Abstract
BACKGROUND
The most striking feature of life is biodiversity. However, mechanisms of biodiversity remain poorly understood, as most protein orthologues of different species are highly homologous in sequence and identical in function. Interestingly, recent evidence has demonstrated heterogeneity for a G protein-coupled AT2 receptor in both ligand binding and induction of arachidonic acid release. The present study investigated the properties of AT2 receptors in closely related species.
METHODS.
AT2 receptors cloned from human, rabbit, rat, and mouse were expressed in CHO-K1, COS-1, and HEK293 cells and characterized in ligand binding and signal transductions. Critical residues in rabbit AT2 receptor attributable to heterogeneity were examined using both gain-of-function and loss-of-function approaches with mutagenesis.
RESULTS
The newly-cloned rabbit AT2 receptor exhibits distinct biochemical and biological properties compared to its highly homologous orthologues (91% in overall amino acid sequence) of rat, mouse, and human. All these orthologues activate SH2 domain-containing phosphatase-1 (SHP-1) and show similar binding affinities for angiotensin II (Ang II) and AT2-specific ligands CGP42112A and PD123319. However, reducing agent dithiothreitol (DTT) inactivates the rabbit orthologue but potentiates the others in ligand binding, a hallmark of AT2 versus AT1 receptor subtypes. Most interestingly, rabbit AT2 receptor, but not the other orthologues, induces arachidonic acid release in various cell systems when stimulated by both Ang II and CGP42112A, the peptide antagonist. Mutagenesis studies and sequence analyses further indicate that residues His106, Asp188, and Thr293 are responsible for the DTT inactivation and residues Val209 and Val249 are partially responsible for arachidonic acid release.
CONCLUSION
These results deny the co-existence of an additional AT2 subtype in rabbit proximal tubule cells and demonstrate for the first time the presence of functional diversity for closely related Eutherian orthologues of a GPCR that are more than 90% homologous in the amino acid sequence.
Keywords: Biodiversity, AT2 receptor, orthologue, GPCR, angiotensin II, arachidonic acid
Abbreviations: GPCR, G protein-coupled receptor; Ang II, angiotensin II (Asp1-Arg2-Val3-Tyr4-Ile5-His6-Pro7-Phe8); TM, transmembrane helix; AA, arachidonic acid; SHP-1; SH2 domain-containing phosphatase-1; rbAT2, rabbit AT2 receptor; rAT2, rat AT2 receptor; mAT2, mouse AT2 receptor; hAT2, human AT2 receptor
Introduction
The AT2 receptor belongs to the superfamily of G protein-coupled receptor (GPCR). It exerts inhibitory actions to counteract the type 1 Ang II receptor, AT1, in blood pressure regulation and cell growth. In humans and mice, AT2 exists as a single copy localized on the X chromosome (region Xq24-q25 and region XA2-A4, respectively) and contains no intron in its coding region. Thus the existence of multiple forms of AT2, encoded by several homologous genes or derived by alternative splicing, is not possible [1-3]. However, earlier studies from several independent groups have suggested the presence of heterogeneous populations of the AT2 receptor. For example, the AT2 receptor in different brain regions exhibited differential sensitivity to GTPγS [4]. In rabbit proximal tubule epithelial cells, Douglas’ laboratory discovered an AT2 population with completely reduced binding in the presence of dithiothreitol (DTT) [5,6]. Activation of this population of AT2 receptors induced arachidonic acid release in rabbit proximal tubular epithelial cells, a finding not observed in primary cells of other species [6]. In N1E-115 murine neuroblastoma cells, Reagan and Yee also observed two biochemically, pharmacologically, and immunologically different subpopulations of AT2 receptors [7,8]. These investigators have therefore suggested the existence of AT2A and AT2B subtypes to explain the observed heterogeneity concerning the AT2 receptors.
It is logical to assume that protein orthologues, particularly orthologues of closely related species, are identical in function because they are highly homologous (>90%) in amino acid sequence. AT2 receptors share >91% amino acid identity and presumably should not be exceptional. In order to determine whether the heterogeneity of the rabbit AT2 receptor results from a new subtype of AT2 receptor, we have cloned an AT2 receptor from rabbit proximal tubular epithelial cells to examine the structural determinants which contribute to the unique phenotypic characteristics of this species [5,6]. We now show that the rabbit AT2 receptor (rbAT2) is a conventional AT2 receptor orthologue rather than a novel subtype of the AT2B receptor. However, this rabbit orthologue exhibits functional diversity in biochemical, pharmacological, and physiological properties.
Methods and materials
Materials
The monoclonal anti-c-Myc antibody (Roche, clone 9E10), oligonucleotides (Sigma-Genosys and MWG Biotech), Ang II and [Sar1,Ile8]Ang II ( Bachem), PD123319 and CGP42112A (RBI), [3H]-arachidonic acid (NEN Life science), Protein Tyrosine Phosphatase Assay Kit (Upstate Biotech), DTT and other chemicals (Sigma) were purchased. 125I-[Sar1,Ile8]Ang II (2200 Ci/mmol) was supplied by Dr. Robert Speth, University of Washington, Pulman, WA.
DNA cloning, mutagenesis, PCR, and expression
The cDNA of rbAT2 was cloned from total RNA isolated from rabbit primary proximal tubule epithelial cells by RT-PCR with degenerate oligonucleotide primer FD1 (GTGGTT/CA/TCACTGTTTTGTTGTCAAAA) and FD2 (CAGCTGTTGGTGAATCCCAG/AGAGGATGGCAAAA), and SMART™ RACE cDNA Amplification Kit (ClonTech). The cloned cDNA was fully sequenced and modified for expression in cell lines: 1) To contain a consensus Marilyn-Kozak sequence and a unique Eco RI site at the 5′ end and Not I site at the 3′ end of the gene; and 2) To encode a decapeptide (EQKLISEEDL) epitope tag for a monoclonal antibody anti-c-Myc at the 5′ end after the start codon. The epitope tagged cDNA was subcloned into a shuttle vector pcDNA6 (Invitrogen). Identical modification was applied to rat, mouse, and human AT2 genes subcloned into the pcDNA6 vector for expression. Mutant AT2 receptors were prepared by the restriction fragment-replacement method and the PCR method. DNA sequence analysis was done to confirm each mutant construct. For expression of receptor proteins, 3 μg of column (Qiagen) purified plasmid DNA per 107 cells was used in transfection. CHO-K1, COS-1, and HEK293 cells (ATCC), cultured in ATCC preferred medium supplemented with 10% fetal bovine serum, were transfected by the GenePORTER™ transfection reagents (GTS Inc). For PCR detection of intronless AT2 receptors, genomic DNAs of human, mouse, rat, and rabbit purchased from ClonTech were used as templates for conventional PCR with respective sense and antisense primers derived from known sequences available in the GeneBank.
Southern blot
A DNA fragment probe of 740bp was prepared by Eco RI digestion of a TOPO TA vector plasmid DNA (Invitrogen) that contains a PCR insert of rbAT2 synthesized with primer FD1 and FD2. This DNA probe (20 ng/μl) was labeled with AlkPhos Direct Kit (Amersham) following the company’s protocol. Genomic DNAs of rabbit, human, rat, and mouse (ClonTech) were digested with Eco RI and visualized by chemiluminescent detection with CDP-Star (Amersham) after gel separation, membrane transfer, and hybridization with the labeled probe.
Western blot of receptors expressed in CHO-K1 cells
Post-nuclear supernatant of CHO-K1 cells solubilized in lysis buffer (50 mM Tris-HCl, pH 6.8, 1% CHAPS, 5 mM EDTA, pH 8.0) containing 50 μg/ml PMSF, 10 μg/ml of Benzamidine, 10 μg/ml of Bacitracine, 10 μg/ml of Leupeptin, and 2 μg/ml of Aprotinine, was used for SDS-polyacrylamide gel electrophoresis (PAGE) and western blotting analysis. Receptor polypeptides were visualized using 1:1000 dilution of mouse monoclonal anti-c-Myc antibody and 1:3000 dilution of secondary anti-mouse antibody coupled to peroxidase and the ECL system (Santa Cruz) as described [9-11].
DTT treatment, ligand binding assay, and AA release assay
DTT treatment of membrane preparations, saturation binding, and competitive binding assay were performed as described earlier [9-11]. AA release was assayed based on published methods [6] with minor modifications. Briefly, CHO-K1 cells and transfected CHO-K1 cells cultured in 6-well plates were allowed to grow for 24 h before they were labeled with [3H]-AA (0.3 μCi/ml or 10 nM [3H]-AA) overnight. The cells were washed three times with basal medium (serum-free medium plus 25 mM HEPES, pH 7.5) and then treated with 1.0 ml Basal medium containing Ang II for 15 min at 37°C. The stimulation was stopped by putting the plates on ice and the medium was transferred into pre-chilled appropriate tube for centrifugation at 15,600xg and at 4°C. Count 0.5 ml medium (half the volume) in 5 ml scintillation tubes. For total cellular radioactivity, cells in each well were solubilized with 1 ml of 1 M NaOH and counted in 5 ml scintillation tubes. [3H]-AA released into the medium is expressed as percent of the total cellular radioactivity and referred to as fractional release: Fractional [3H]-AA release = (count in the medium /count in the cells)*%.
Immunoprecipitation and SHP-1 activity assay
Twenty-four hours after co-transfection with AT2 and SHP-1 (kindly provided by M. Thomas, Washington University, St. Louis, USA), CHO-K1 cells were starved overnight and then stimulated with Ang II for 3 minutes. After treatment, cells were rinsed with ice-cold PBS, then solubilized for 30 min at 4°C in lysis Buffer (50 mM Tris-HCl, pH 7.5, 1.5% CHAPS, 150 mM NaCl, 5 mM EDTA), containing 0.5 mM Sodium orthovanadate, 10 μg/ml of Benzamidine, 10 μg/ml of Bacitracine, 10 μg/ml of Leupeptin, and 2 μg/ml of Aprotinine, and 50 μg/ml PMSF. After centrifugation at 15,000g for 15 min, the resulting supernatant (1-1.5 mg of proteins) was incubated for 2 h at 4°C with 10 μl of monoclonal anti-SHP-1 antibody prebound to Sepharose Protein A (Santa Cruz). The immunocomplexes were washed five times with washing buffer (identical to the lysis buffer but without 1.5% CHAPS and 0.5 mM Sodium orthovanadate). At the end of washing, the immunocomplexes were resuspended in PTPase buffer (50 mM Tris-HCl, pH 7.0, 1 mg/ml Bovine serum albumin, 5 mM DTT). The PTP activity was assayed by measuring the release of inorganic phosphate from phosphopeptides based on a malachite green detection system [12] using the PTPase assay Kit (Upstate) following the company’s protocol.
Statistical Analysis
The results are expressed as the mean ± S.E.M. of two to five independent determinations. The significance of measured values was evaluated with an unpaired Student’s t-test.
Results
Cloning and sequence analysis of a rabbit AT2 receptor
To determine whether an AT2B subtype exists in the rabbit kidney cortex and to elucidate the mechanism of functional diversity of the renal AT2, we cloned the renal AT2 from rabbit kidney cortex (GeneBank accession number AF451328). The polypeptide encoded by the open reading frame consists of 363 amino acids and shares a high identity of 91% in overall amino acid sequence with its human, rat, and mouse AT2 orthologues (Fig. 1). This high degree of identity falls within the range for many other protein orthologues including AT1. The rbAT2 bears all the structural features of the other AT2 orthologues, including seven transmembrane helices (TM) and four cysteine residues located on the extracellular domains. Sequence alignment identified 19 non-conserved unique residues for rbAT2 when compared to the human, rat and mouse orthologues as shown in Figure 1. Of these 19 residues, 63.2% located in the N-terminal and TM I, 10.5% in the TM II-VII, 10.5% in intracellular domains, and 15.8% in extracellular domains. In addition, the rbAT2 is short of one potential N-glycosylation site at Asn24 due to the Phe26 substitution in comparison to the other three orthologues.
Fig. 1.

ClustalW alignment of AT2 receptor orthologues of rat, mouse, human, and rabbit (GeneBank accession number AF451328). Putative transmembrane domains (TM I-VII) are underlined. The * indicates 19 non-conserved unique residues for rabbit AT2 in comparison to rat, mouse, and human orthologues. Of the 19 residues, 12 residues (63.2%) are located in the N-terminal and TM I, 3 residues (15.8%) in extracellular domains, 2 residues (10.5%) in TM II-VII, and 2 residues (10.5%) in intracellular domains. Mark Δ indicates four potential glycosylation sites (NxS/T) present in all four orthologues. Mark ♦ at Asn24 shows the potential glycosylation site present in all three orthologues but not in the rabbit AT2 (NxF). § indicates four highly conserved Cys residues that form two disulfide bonds of Cys35-Cys290 and Cys117-Cys195 in AT2 receptor. Computer software: MacVector (Oxford Molecular, Inc.).
The rbAT2 is a conventional AT2 orthologue
Polymerase chain reaction (PCR) using rabbit genomic DNA as templates for amplification produced a single band of 1.1kb (data not shown). This PCR product is identical to the complete coding region of the rbAT2 gene as confirmed by DNA sequencing, indicating its intronless property of the coding region similar to the other three orthologues. Southern blot using an 800bp fragment of rbAT2 DNA as probe detected a single band, suggesting little possibility of existence of other AT2 subtypes (Fig. 2). Thus, we conclude that this rbAT2 is an orthologue of the human, rat, and mouse AT2, rather than a new AT2B subtype.
Fig. 2.
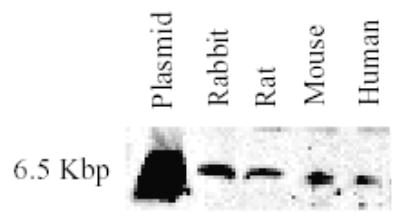
Southern blot detection of AT2 receptor gene subtypes or isoforms. Plasmid, rat AT2 receptor cDNA subcloned in pcDNA3 vector.
Identical ligand binding affinity but distinct DTT sensitivity of rbAT2
The rbAT2 must reproduce the similar function observed in rabbit kidney cortex in a surrogate cell system since the data above has ruled out the presence of AT2 heterogeneity [5,6]. To first examine the ligand binding property, expression of rabbit, human, rat, and mouse AT2 orthologues in COS-1 and CHO-K1 cell lines was verified by Western blot [9-11] with a specific anti-c-Myc antibody which recognizes the c-Myc tag at the N-terminal of the receptors (Fig. 3). Saturation binding of 125I-[Sar1,Ile8]Ang II and competitive binding of 125I-[Sar1,Ile8]Ang II against [Sar1,Ile8]Ang II, and AT2-specific ligand CGP42112A and PD123319 were applied to the four AT2 orthologues, respectively [9-11]. Table I summarizes the data and shows little difference in binding affinity among four orthologues in the presence or absence of DTT treatment. Analogous to the human, rat, and mouse AT2, the rabbit orthologue also exhibits an 8-fold increase in affinity in the presence of 5 mM DTT [9]. However, distinct from the human, rat, and mouse AT2, the rabbit orthologue shows an 80% (4.8 fold) decrease in Bmax in the presence of 5 mM DTT as compared to no DTT treatment. As a result, the binding curve of 125I-[Sar1,Ile8]Ang II for rbAT2 is in sharp contrast to the curves for the human, rat, and mouse orthologues (Fig. 4). Of 19 non-conserved unique residues, His106, Asp188, and Thr293 are the amino acids that locate in the extracellular domains of the rbAT2. Deletion of the N-terminal (the first 33 amino acids) of the rbAT2 (mutant rbAT2N) does not alter the property of DTT inactivation (Table I).
Fig. 3.
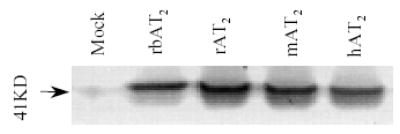
Western blot detection of AT2 receptor orthologues expressed in CHO-K1 cells with specific anti-c-Myc antibody.
Table I.
Binding affinity and Bmax of AT2 receptor orthologues
| [Sar1,Ile8]Ang II, (Ki, nM) |
CGP42112A, (Ki, nM) |
PD123319, (Ki, nM) |
Bmax, (pmol/mg) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | DTT (5mM) | -fold | Control | DTT (5mM) | -fold | Control | DTT (5mM) | -fold | Control | DTT (5mM) | -fold | |
| rbAT2 | 0.89±0.15 | 0.09±0.02 | 9.89 | 0.61±0.13 | 0.07±0.01 | 8.71 | 17.7±2.2 | 2.4±0.43 | 7.38 | 4.05±0.35 | 0.85±0.38 | 4.765 |
| rAT2 | 0.82±0.14 | 0.08±0.01 | 10.3 | 0.65±0.11 | 0.07±0.01 | 9.29 | 17.2±2.5 | 2.1±0.37 | 8.19 | 4.11±0.32 | 3.95±0.34 | 1.041 |
| mAT2 | 0.83±0.12 | 0.08±0.01 | 10.4 | 0.63±0.10 | 0.08±0.01 | 7.88 | 18.4±2.8 | 2.5±0.31 | 7.36 | 4.08±0.36 | 4.09±0.41 | 0.998 |
| hAT2 | 0.87±0.11 | 0.09±0.01 | 9.67 | 0.57±0.09 | 0.07±0.01 | 8.14 | 16.7±2.6 | 2.3±0.36 | 7.26 | 3.93±0.33 | 3.91±0.37 | 0.995 |
| rbAT2N | 2.98±0.76 | 0.11±0.02 | 27.1 | 0.69±0.12 | 0.08±0.01 | 8.63 | 17.6±2.3 | 2.3±0.38 | 7.69 | 3.78±0.32 | 0.62±0.35 | 6.096 |
Cell membranes were pre-treated with and without 5mM DTT at 22°C for 20 minutes. Here rbAT2, rAT2, mAT2, and hAT2 represent rabbit, rat, mouse, and human AT2 receptor orthologues, respectively. The rbAT2N is a rbAT2 mutant with deletion of the first 33 amino acids. This mutant is very similar to the wild type rbAT2 except for a reduced binding affinity (3.3 fold) for [Sar1,Ile8]Ang II in the absence of DTT treatment. Data represent results of three or more experiments of binding isotherms followed by Scatchard analysis. Results are presented as mean ± s.e.m.
Fig. 4.
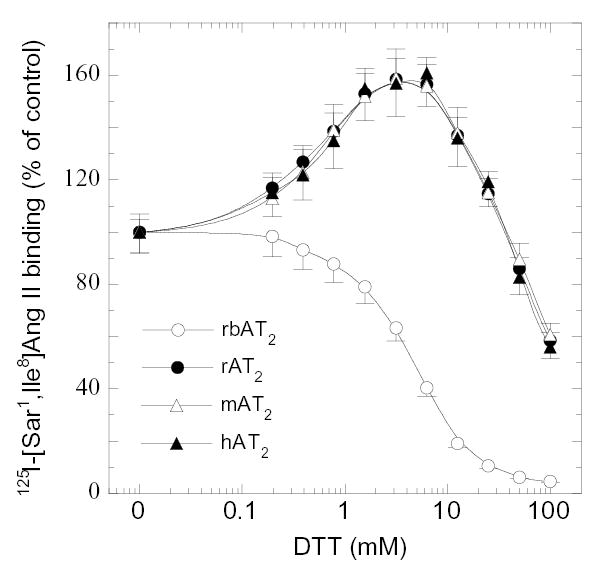
Effect of varying concentrations of DTT on 125I-[Sar1,Ile8]Ang II binding to AT2 receptor orthologues. The specific binding of 125I-[Sar1,Ile8]Ang II in each sample was adjusted to be < 10 ± 1% (≈ 20,000) of total input cpm without any treatment. This value is represented as 100%. Samples were first exposed to DTT at 22°C for 20 min and then the ligand was added.
All four AT2 orthologues activate SHP-1 but only the rbAT2 mediates AA release
Activation of an AT2 by Ang II in rabbit kidney epithelial cells has been shown to induce arachidonic acid (AA) release [5,6]. Consistently, Ang II stimulation induced marked [3H]-AA release in CHO-K1 cells transiently transfected with rbAT2. The addition of an AT2-specific antagonist PD123319 inhibited the [3H]-AA release. However, [3H]-AA release was not observed in CHO-K1 cells transiently transfected with human, rat, and mouse AT2 orthologues (Fig. 5). These results were reproduced using COS-1 and HEK293 cell lines (data not shown).
Fig. 5.
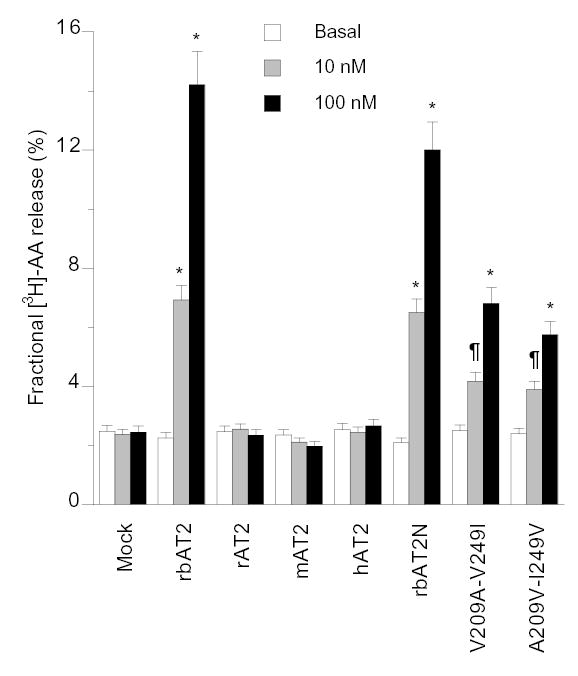
[3H]-AA release induced by Ang II stimulation of mock-transfected and AT2-transfected CHO-K1 cells. The mutant V209A-V249I of rbAT2 and the mutant A209V-I249V of rat AT2 maintained all binding properties of their own wild type as shown in Table 1 (data not shown). ¶, p<0.05, and *, p<0.01 by t-test in comparison to the basal of any group. No significant elevation of basal [3H]-AA release was detected in rbAT2 group in comparison to the basal levels of other groups. Results shown are mean ± s.e.m. of at least three independent experiments.
Ang II stimulation of human AT2 in CHO-K1 and PC12 cells has been shown to activate SHP-1, a SH2 domain-containing phosphatase [13,14]. Contrary to the [3H]-AA release, all four AT2 orthologues activated SHP-1 with no appreciable differences in CHO-K1 cells as detected by tyrosine phosphatase assay of SHP-1 specific immunoprecipitates (Fig. 6).
Fig. 6.
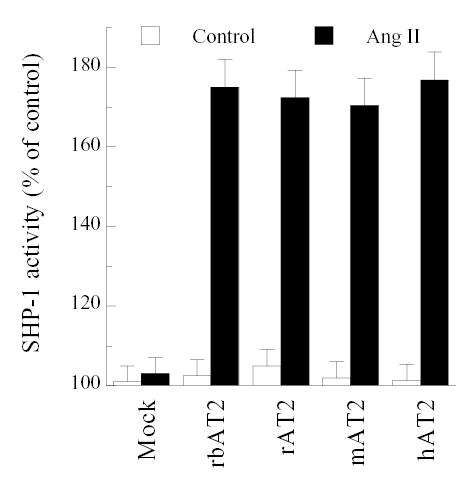
SHP-1 activity induced by Ang II (100 nM) stimulation of CHO-K1 cells co-transfected with SHP-1 and AT2 receptor orthologues.
Key residues determine AA release
The most critical regions directly specifying G protein selectivity are the cytoloop 2 and 3 domains [15]. To understand the molecular mechanism of rbAT2-mediated [3H]-AA release, rbAT2 mutant, V209A-V249I, and its rat reciprocal counterpart, A209V-I249V, were constructed simply by switching a DNA fragment between rabbit and rat wild-type AT2 receptors. The two mutants maintained all the binding properties of their own wild type (data not shown). The mutant V209A-V249I of rbAT2 lost 64% activity in [3H]-AA release whereas the mutant A209V-I249V of rat AT2 gained 30% activity (Fig. 5), indicating that Val209 and Val249 of the rbAT2 contribute to [3H]-AA release. Substitution of amino acid in the N-terminus of AT2 receptor shows no effect on AA release since rbAT2N maintains full activity (Fig. 5).
Variations in potential phosphorylation sites
Phosphorylation is a fundamental mechanism in regulation of GPCR functions. We analyzed potential sites of Ser, Thr, and Tyr phosphorylation for these four orthologues by NetPhos 2.0 and detected several unique sites for each orthologue as summarized in Table II [16]. This suggests another potential mechanism to understand how substitution of a seemingly non-critical amino acid in the microenvironment could impose profound impact on receptor function through alteration of phosphorylation.
Table II.
Phosphorylation sites of AT2 orthologues
| rbAT2 | rAT2 | mAT2 | hAT2 | |
|---|---|---|---|---|
| Ser6* | NA | x | x | NA |
| Ser16* | NA | --- | x | --- |
| Ser36* | x | x | x | x |
| Ser40* | x | NA | x | x |
| Ser145 | x | x | x | x |
| Ser152 | x | x | x | x |
| Ser346 | x | NA | NA | x |
| Ser348 | x | x | x | x |
| Ser352 | x | x | NA | x |
| Ser353 | x | x | x | x |
| Ser354 | x | x | x | x |
| Thr10* | NA | x | x | --- |
| Tyr143 | x | x | x | x |
| Tyr189* | x | --- | --- | --- |
| Tyr204* | x | x | x | x |
| Total | 12(61) | 11(63) | 12(63) | 11(64) |
Potential sites of Ser, Thr and Tyr phosphorylation are predicted by NetPhos 2.0 Internet program(16). Here x means the potential phosphorylation site is predicted;
---, no potential phosphorylation is predicted;
NA, residue not available.
Located in the extracellular domains and extracellular half region of TM domains. The superscript numbers in the brackets represent the total numbers of Ser, Thr, and Tyr residues available in the receptor orthologues, respectively.
Discussion
Almost all receptors including the AT2 may act differently in different locations of the same species. This exciting and complex phenomenon is beyond the sphere of this study because this type of difference displayed by an identical receptor is unlikely caused by a receptor itself. In contrast, this study deals with functional differences caused by a receptor itself due to structural differences. In the AT2 receptor, most variations (~50%) of amino acids are located within the short N-terminal region of the receptor orthologues (Fig. 1). However, these variations reveal little functional diversity in the study as demonstrated by the rbAT2N with comparison to the wild-type rbAT2. Thus, we speculate that the amino acid sequence in the N-terminus may specify the species signal or origin for the AT2 receptors.
Substitution of His106, Asp188, and Thr293 in the rbAT2 may increase the DTT accessibility to the highly conserved disulfide bond of Cys117-Cys195 of the receptor [9]. Breakage of this disulfide bond in a G protein-coupled receptor results in complete loss of function due to mis-folding [9].
Residue Ala63 located in the cytoloop 1 also may directly attribute to AA release [12] since the gain-of-function and loss-of-function were not equivalent and not close to 100%. In addition, residues located in the TM domains (Tyr37, Gln43, Arg48, Phe53, Ser59, and Val94) may indirectly and collectively affect G protein coupling selectivity and efficiency through alteration of critical GPCR conformations [10,11,15].
Although properties of ligand binding to AT2 receptors are well established, the full impact on biological function(s) and many other properties of the AT2 receptor remain elusive. This report on one hand explains why two types of AT2 receptors were detected previously in the presence and absence of DTT and indicates that no AT2 subtype receptors exist in rabbit. The previously observed heterogeneity likely resulted from functional diversity. However, it is unclear to us at this stage as to what the physiological significance of the functional diversity, predominantly, the DTT sensitivity and induction of arachidonic acid release can be. Given the observation that redox potential can also serve as an important regulation mechanism for protein activation [17,18], the distinct DTT property of rabbit AT2 may provide an alternative mechanism for receptor activation under pathological states in which redox potential is altered. However, it is unclear whether the cell surface receptor AT2 could be influenced intracellularly by the redox potential during the receptor folding, maturation, and intracellular trafficking since alteration of redox potential may occur only inside the cell under physiological conditions. Under pathological conditions such as myocardial infarction, injury, and inflammation, interstitial redox potential may change significantly. Interestingly, the expressions of AT2 receptors were increased significantly under these pathological conditions [1-3].
For protein orthologues with significantly less amino acid identity, functional diversity has been observed. For example, the human P2 nucleotide receptor P2X7 binds to ligand KN-62 and KN-04 whereas rat P2X7 (80% identity) does not [19]. Mammalian AT1 orthologues (human, rat, mouse, rabbit, etc., with 95% identity) bind to AT1-specific ligand losartan whereas amphibian AT1 receptor (Xenopus, 60% identity) does not [20]. Functional diversity for protein orthologues with amino acid identity greater than 90% is rare. The collective or synergistic effect of limited substitutions of seemingly unimportant amino acids may play a more important role in functional diversity than previously appreciated. Variation of phosphorylation sites (as shown in Table II), conformation of interaction motifs, and accessibility of interaction motif or domains are potential factors that also determine functional diversity of receptor orthologues.
For G protein-coupled receptors, Granneman et al reported different pharmacological profiles between human and rat β3-adrenergic receptors [21,22]. However, the human and rat β3-adrenergic receptors share only 79% identity in amino acid sequence. Mallee et al. also documented different species selectivity for non-peptide antagonists between human and rat CGRP receptors [23], an atypical GPCR in that it requires heterodimerization of CRLR, a classical seven transmembrane receptor, with the accessory protein RAMP1. However, this difference resulted from variations of a single residue at position 74 in the accessory protein RAMP1 that bears 71% homology. Replacement of this single residue in CGRP receptor produced little difference in binding to the native peptide ligand CGRP. The mechanism of an accessory protein-mediated modulation in receptor pharmacology could be very different and complex. One may find different pharmacological properties for receptor orthologues of closely related species. However, differences in intrinsic properties such as signal transduction are not documented.
The mechanism of biodiversity is largely unknown. Functional diversity of many individual genes may serve as one of the potential mechanisms. Most recently, identification of the human-specific gene family provided another exciting potential mechanism to explain what makes chimpanzees and humans different [24].
Biodiversity is an intriguing concept for biomedical research. Mouse, rat, and rabbit are the most widely-used animal species for man’s study in biomedicine. For example, more and more genetically modified animals with diseases or disorders are used to study gene functions, produce gene products, conduct gene therapies and test new drugs. Animal genes and cells are widely used as objects or tools of molecular and cellular studies on fundamental mechanisms such as protein-protein interaction and signal transduction. In many studies, species compatibility is not considered for heterologous expression of a gene in cells or even animals. These have posed an unprecedented challenge to biomedical scientists because in many cases results from one study are not completely reproducible in another application. In the field of GPCR research, the presence of multiple subtypes of many receptors with yet no detectable difference in function represents just another example of the current challenge [25]. Elucidation of functional diversity for these GPCR subtypes would improve drug efficacy and reduce side effects. In the field of evolution and comparative biology, phylogenetic methods based on 1D sequence analysis may not be accurate in predicting the evolutionary positions of branching orders of organisms [26] if functional diversity as reported here is not considered.
Acknowledgments
We gratefully acknowledge the insightful comments of anonymous reviewers of the manuscript. We thank Drs. Yaxian Ding and Nnenna Nkeme for assistance with the preparation of primary RTE cells and rabbit kidney cortex. We thank Ms. Justine Cowan and Dr. Shiv Srivastava for critical reading of the manuscript. This work was supported by a SDG grant (YHF) of the American Heart Association, NIH HL65492 (YHF), and NIH HL41618 grant (to YHF and JGD).
References
- 1.Feng YH, Douglas JG. Angiotensin receptors: an overview, in Angiotensin II receptor antagonists, edited by Epstein M, Brunner H, Philadelphia, Hanley & Belfus, 2000, pp 29-48
- 2.Gallinat S, Busche S, Raizada MK, Sumners C. The angiotensin II type 2 receptor: an enigma with multiple variations. Am J Physiol Endocrinol Metab. 2000;278:E357–E374. doi: 10.1152/ajpendo.2000.278.3.E357. [DOI] [PubMed] [Google Scholar]
- 3.Horiuchi M, Akishita M, Dzau VJ. Recent progress in angiotensin II type 2 receptor research in the cardiovascular system. Hypertension. 1999;33:613–621. doi: 10.1161/01.hyp.33.2.613. [DOI] [PubMed] [Google Scholar]
- 4.Tsutsumi K, Saavedra JM. Heterogeneity of angiotensin II AT2 receptors in the rat brain. Mol Pharmacol. 1992;41:290–297. [PubMed] [Google Scholar]
- 5.Dulin NO, Ernsberger P, Suciu DJ, Douglas JG. Rabbit renal epithelial angiotensin II receptors. Am J Physiol. 1994;267:F776–F782. doi: 10.1152/ajprenal.1994.267.5.F776. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs LS, Douglas JG. Angiotensin II type 2 receptor subtype mediates phospholipase A2-dependent signaling in rabbit proximal tubular epithelial cells. Hypertension. 1996;28:663–668. doi: 10.1161/01.hyp.28.4.663. [DOI] [PubMed] [Google Scholar]
- 7.Reagan LP, Yee DK, He PF, Fluharty SJ. Heterogeneity of angiotensin type 2 (AT2) receptors. Adv Exp Med Biol. 1996;396:199–208. doi: 10.1007/978-1-4899-1376-0_21. [DOI] [PubMed] [Google Scholar]
- 8.Yee DK, et al. Cloning and expression of angiotensin II type 2 (AT2) receptors from murine neuroblastoma N1E-115 cells: evidence for AT2 receptor heterogeneity. Brain Res Mol Brain Res. 1997;45:108–116. doi: 10.1016/s0169-328x(96)00242-2. [DOI] [PubMed] [Google Scholar]
- 9.Feng YH, Saad Y, Karnik SS. Reversible inactivation of AT2 angiotensin II receptor from cysteine-disulfide bond exchange. FEBS Letter. 2000;484:133–138. doi: 10.1016/s0014-5793(00)02141-4. [DOI] [PubMed] [Google Scholar]
- 10.Feng YH, Karnik SS. Role of transmembrane helix IV in G-protein specificity of the angiotensin II type 1 receptor. J Biol Chem. 1999;274:35546–35552. doi: 10.1074/jbc.274.50.35546. [DOI] [PubMed] [Google Scholar]
- 11.Feng YH, Miura S, Husain A, Karnik SS. Mechanism of constitutive activation of the AT1 receptor: influence of the size of the agonist switch binding residue Asn(111) Biochemistry. 1998;37:15791–15798. doi: 10.1021/bi980863t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burshtyn DN, Yang W, Yi T, Long EO. A novel phosphotyrosine motif with a critical amino acid at position -2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J Biol Chem. 1997;272:13066–13072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- 13.Bedes K, Elbaz N, Sutren M. Angiotensin II type 2 receptors mediate inhibition of mitogen-activated protein kinase cascade and functional activation of SHP-1 tyrosine phosphatase. Biochem J. 1997;325:449–454. doi: 10.1042/bj3250449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng YH, Sun Y, Douglas JG. Gβγ -Independent Constitutive Association of Gsα with SHP-1 and Angiotensin II Receptor AT2 Is Essential in AT2-mediated Activation of SHP-1. Proc Natl Acad Sci USA. 2002;99:12049–12054. doi: 10.1073/pnas.192404199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wess J. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol Ther. 1998;80:231–264. doi: 10.1016/s0163-7258(98)00030-8. [DOI] [PubMed] [Google Scholar]
- 16.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 17.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 18.Choi H, Kim S, Mukhopadhyay P, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–13. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 19.Humphreys BD, Virginio C, Surprenant A, et al. Isoquinolines as antagonists of the P2X7 nucleotide receptor: high selectivity for the human versus rat receptor homologues. Mol Pharmacol. 1998;54:22–32. doi: 10.1124/mol.54.1.22. [DOI] [PubMed] [Google Scholar]
- 20.Ji H, Zheng W, Zhang Y, Catt K, Sandberg K. Genetic transfer of a nonpeptide antagonist binding site to a previously unresponsive angiotensin receptor. Proc Natl Acad Sci U S A. 1995;92:9240–4. doi: 10.1073/pnas.92.20.9240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granneman JG, Lahners KN, Chaudhry A. Molecular cloning and expression of the rat beta 3-adrenergic receptor. Mol Pharmacol. 1991;40:895–9. [PubMed] [Google Scholar]
- 22.Granneman JG, Lahners KN, Rao DD. Rodent and human beta 3-adrenergic receptor genes contain an intron within the protein-coding block. Mol Pharmacol. 1992;42:964–70. [PubMed] [Google Scholar]
- 23.Mallee JJ, Salvatore CA, Lebourdelles B, et al. Receptor activity-modifying protein 1 determines the species selectivity of non-peptide CGRP receptor antagonists. J Biol Chem. 2002;277:14294–14298. doi: 10.1074/jbc.M109661200. [DOI] [PubMed] [Google Scholar]
- 24.Johnson ME, Viggiano L, Bailey JA, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–9. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald E, Kobilka BK, Scheinin M. Gene targeting--homing in on alpha 2-adrenoceptor-subtype function. Trends Pharmacol Sci. 1997;18:211–219. doi: 10.1016/s0165-6147(97)01063-8. [DOI] [PubMed] [Google Scholar]
- 26.Thornton JW, DeSalle R. A new method to localize and test the significance of incongruence: Detecting domain shuffling in the nuclear receptor superfamily. Syst Biol. 2000;49:183–201. [PubMed] [Google Scholar]


