Abstract
Virulent Staphylococcus aureus strains typically produce and secrete large quantities of many extracellular proteins involved in pathogenesis. Such strains cause the classical staphylococcal lesion—local tissue destruction and aggressive inflammation accompanied by the massive influx of polymorphonuclear leukocytes, leading to the formation of pus. Most strains causing toxic shock syndrome, however, produce and secrete very small quantities of most exoproteins although they elaborate high levels of toxic shock syndrome toxin-1 (TSST-1). These strains cause local infections that are remarkably apurulent although potentially fatal owing to the superantigen. We have analyzed this disparity and have found that TSST-1 itself is a negative global regulator of exoprotein gene transcription. TSST-1 not only represses most exoprotein genes but determines its own high expression level by autorepression. We report also that a second superantigen, enterotoxin B, has similar regulatory properties.
The pathogenicity of Staphylococcus aureus is multifactorial, generally involving a large number of extracellular proteins. Some of these proteins, including cytotoxins and exoenzymes, are secreted; others, including protein A and various adhesins, remain attached to the cell wall. Together, these proteins enable the organism to evade host defenses, adhere to host cells and intercellular matrix molecules, invade or destroy host cells, and spread within the tissues. Their production is governed by a complex network of regulatory functions whose expression in vitro is temporally programmed and largely depends, directly or indirectly, on environmental signals. It is assumed that the regulatory functions tune the expression of pathogenicity factors to achieve patterns that are optimal for local spatial and temporal adaptations.
At the same time, there is a subset of staphylococcal diseases whose primary cause is a single toxin. The most important of these toxinoses is toxic shock syndrome (TSS), caused most often by TSS toxin-1 (TSST-1) and less frequently by other superantigens (SAgs) such as enterotoxin A, enterotoxin B (SEB), enterotoxin C, or enterotoxin D. These and other staphylococcal SAgs are encoded by accessory genetic elements, including plasmids, prophages, and mobile pathogenicity islands (SaPIs) (1) and often are produced at relatively high levels (2). Remarkably, however, TSS-producing strains generally produce very little in the way of other extracellular toxins and other proteins (2).
Although TSS occurs most commonly in association with menstrual tampons, it was first described in connection with skin infections in children (3) and continues to be seen in extra-vaginal infections, often postsurgical (4). A remarkable feature of infections caused by TSST-1-producing staphylococcal strains is that they are largely apurulent and lack inflammation and tissue destruction characteristic of staphylococcal infections. Consequently, these cases often go unrecognized until several days after the symptoms of TSS have developed (5).
These features of TSS and the producing organisms prompted us to investigate the regulation of extracellular protein production in TSS strains. We report here that TSST-1 strongly represses the production of essentially all other exoproteins, acting at the level of transcription, and that in addition, it controls its own synthesis, acting as an autorepressor. We find also that SEB behaves very similarly. The possible relation of these findings to the clinical features of infections caused by S. aureus strains producing TSST-1 and possibly those producing other SAgs is discussed.
Materials and Methods
Bacterial Strains and Plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
Table 1.
Strains and plasmids used in this study
| Relevant characteristics | Reference/ source | |
|---|---|---|
| Strains | ||
| S6 | seb+ strain | 31 |
| RN4220 | Restriction-deficient mutant of strain 8325-4 | 32 |
| RN4282 | Clinical isolate, contains SaPI1, tst+ | 32 |
| RN6734 | agr+, 8325-4 derivative | |
| RN6938 | RN4282 derivative, SaPI1∷tetM, tst− | 14 |
| RN9131 | RN6734 derivative containing SaPI1 and tst gene | |
| Plasmids | ||
| pRN5543 | pC194 derivative, pUC19 polylinker, Cmr | 12 |
| pRN7034 | Shuttle vector containing promoterless blaZ gene, Emr | Unpublished |
| pRN7040 | pRN7034∷lukS-blaZ transcriptional fusion | This work |
| pRN7041 | pRN7034∷spr-blaZ transcriptional fusion | This work |
| pRN7044 | pRN7034∷tst(a.1)blaZ transcriptional fusion | This work |
| pRN7045 | pRN7034∷tst(a.8)blaZ transcriptional fusion | This work |
| pRN7056 | pRN7034∷tst(a.5)blaZ transcriptional fusion | This work |
| pRN7112 | pRN7034∷seb(b.1)blaZ transcriptional fusion | This work |
| pRN7114 | pRN5543∷seb, intact seb gene | This work |
| pRN7116 | pRN5543∷seb(b.2), truncated seb gene | This work |
| pRN7118 | pRN5543∷tst(a.5), truncated tst gene | This work |
| pRN7119 | pRN5543∷tst(a.8), intact tst gene | This work |
| pRN7123 | pRN5543∷tst(FSh), tst gene with frame-shift mutation | This work |
| pRN7126 | pRN7034∷tst(FSh)blaZ transcriptional fusion | This work |
Media and Growth Conditions.
S. aureus cultures were grown in CY broth (6) without glucose, supplemented with glycerol phosphate (0.1 M), or on GL agar (6) with suitable antibiotic. Liquid cultures were shaken at 37°C and monitored turbidometrically with a Klett–Summerson (New York) photoelectric colorimeter at 540 nm.
β-Lactamase Measurements.
Starting at low bacterial density, S. aureus cultures were grown in CY broth (6) without glucose. Samples were collected hourly. One portion of the sample was used to determine bacterial density and the other was quickly frozen and kept for β-lactamase measurements. β-Lactamase measurements were done on equalized samples by using nitrocefin as a substrate (7). Growth rates of the strains under study were indistinguishable within experimental error.
Generation of Murine Subcutaneous Abscess.
Bacterial strains RN4282 and RN6938 were grown to midexponential phase, ≈109 cells/ml, washed, and resuspended in PBS to 109 cells/ml. Bacterial suspensions were mixed with cytodex beads as described by Barg et al. (8), and 140 ml of beads + cells, containing 108 cells, was injected s.c. in the flank area of hairless mice, strain SKH 1. Mice were monitored daily by visual inspection of the injection site.
DNA Procedures.
DNA fragments were amplified by PCR (see Table 2). Primers contained either PstI or KpnI restriction sites at 5′ ends. Templates were chromosomal DNA isolated from strain RN4282 in the case of tst gene, S6 in the case of seb gene, and RN6734 in the case of lukS and spr genes. Plasmid and chromosomal DNA were isolated by using a QIAprep Spin Miniprep Kit with minor modifications. PCR-amplified DNA fragments were purified by using a QIAquick PCR Purification Kit. Both kits were obtained from Qiagen (Valencia, CA). PCR products were cut with KpnI and PstI and cloned into the multiple cloning sites of pRN5543 or pRN7034. Clones in pRN7034 shuttle vector were electroporated into Escherichia coli and afterward transformed into RN4220 (9). Clones in pRN5543 were transformed to S. aureus RN4220. All plasmids were transferred from RN4220 to other strains by transduction with staphylococcal phage 80α (9). Frame-shift mutagenesis was performed by using a QuikChange XL Site-Directed Mutagenesis Kit according to the manufacturer's instructions (Stratagene). Primers were obtained from Integrated DNA Technologies (Coralville, IA). Primer sequences are listed in Table 2.
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| tst a | GTTGCTGCAGACTCACACTTTGTTTTTTGC |
| tst 1 | GTTGGGTACCAAAATCTGTAGCGATTGTC |
| tst 2 | GTTGGGTACCGTTTGTAGATGCTTTTGC |
| tst 3 | GAAGGGTACCAAGGAATTATCTAAAACTTCAC |
| tst 4 | GTTGGGTACCAGGGCTATAATAAGG |
| tst 5 | GTTGGGTACCTTCAGTATTTGTAACGCCAC |
| tst 6 | GTTGGGTACCTTTTTATCGAACTTTGGC |
| tst 7 | GTTGGGTACCGTCATTCATTGTTATTTTCC |
| tst(FSh)1 | GGGGAAAAAGTGACTTAAACACAAAAAGAACTAAAAAAAGC |
| tst(FSh)2 | GCTTTTTTTAGTTCTTTTTGTGTTTAAGTCACTTTTTCCCC |
| lukS 1 | GTTGCTGCAGGCCTCATAACATTAAATTATTTTATCG |
| lukS 2 | AAAAAAGAATTCATAACGGATTGGCAAGAGGG |
| seb b | GTTGCTGCAGTGTTGTTAAAGATGTTTTCG |
| seb 1 | GTTGGGTACCATTTTTTATCTCCTTTATCC |
| seb 2 | GTTGGGTACCGTTTGTCAGTTTGATGCG |
| spr 1 | GTTGCTGCAGATTGTTCTTCGAAACTTAAGCACTC |
| spr 2 | GTTGGAATTCTTGCTGTTTGCTTGACTGCG |
Protein Procedures.
PAGE with SDS was performed according to Laemmli (10). β-Lactamase was assayed by the nitrocefin method (11) adapted for the microtiter format (7).
Results
Effect of tst on Exoprotein Patterns.
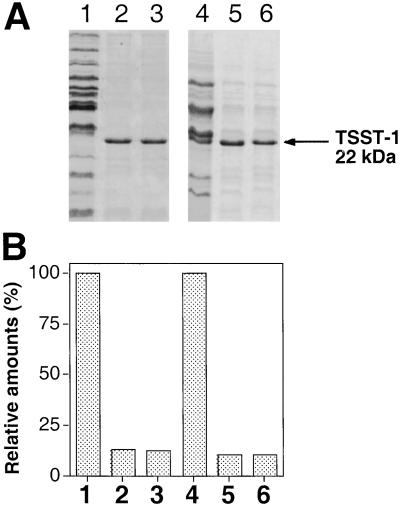
To test for the effect of tst on the overall pattern of exoprotein production by staphylococci, we compared postexponential phase culture supernatants of TSST-1-producing and nonproducing strains as shown in Fig. 1. As can be seen, the supernatant of an RN4282 (TSST-1-producing) culture (Fig. 1, lanes 3) contains very little in the way of exoproteins, with the exception of the strong TSST-1 band, whereas the RN6734 (non-TSST-1-producing) supernatant (Fig. 1, lanes 4) contains many exoproteins in considerable quantities, as has been observed (12), but lacks TSST-1. Exoprotein patterns similar to that of RN4282 have been observed for several other naturally occurring TSS strains (not shown). SaPI1, encoding TSST-1, is responsible for this effect. Thus transfer of SaPI1 to RN6734, resulting in strain RN9131 (13), caused a reduction in overall exoprotein production comparable to that seen with RN4282 (Fig. 1, lanes 5). In contrast, inactivation of tst in RN4282 (RN6938) by the insertion of tetM (14) had the opposite effect, sharply enhancing exoprotein production in that strain (Fig. 1, lanes 1). When SaPI1tst∷tetM was moved to RN6734, it had no effect on the exoprotein pattern (not shown). The SaPI1-specific inhibition of exoprotein synthesis could represent repression by TSST-1 itself or, because SaPI1 contains some 25 other ORFs, a polar effect of the tetM insertion on a downstream gene. The latter possibility was ruled out by complementation tests with a plasmid containing the cloned tst but no other SaPI1 material. For this test, a plasmid containing the cloned tst gene, pRN7119, was introduced into two strains, the tst-negative RN6734 (Fig. 1, lanes 6) and a strain containing SaPI1 with the above-mentioned tetM insertion in tst, RN6938 (Fig. 1, lanes 2). In each case, the cloned tst gene caused the same reduction in exoprotein production as that caused by the intact pathogenicity island, showing that the tst gene and not some downstream SaPI1 gene was responsible. We note that SaPI1 encodes two other SAgs, SEK and SEL (15), and conclude that neither of these has a demonstrable effect on exoprotein production. These results demonstrate additionally that the inhibitory effect of TSST-1 does not significantly depend on gene dosage.
Figure 1.
Effect of tst on exoprotein patterns. (A) Bacteria were grown to early stationary phase in CYGP in the absence of glucose, and supernatants were trichloroacetic acid-precipitated and analyzed by SDS/PAGE. Gels were stained with Coomassie brilliant blue and scanned. The following samples are shown: RN4282 with SaPI1 tst∷tetM insertional inactivation (RN6938) (lane 1), RN4282 tst∷tetM containing pRN5543∷tst(a.8) (pRN7119), expressing full-length tst (lane 2), RN4282 with SaPI1 carrying intact tst gene (lane 3), RN6734 (lane 4), RN6734 (SaPI1) strain (RN9131), expressing tst (lane 5), and RN6734 containing pRN5543∷tst(a.8) (pRN7119) and expressing tst (lane 6). Location of TSST-1 protein is indicated by the arrow. (B) Scanning data were used to compare the amounts of exoproteins produced by the various strains. Values for RN6938 (lane 1) and RN6734 (lane 4) were set to 100%. Amounts of exoproteins produced by derivative strains were normalized to this value, after subtracting the amount of TSST-1.
Mapping the Locus of Inhibition.
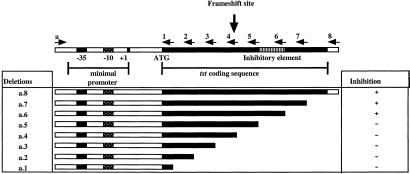
The region of tst responsible for this inhibition was localized by deletion analysis. The cloned tst gene and a set of PCR-generated 3′ deletion derivatives were subcloned to a pC194-based vector, pRN5543 (9), transduced to the standard TSST-1-negative strain, RN6734, and analyzed for their effects on overall exoprotein synthesis. The results of this analysis are shown in Fig. 2. The column at the right indicates whether a particular deletion derivative inhibited overall exoprotein synthesis (+) or did not (−). With this set of deletions, there was an all-or-none effect with respect to the inhibition phenomenon—the exoprotein patterns observed resembled that of the standard TSST-1-producing strain, RN4282, or that of the TSST-1-negative RN6734. On the basis of these results, it is concluded that a 30-aa region of the tst gene, from 91–120 of the mature protein, shown in gray on the map in Fig. 2, is essential for the inhibition of exoprotein synthesis.
Figure 2.
Mapping the locus of inhibition. At the top is a diagram of the SaPI1 tst locus, showing the promoter, the coding sequence (heavy black line), the inhibitory region (gray hatching), and the site of a frame-shift mutation. Deletions, introduced by the PCR, using primers listed in Table 2, are shown below, with the heavy black line indicating the extent of the remaining tst DNA. The effect of the progressive 3′ deletions on exoprotein expression is indicated by + for exoprotein inhibition or − for loss of the inhibitory activity. Numbers at the left, designating the individual deletions, are used throughout the text.
Nature of the Effector Molecule.
We next addressed the question of whether the inhibitory element was the gene itself, the mRNA, or the protein product. Here, we introduced a frame shift at nucleotide position 305 of the tst coding sequence, 5′ to the inhibitory region, using PCR (see Fig. 2). This frame shift created a nonsense codon 6 nt downstream. The mutated gene was cloned to vector plasmid pRN5543, generating pRN7123, which was transduced to RN6734 for testing. The frame-shift mutation eliminated the inhibitory effect of the gene on overall exoprotein production (not shown), suggesting that the protein itself is responsible for the inhibition. This notion was confirmed by studies with tst∷blaZ fusions (see below).
Effect of tst on Exoprotein Gene Transcription.
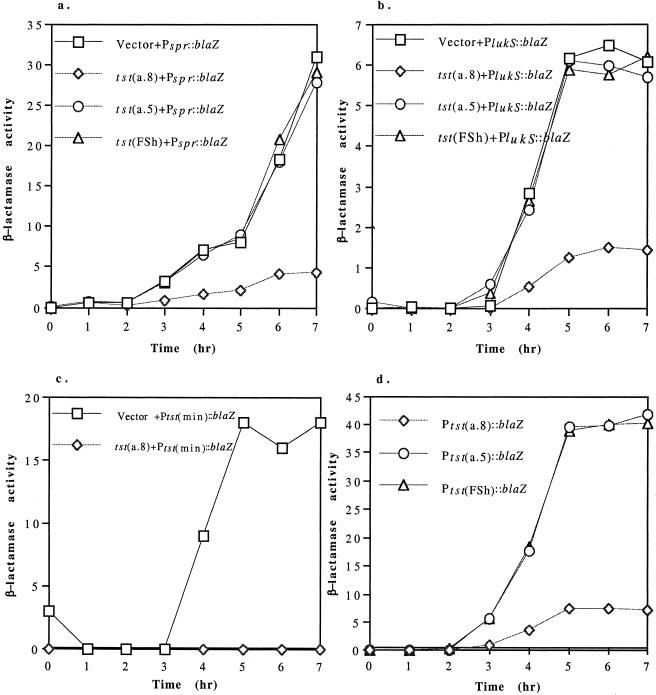
To test the possibility that the tst-specific inhibition of exoprotein synthesis was at the level of transcription, we made (in E. coli) transcriptional fusions of a staphylococcal β-lactamase gene (blaZ) to the promoters of two of the individual exoprotein genes, lukS (encoding the S subunit of leukocidin; ref. 16) and spr (encoding V8 serine protease; ref. 17), using a shuttle vector pRN7034, which contains blaZ lacking its promoter. The resulting plasmids, pRN7040 and pRN7041, respectively, were transferred to S. aureus strain RN4220 by protoplast transformation, and thence to RN6734 derivatives containing an inhibitory tst fragment, a noninhibitory fragment, and the frame-shifted tst, all cloned to pRN5543. As shown in Fig. 3 a and b, the larger tst fragment inhibited both of the exoprotein gene promoters, whereas neither the shorter tst fragment nor the frame-shifted tst had any effect in comparison to the vector alone. Therefore, tst acts by inhibiting transcription of the exoprotein genes.
Figure 3.
Effect of tst on exoprotein gene transcription. pRN5543 derivatives containing the intact tst gene (a.8) (pRN7119), a tst deletion, tst(a.5) (pRN7118), or tst with the frame shift mutation tst(FSh) (pRN7123) were transduced to separate RN6734 clones each with one of the following blaZ transcriptional fusions: (a) Pspr∷blaZ (pRN7041) and (b) PlukS∷blaZ (pRN7040), with promoter-containing fragments of spr and lukS, respectively, or (c) Ptst(min)∷blaZ (with minimal promoter sequence of tst, defined as a +1 to −45 fragment, in respect to start of transcription). Expression of the reporter gene was monitored during growth at 37°C. (d) β-Lactamase expression kinetics for blaZ fusions with the intact tst, Ptst(a.8)∷blaZ (pRN7045), truncated tst, Ptst(a.5)∷blaZ (pRN7056), and frame-shifted tst, Ptst(FSh)∷blaZ (pRN7126).
tst Autoregulation.
We had noted earlier that strains containing SaPI1 produced approximately the same level of TSST-1 as strains containing tst cloned to a high copy plasmid (see Fig. 1, lanes 5 and 6), suggesting that TSST-1 is an autorepressor as well as a repressor of other exoprotein genes. To test for autorepression, we prepared plasmids in which intact tst (a.8, see Fig. 2), a 3′ deletion derivative (a.5) and the frame-shift mutant (FSh) were transcriptionally fused to the β-lactamase reporter by using the vector plasmid, pRN7034, resulting in pRN7045, pRN7056, and pRN7126, respectively. Measurements of the β-lactamase activities of RN6734 derivatives containing these plasmids, during growth in CY medium (9) (omitting glucose, which has been reported to inhibit toxin production; ref. 18) are shown in Fig. 3d. As can be seen, the β-lactamase activity of the plasmid containing the truncated tst gene (a.5) was sharply elevated in comparison to that of the plasmid with tst intact (a.8). Similarly, the frame-shifted derivative (FSh) also showed a sharp elevation of β-lactamase activity. Because β-lactamase production with the frame-shift mutant was stimulated to the same extent as with the short tst fragment (a.5), the loss of inhibitory activity by the mutant could have resulted from rapid degradation of the frame-shifted tst mRNA only in the very unlikely event that there was a specific cleavage of the fused transcript just upstream of the blaZ translational start, followed by rapid degradation of the upstream (tst) fragment only. We conclude, therefore, that the inhibitory factor is the TSST-1 protein itself, and not the mRNA or DNA. The addition of pure TSST-1 protein to a growing culture had no effect on β-lactamase production by a short tst fragment (a.1) fused to blaZ, pRN7044, (not shown), indicating that the inhibitory behavior of the tst product is intracellular and presumably represents the TSST-1 precursor, because the mature form is not normally free in the cytoplasm.
The tst Promoter Is the Target of TSST-1 Repression.
We have mapped the tst transcription start point by the standard primer extension method. Assuming that the transcript is not posttranscriptionally processed or cleaved in vitro during the extraction procedure, this method has localized the tst promoter to the position shown in Fig. 2. Accordingly, we cloned a synthetic 45-mer (−45 to +1) to pRN7034, creating a Ptst∷blaZ fusion. Transcriptional activity of this 45-mer was regulated similarly to the other tst∷blaZ fusions, but had a maximum activity level of about 50% that of larger fragments cloned to the same vector (not shown). This finding suggests that one or more regulatory elements are missing from the 45-nt segment. Nevertheless, as shown in Fig. 3c, the intact tst gene (a.8) caused a substantial reduction in the β-lactamase activity of the cloned promoter in trans. These results establish that TSST-1 is an autorepressor as well as a transinhibitor of the production of other exoproteins, the autorepression effect is at the level of transcription, and the autorepression target lies within a 45-nt segment containing the tst promoter.
Effect of seb on Expression of Exoprotein Genes.
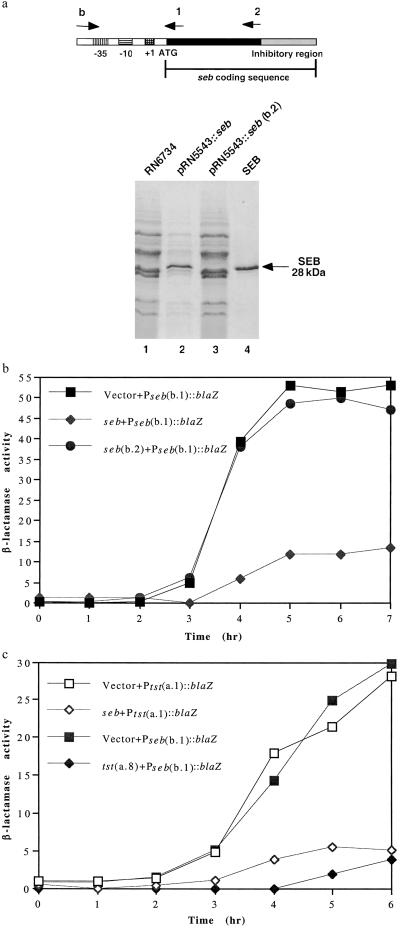
A further question was whether other SAgs have inhibitory activity similar to that of TSST-1. An obvious candidate was SEB, which is encoded by SaPI3, closely related to SaPI1 (15), occupies the same chromosomal site (see the COL genome, www.tigr.org), causes TSS, and has been observed, when cloned to a high copy plasmid, to inhibit production of other exoproteins (19). The seb gene and a derivative with a large 3′ deletion (b.2) were each cloned to pRN5543, generating pRN7114 and pRN7116, respectively, and tested for their effects on overall exoprotein synthesis in RN6734. As can be seen (Fig. 4a), the intact seb gene (lane 2) caused a dramatic decrease in overall exoprotein production, similar to that seen with tst, whereas the deletion derivative (lane 3) had no effect in comparison to the parental seb-negative strain, RN6734 (lane 1).
Figure 4.
Effect of seb on expression of exoprotein genes. (a) Cultures of RN6734 containing the intact cloned seb, pRN5543∷seb (pRN7114) or a derivative with a large 3′ deletion, pRN5543∷seb(b.2) (pRN7116), were grown to early stationary phase, and the culture supernatants were analyzed for exoprotein patterns as in Fig. 1. Location of SEB protein is indicated by the arrow. (b) The plasmids used in a were introduced into a RN6734 derivative containing the Pseb(b.1)∷blaZ transcriptional fusion (pRN7112), and the kinetics of β-lactamase synthesis were monitored throughout growth. (c) pRN5543 derivatives containing either tst(a.8) (pRN7119) or intact seb (pRN7114) were introduced into RN6734 derivatives containing either Pseb(b.1)∷blaZ (pRN7112) or Ptst(a.1)∷blaZ (pRN7044), and β-lactamase activity was measured as in b.
Autoregulatory Activity of seb Gene.
Using a third derivative in which a promoter-containing segment with no seb coding sequence (b.1) was cloned to the β-lactamase fusion vector (pRN7034), generating pRN7112. We note that the full-length seb gene inhibited β-lactamase synthesis in trans in comparison to the (b.2) deletion (see Fig. 4b), confirming that seb is also an autorepressor and showing that the 3′ half of the gene is essential for autorepression. Again, as with tst, the cross-repression of other exoprotein genes seems to be colocalized with the autorepression activity, although in this case, the resolution is low, because only one 3′ deletion has so far been made.
Finally, tst and seb are mutual cross-repressors (Fig. 4c), although cross-repression was not seen when both proteins are intact, presumably because autorepression sets a level of expression that is not affected by a second, cross-reacting autorepressor.
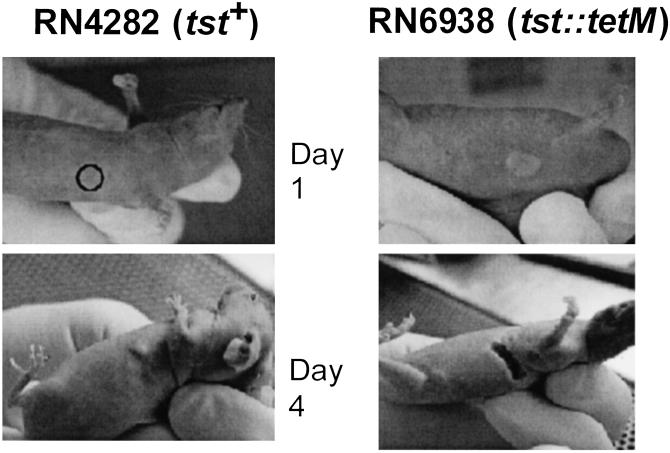
Inhibition of Inflammation by TSST-1 in Vivo.
As noted, superficial infections with TSST-1-producing strains are often apurulent; in view of the profound inhibition of exoprotein production by our standard TSST-1-producing strain in vitro, we sought to correlate these observations by using a simple experimental murine infection, namely the skin abscess model of Barg et al. (8). In this experiment, hairless mice were injected s.c., in the flank region, with 108 midexponential phase cells of either RN4282 (tst+) or RN6938 (tst∷tetM) and the site of injection inspected daily. As can be seen in Fig. 5, the tst-knockout strain, RN6938, produced the usual s.c. abscess, that was readily apparent after 1 day, and by the fourth day had sloughed and drained. In contrast, the tst+ strain, RN4282, produced no visible lesion after 1 day, and by the fourth day, had produced a small swelling in one of three mice only, consistent with the clinical observations. These results are not consistent with the results of Molne and Tarkowski (20) who have observed a strong inflammatory response after intracutaneous injection of 108 RN4282 organisms in mice.
Figure 5.
Inhibition of inflammation by TSST-1-producing strain in vivo. Three mice were injected with either TSST-1-producing strain (RN4282) or isogenic tst− strain (RN6938). Mice were inspected daily. Results shown are representative of each group.
Discussion
In this report, we describe a paradigm for the regulation of toxin gene expression in bacteria, namely down-regulation by the toxins themselves. The consequences of this regulation are that with strains producing certain SAgs there is a virtual absence of extracellular proteins in culture supernatants, whereas the SAg itself is present at a high and constant level. We have found that two of the major staphylococcal SAgs, TSST-1 and SEB, display this behavior, and that they block or strongly repress exoprotein production at the level of transcription, while regulating their own production as autorepressors. We have established that the cytoplasmic (precursor) form of each protein, rather than the DNA or mRNA is the regulatory effector for transrepression of the other exoproteins as well as for its own repression and that a short internal segment of the protein is required. For the tst gene, we have shown that a 45-nt segment containing the gene's promoter contains the target of repression. The target for seb has been localized to a somewhat larger segment, which contains the promoter plus the region from −35 to −90, a region that apparently binds a positively acting transcription factor and is required for expression (19). The inhibitory effect of seb on exoprotein production has previously been seen, in strain S6, with the gene cloned to a multicopy vector but not with a single chromosomal copy (21). This effect was attributed by the authors to titration of a positive transcription factor by the 5′ region of the cloned gene; our results rule this out because 3′ deletions eliminate the effect without altering the gene dosage of the 5′ region. Because S6 is a hyperproducer of RNAIII (21, 22), the agr effector, RNAIII overproduction may override the repressive effect of seb in single copy. Although TSST-1 and SEB are likely to act by the same mechanism, and TSST-1 shows 20–30% identity to SEB, sequence comparison has not suggested any obvious commonality. However, SAgs show remarkably similar folds and we believe that a fine-structure map of the inhibitory regions will reveal whether or not a common structure is involved.
Not all SAgs have this inhibitory activity. Thus the expression of enterotoxins K and L has no visible effect on the exoprotein patterns of the producing organisms. Because neither of these has been shown to have any clinically relevant activity, the significance of their lack of regulatory activity is uncertain. However, preliminary observations suggest that enterotoxin A, a well-characterized SAg that is an important cause of food poisoning as well as of extra-vaginal TSS, also lacks autorepressive and cross-repressive activity. It may be relevant that neither enterotoxin A (23) nor SEK (24) is regulated by agr. Perhaps the regulatory properties of tst and seb, which are agr-regulated, are related to the larger regulatory network to which they belong. Consistent with this possibility is our inability to demonstrate direct binding of purified TSST-1 to the tst promoter, suggesting that one or more intermediary factors may be involved. Any such intermediary factor(s) could well belong to the overall regulatory network that governs exoprotein production in S. aureus. Identification of these factors is currently a high priority.
As the target sequences responsive to the newly observed self-inhibition of TSST-1 synthesis and to the well-established agr activation (unpublished results) are both located within the promoter regions of the toxin gene, we have not yet been able to separate them genetically, either from each other or from function of the promoter itself. Indeed, the agr-responsive element of sed, which encodes enterotoxin D, has recently been mapped also to within the minimal sed promoter (25). Whether the agr and autorepression targets coincide for seb remains to be determined.
In sum, we have observed that at least two of the important staphylococcal SAgs, TSST-1 and SEB, strongly inhibit their own synthesis as well as that of many other exoproteins. Although the autorepression and trans inhibition functions of the two SAgs seem to colocalize within the respective coding sequences, there is reason to believe that they are biologically distinct and may involve different functional pathways. Thus, trans inhibition could be relevant to the pathogenic strategy of the organism: it will be recalled that infections with TSST-1-producing staphylococci are rapidly fatal in several animal models as well as in susceptible humans (26, 27), and that local infections with TSST-1-producing strains typically lack the ebullient inflammatory response that is highly characteristic of staphylococcal disease (28, 29). We have confirmed this effect in a simple murine s.c. abscess model, where the TSST-1-producing strain produced essentially no significant lesion whereas the knockout strain produced the typical S. aureus abscess. Because among the affected exoproteins are proteases, the inhibitory effect of the SAg might lead to a higher effective tissue concentration of TSST-1 owing to reduced proteolysis. The possible relevance of this to pathogenesis remains to be determined.
The suppression of inflammation by TSST-1 has been attributed to the induction of tumor necrosis factor α by the SAg (29). Alternatively (or additionally) it could be a consequence of the sharp reduction in exoproteins, which have an important role in excitation of the inflammatory response, thus retarding elimination of the organism and permitting the elaboration of a lethal dose of the SAg. We note that one of these proteins, lipase, is cytotactic for polymorphonuclear leukocytes (30). Whether other staphylococcal exoproteins are also cytotactic is presently unknown.
Regarding autorepression, perhaps the precursors of these two SAgs are toxic to the organism at elevated levels and autorepression prevents their accumulation. Study of the effects of TSST-1 and SEB on the comparative histopathology of experimental infections, in comparison with SAgs that do not repress other exoproteins, is expected to clarify the role of these toxins and their regulatory properties with respect to the inflammatory response and to the biology and/or pathogenicity of the organism.
Acknowledgments
We thank Yuan Fang, Jesse Wright, and Emmanuelle Charpentier for help and suggestions. This work was supported by National Institutes of Health Grants AI22159 and A130138 (to R.P.N.) and Graduate Program in Cellular and Molecular Biology Training Grant 5T32GH07238 (to N.V.).
Abbreviations
- TSS
toxic shock syndrome
- TSST-1
TSS toxin-1
- SAg
superantigen
- SEB
enterotoxin B
- SaPI
mobile pathogenicity island
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McCormick J K, Yarwood J M, Schlievert P M. Annu Rev Microbiol. 2001;55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Schlievert P, Osterholm M, Kelly J, Nishimura R. Ann Intern Med. 1982;96:937–940. doi: 10.7326/0003-4819-96-6-937. [DOI] [PubMed] [Google Scholar]
- 3.Todd J, Fishaut M, Kapral F, Welch T. Lancet. 1978;ii:1116–1118. doi: 10.1016/s0140-6736(78)92274-2. [DOI] [PubMed] [Google Scholar]
- 4.Hajjeh R A, Reingold A, Weil A, Shutt K, Schuchat A, Perkins B A. Emerg Infect Dis. 1999;5:807–810. doi: 10.3201/eid0506.990611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresler M J. West J Med. 1983;139:710–713. [PMC free article] [PubMed] [Google Scholar]
- 6.Novick R P, Brodsky R J. J Mol Biol. 1972;68:285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- 7.Ji G, Beavis R, Novick R. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barg N, Bunce C, Wheeler L, Reed G, Musser J. Infect Immun. 1992;60:2636–2640. doi: 10.1128/iai.60.7.2636-2640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick R P. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.O'Callaghan C H, Morris A, Kirby S M, Shingler A H. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruzin A, Lindsay J, Novick R P. Mol Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 14.Sloane R, deAzavedo J C S, Arbuthnott J P, Hartigan P J, Kreiswirth B, Novick R, Foster T J. FEMS Microbiol Lett. 1991;78:239–244. doi: 10.1016/0378-1097(91)90164-6. [DOI] [PubMed] [Google Scholar]
- 15.Novick R P, Schlievert P, Ruzin A. Microbes Infect. 2001;3:585–594. doi: 10.1016/s1286-4579(01)01414-9. [DOI] [PubMed] [Google Scholar]
- 16.Cooney J, Kienle Z, Foster T J, O'Toole P W. Infect Immun. 1993;61:768–771. doi: 10.1128/iai.61.2.768-771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmona C, Gray G L. Nucleic Acids Res. 1987;15:6757. doi: 10.1093/nar/15.16.6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iandolo J J, Shafer W M. Infect Immun. 1977;16:610–616. doi: 10.1128/iai.16.2.610-616.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahmood R, Khan S A. J Biol Chem. 1990;265:4652–4656. [PubMed] [Google Scholar]
- 20.Molne L, Tarkowski A. J Invest Dermatol. 2000;114:1120–1125. doi: 10.1046/j.1523-1747.2000.00973.x. [DOI] [PubMed] [Google Scholar]
- 21.Compagnone-Post P, Malyankar U, Khan S A. J Bacteriol. 1991;173:1827–1830. doi: 10.1128/jb.173.5.1827-1830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart M E, Smeltzer M S, Iandolo J J. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tremaine M T, Brockman D K, Betley M J. Infect Immun. 1993;61:356–359. doi: 10.1128/iai.61.1.356-359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orwin P M, Leung D Y, Donahue H L, Novick R P, Schlievert P M. Infect Immun. 2001;69:360–366. doi: 10.1128/IAI.69.1.360-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Stewart G C. J Bacteriol. 2000;182:2321–2325. doi: 10.1128/jb.182.8.2321-2325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlievert P M. Infect Immun. 1982;36:123–128. doi: 10.1128/iai.36.1.123-128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee P, Deringer J, Kreiswirth B, Novick R, Schlievert P. Infect Immun. 1991;59:879–884. doi: 10.1128/iai.59.3.879-884.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreiswirth B N, Kravitz G R, Schlievert P M, Novick R P. Ann Intern Med. 1986;105:704–707. doi: 10.7326/0003-4819-105-5-704. [DOI] [PubMed] [Google Scholar]
- 29.Fast D J, Schlievert P M, Nelson R D. J Immunol. 1988;140:949–953. [PubMed] [Google Scholar]
- 30.Tyski S, Tylewska S, Hryniewicz W, Jeljaszewicz J. Zentralbl Bakteriol Mikrobiol Hyg [A] 1987;265:360–368. doi: 10.1016/s0176-6724(87)80254-7. [DOI] [PubMed] [Google Scholar]
- 31.Dyer D W, Iandolo J J. Infect Immun. 1981;33:450–458. doi: 10.1128/iai.33.2.450-458.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreiswirth B, Lofdahl S, Betley M, O'Reilly M, Schlievert P, Bergdoll M, Novick R P. Nature (London) 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]