintain the bulk of methylation in cells. In the presence of LMP1, all three enzyme levels are elevated, indicating that LMP1 activates de novo methylation.
In this study, we used methylation-specific PCR to access the LMP1-mediated methylation activity. This method is simple, sensitive, and specific for measuring the E-cadherin promoter hypermethylation status (23). The E-cadherin promoter region amplified with methylation-specific primers covers the specific CpG island 1, which is located between −161 and −62 relative to the transcription initiation site of the gene and is also included in the promoter construct pEcad−164/+49 examined in this study. Further confirmation of methylated CpG sequences within E-cadherin promoter in LMP1-expressing cells by bisulfite-modified DNA sequencing revealed that hypermethylation was detected in a low proportion of cells (data not shown). The proximal E-cadherin promoter (−164 to −74) that is hypermethylated in LMP1-expressing cells is seldom methylated in normal cells or early stages of cancers. In contrast, significant methylation is noted in neoplastic cells of various cancers, such as oral squamous cell carcinomas (33), prostate cancer (34), breast carcinoma (32, 35), gastric carcinoma (36), and lung cancer (37). Hypermethylation of the promoter is additionally critical for reducing E-cadherin expression during the metastasis of carcinoma (23) and enhancement of the cell migration ability of epithelial tumor cell lines. In NPC cells, the promoters of two other tumor suppressor genes, p16 and RASSF1A, are also hypermethylated in addition to the E-cadherin gene (38, 39). It is currently unclear whether LMP1 also affects the regulation of p16 and RASSF1A genes through this mechanism. These data strongly suggest that DNA methylation of critical genes plays an important role in tumorigenesis of NPC, and that LMP1 contributes to tumorigenesis at this level.
Our study elucidates the mechanism of LMP1-mediated repression of E-cadherin in human epithelial cells. This report shows that LMP1 activates de novo DNMTs to induce DNA methylation. LMP1 affects another cellular protein, matrix metalloproteinase 9 (MMP9), which is associated with metastasis in cultured cells (40) and NPC tissue (41). Induction of MMP-9 expression is mediated through the LMP1-mediated nuclear factor (NF)-κB signaling pathway (40). The CTAR2 of LMP1, which is critical for E-cadherin repression, contributes ≈70% of LMP1-mediated NF-κB activation and is also the sole region triggers LMP1-mediated AP-1 activity specifically via the c-Jun N-terminal kinase 1 pathway (42). DNMT1 promoter contains the binding motifs of the AP-1 complex, suggesting LMP1 may activate the DNMT1 gene through this signaling pathway. Thus, it would be interesting to explore the potential cooperation between DNA methylation and signaling pathways mediated by LMP1 that facilitate cell migration activity. Data from this study further implicates aberrant methylation as a mechanism of tumorigenesis in EBV-associated cancers, suggesting methylation machinery as a novel target in cancer therapy.
Acknowledgments
We thank Professor James C. K. Shen, Institute of Molecular Biology, Academia Sinica, Taiwan for critically reviewing the manuscript. This work was supported by National Science Council Grants NSC 88-2318-B-182-001-M51 and NSC 89-2318-B-182-005-M51, Striving for Academic Excellency Program of Universities, Ministry of Education Grant 89-B-FA04-1-4, and National Health Research Institute (Taiwan) Grant NHRI-EX90-8703SL. C.-N.T. is the recipient of a postdoctoral fellowship from the National Health Research Institute.
Abbreviations
- NPC
nasopharyngeal carcinoma
- EBV
Epstein–Barr virus
- LMP1
latent membrane protein 1
- MDCK
Madin–Darby canine kidney
- CMV
cytomegalovirus
- RT
reverse transcription
- PCNA
proliferating cell nuclear antigen
- DNMT
DNA methyltransferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Takeichi M. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 2.Momparler R L, Bovenzi V. J Cell Physiol. 2000;183:145–154. doi: 10.1002/(SICI)1097-4652(200005)183:2<145::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.Lou P J, Chen W P, Sheen T S, Ko J Y, Hsu M M, Wu J C. Oncol Report. 1999;6:1065–1071. doi: 10.3892/or.6.5.1065. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Z, Pan J, B, Chu B, Wong Y C, Cheung A L, Tsao S W. Hum Pathol. 1999;30:458–466. doi: 10.1016/s0046-8177(99)90123-5. [DOI] [PubMed] [Google Scholar]
- 5.Frixen U H, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeier W. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, van Roy F. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 7.Perl A K, Wilgenbus P, Dahl U, Semb H, Christofori G. Nature (London) 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 8.Klein G, Giovanella B C, Lindahl T, Fialkow P J, Singh S, Stehlin J S. Proc Natl Acad Sci USA. 1974;71:4737–4741. doi: 10.1073/pnas.71.12.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata D, Weiss L M. Am J Pathol. 1992;140:769–774. [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet M, Guinebretiere J M, Kremmer E, Grunewald V, Benhamou E, Contesso G, Joab I. J Natl Cancer Inst. 1999;9:1367–1381. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- 11.Hsu M M, Tu S M. Cancer. 1983;52:362–368. doi: 10.1002/1097-0142(19830715)52:2<362::aid-cncr2820520230>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Liebowitz D, Kieff E. Cell. 1985;34:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 13.Fahraeus R, Rymo L, Rhim J-S, Klein G. Nature (London) 1990;345:447–449. doi: 10.1038/345447a0. [DOI] [PubMed] [Google Scholar]
- 14.Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N, Kikutani H. Science. 1999;286:300–303. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- 15.Wilson J-B, Weinberg W, Johnson R, Yuspa S, Levine A J. Cell. 1990;61:1315–1327. doi: 10.1016/0092-8674(90)90695-b. [DOI] [PubMed] [Google Scholar]
- 16.Fahraeus R, Chen W, Trivedi P, Klein G, Obrink B. Int J Cancer. 1992;52:834–838. doi: 10.1002/ijc.2910520527. [DOI] [PubMed] [Google Scholar]
- 17.Kim K R, Yoshizaki T, Miyamori H, Hasegawa K, Horikawa T, Furukawa M, Harada S, Seiki M, Sato H. Oncogene. 2000;19:1764–1771. doi: 10.1038/sj.onc.1203502. [DOI] [PubMed] [Google Scholar]
- 18.Horikawa T, Sheen T Z, Takeshita H, Sato H, Furukawa M, Yoshizaki T. Am J Pathol. 2001;159:27–33. doi: 10.1016/S0002-9440(10)61669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller W E, Earp H S, Raab-Traub N. J Virol. 1995;69:4390–4398. doi: 10.1128/jvi.69.7.4390-4398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M L, Tsai C N, Liang C L, Shu C H, Huang C R, Sulitzeanu D, Liu S T, Chang Y S. Oncogene. 1992;7:2131–2140. [PubMed] [Google Scholar]
- 22.Rhee I, Jair K W, Yen R W, Lengauer C, Herman J G, Kinzler K W, Vogelstein B, Baylin S B, Schuebel K E. Nature (London) 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 23.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 25.Moorthy R K, Thorley-Lawson D A. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh T S, Li S N, Wu C J, Liu S T, Meng C L, Chang Y S. DNA Cell Biol. 1997;16:1311–1319. doi: 10.1089/dna.1997.16.1311. [DOI] [PubMed] [Google Scholar]
- 27.Robertson K D. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 28.Hirohashi S. Am J Pathol. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, Garcia de Herreros A. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 30.Cano A, Pérez-Moreno M A, Rodrigo I, Locascio A, Blanco M J, del Barrio M G, Portillo F, Nieto M A. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 31.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 32.Robertson K D, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales F A, Jones P A. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama S, Sasaki A, Mese H, M, Alcalde R E, Tsuji T, Matsumura T. Int J Cancer. 2001;93:667–673. doi: 10.1002/ijc.1386. [DOI] [PubMed] [Google Scholar]
- 34.Li L C, Zhao H, Nakajima K, Oh B R, Filho L A, Carroll P, Dahiya R. J Urol. 2001;166:705–709. [PubMed] [Google Scholar]
- 35.Cheng C W, Wu P E, Yu J C, Huang C S, Yue C T, Wu C W, Shen C Y. Oncogene. 2001;20:3814–3823. doi: 10.1038/sj.onc.1204505. [DOI] [PubMed] [Google Scholar]
- 36.Leung W K, Yu J, Ng E K, To K F, Ma P K, Lee T L, Go M Y, Chung S C, Sung J J. Cancer. 2001;91:2294–2301. [PubMed] [Google Scholar]
- 37.Toyooka K O, Toyooka S, Virmani A K, Sathyanarayana U G, Euhus D M, Gilcrease M, Minna J D, Gazdar A F. Cancer Res. 2001;61:4556–4560. [PubMed] [Google Scholar]
- 38.Lo K W, Cheung S T, Leung S F, van Hasselt A, Tsang Y S, Mak K F, Chung Y F, Woo J K, Lee J C, Huang D P. Cancer Res. 1996;56:2721–2725. [PubMed] [Google Scholar]
- 39.Lo K W, Kwong J, Hui A B, Chan S Y, To K F, Chan A S, Chow L S, Teo P M, Johnson P J, Huang D P. Cancer Res. 2001;61:3877–3881. [PubMed] [Google Scholar]
- 40.Yoshizaki T, Sato H, Furukawa M, Pagano J S. Proc Natl Acad Sci USA. 1998;95:3621–3626. doi: 10.1073/pnas.95.7.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horikawa T, Yoshizaki T, Sheen T S, Lee S Y, Furukawa M. Cancer. 2000;89:715–723. doi: 10.1002/1097-0142(20000815)89:4<715::aid-cncr1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 42.Kieser A, Kaiser C, Hammerschmidt W. EMBO J. 1999;18:2511–2521. doi: 10.1093/emboj/18.9.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]