Abstract
Neuronal degeneration in spinal muscular atrophy is caused by reduced expression of the survival motor neuron (SMN) protein. SMN and the tightly interacting Gemin2 form part of a macromolecular complex (SMN complex) that mediates assembly of spliceosomal small nuclear ribonucleoproteins (U snRNPs). We used mouse genetics to investigate the function of this complex in motoneuron maintenance. Reduced Smn/Gemin2 protein levels lead to disturbed U snRNP assembly as indicated by reduced nuclear accumulation of Sm proteins. This finding correlates with enhanced motoneuron degeneration in Gemin2+/−/Smn+/− mice. Our data provide in vivo evidence that impaired production of U snRNPs contributes to motoneuron degeneration.
Autosomal recessive spinal muscular atrophy (SMA) is one of the most frequent monogenic disorders leading to morbidity in childhood and death in infancy. The disease is characterized by progressive degeneration of motoneurons resulting in atrophy and weakness of voluntary muscles (1). Positional cloning strategy showed that mutations or loss of the survival motor neuron (SMN) gene cause SMA (2). The SMN gene exists in two copies, termed SMN1 and SMN2, on human chromosome 5q13 (2, 3). Whereas the SMN1 gene allows expression of the functional (i.e., full-length) protein, the major product of the SMN2 gene is a differentially spliced and therefore truncated and nonfunctional protein that lacks exon 7 (4). Only homozygous absence of SMN1 is responsible for SMA, whereas homozygous absence of SMN2, found in about 5% of controls, has no clinical phenotype (5). As a consequence, SMA patients express only low levels of functional SMN protein, resulting in the disease phenotype.
Except for humans and primates (6), all other organisms analyzed so far, including rodents, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces pombe contain only one copy of the Smn gene which is equivalent to SMN1 and, hence, express the full-length Smn protein (7–11). Gene targeting studies in mice revealed that the Smn protein is essential for cellular viability in general (8). However, mice that express reduced levels of Smn, as observed in Smn heterozygous deficient mice, or transgenes that harbor the human SMN2 gene in a Smn null background develop motoneuron disease similar to SMA (12, 13).
The SMN1 gene encodes an ubiquitously expressed protein of 294 amino acids that is located in the cytoplasm and the nucleus, where it is concentrated in specific nuclear structures called gems (gemini of coiled bodies) (14). Previous studies have shown that SMN is a component of one or several large complexes (termed SMN complexes) (15, 16). Proteins that interact directly or indirectly with SMN, and may hence be part of the aforementioned complexes, have been identified. These include the SMN interacting protein 1 (Gemin2) (14), the putative DEAD box helicase dp103/Gemin3 (17–19), the Gemin3 interacting protein Gemin4/GIP1 (16, 20) p175/Gemin5, unrip, hsc70 (21, 22) and Gemin6 (23). Interestingly, SMN has also been shown to interact directly with Sm proteins, i.e., common components of the small nuclear ribonucleoproteins (U snRNPs) (24, 25). This finding suggested a functional link between SMN and the cellular splicing machinery. Indeed, studies in Xenopus laevis oocytes and in vitro revealed a role of SMN in the biogenesis of spliceosomal snRNPs U1, U2, U4, and U5 (26, 27). This process involves the transient export of the U snRNAs to the cytoplasm where the seven related Sm proteins, B/B′, D1, D2, D3, E, F, and G are stored. The Sm proteins assemble onto U snRNAs and form the Sm core, a ring-like structure common to all spliceosomal U snRNPs. The assembled particles are then targeted to the nucleus where they function in splicing (28). SMN, as part of a macromolecular complex, has been shown to facilitate the formation of the Sm core, most likely by regulating the proper binding of Sm proteins onto U snRNAs (21, 24, 26). Hence, the SMN complex has a function in the assembly of spliceosomal U snRNPs and possibly other RNP complexes.
Only little is known about genetic defects in other components of the SMN complex, and whether they are of pathophysiological relevance for SMA. A first study investigating mutations in the Gemin2 gene did not reveal any alteration in 26 patients from 11 SMA families with homozygous SMN1 mutations. Although patients within families exhibited identical alterations in the SMN gene, they showed variations in disease phenotype indicative of other disease modifying genes. Thus, it was concluded that Gemin2 mutations are rare and they do not function as a disease modifier in SMA (29). However, this study did not address the point whether levels of Gemin2 protein are altered in these patients by mechanisms such as, for example, reduced levels of SMN protein (30).
A model arising from the data described above suggests that patients expressing only low levels of SMN, exhibit defects in the biogenesis of snRNPs, and hence are suffering from SMA. To test this model, we have used a genetic strategy to investigate the consequence of reduced amounts of Smn complex in mice. By gene targeting, we generated mice that are deficient for one or both copies of Gemin2. Whereas the homozygous knockout of the gene resulted in early embryonic lethality, Gemin2 heterozygous deficient mice were viable and were crossbred with Smn heterozygous deficient mice. The resulting double heterozygous mice showed a degeneration of lumbar spinal motoneurons, which is significantly higher than in Smn heterozygous deficient mice. Most interestingly, degenerating motoneurons exhibited a marked defect in U snRNP assembly, as monitored by the reduced accumulation of Sm proteins in the nucleus. This finding indicates that reduced amounts of Smn complex leads to impaired production of spliceosomal U snRNPs and thus to motoneuron degeneration. Our data favor the idea that impaired assembly of spliceosomal U snRNPs contributes to the pathophysiological mechanisms in SMA.
Materials and Methods
Generation of Gemin2 Gene Knockout Mice.
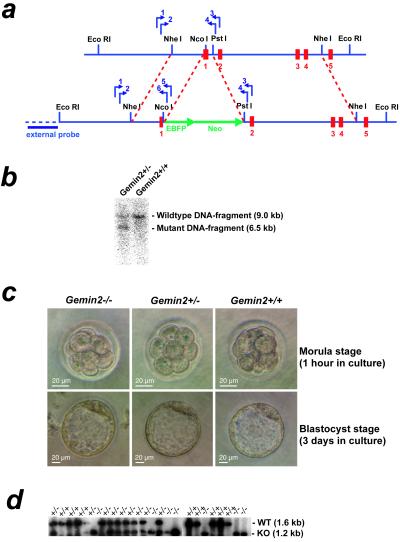
A targeting construct containing a 1,149-bp fragment of the promoter region including the start codon of the Gemin2 gene (NheI–NcoI fragment) was fused in-frame to the EBFP gene. The 3′ flanking region consisted of a 4,016-bp fragment of the Gemin2 genomic sequence (BglII–PstI fragment). The construct was electroporated into E14Tg2a-IV embryonic stem cells, and clones were selected in the presence of G418. Genotyping was done by nested PCR using primers corresponding to the 5′ flanking region of the targeting construct and reverse primers corresponding to the EBFP region. The wild-type allele was detected with primers corresponding to exon 2 and intron 1. A total of 516 clones were picked after transfection and G418 selection, and 497 clones were tested by PCR. After the first screening, 38 clones were identified as positive and subjected to Southern blot analysis, as shown in Fig. 1b. Nine of these clones showed a wild-type (9 kb) and a mutant (6 kb) band with similar intensities, and two clones were used for blastocyst injection. Chimeric mice were bred with female C57BL/6 mice, and germ-line transmission of the mutant Gemin2 allele was tested by coat color and genotyping as described above.
Figure 1.
Homologous recombination of the Gemin2 locus in mice and characterization of wild-type and Gemin2 targeted preimplantation embryos. (a) An EBFP-Neo-cassette (large green arrows) was inserted into exon 1 of the Gemin2 gene by homologous recombination (dashed lines in red) in mouse embryonic stem cells. Small blue arrows indicate the position of primers used for genotyping and the horizontal blue bar indicates the DNA probe used for Southern blot analysis of EcoRI-digested genomic DNA. Exons are marked in red. (b) Southern blot analysis of Gemin2+/− and wild-type mice with EcoRI-digested genomic DNA. All Gemin2+/− clones identified by PCR showed an EcoRI fragment of 6.5 kb indicative for the mutated allele along with the 9.0-kb wild-type allele fragment. (c) No morphological alterations were observed at the morula (Upper) and blastocyst stages (Lower) in Gemin2−/−, Gemin2+/−, and wild-type embryos. (d) Genotyping by PCR and subsequent Southern blotting showed products corresponding to the wild type (WT, 1.6 kb) and the mutant allele (KO, 1.2 kb).
Analysis of Gemin2 Knockout Embryos.
Gemin2+/− female mice were superovulated and bred with Gemin2+/− males. At approximately 56 h after mating early morulae were isolated and transferred to M16 medium overlaid with mineral oil and incubated at 37°C and 5% CO2 as described (31). Embryos were photographed at 1 h and 72 h after initiation of culture before they were harvested for DNA analysis by PCR. Each embryo was transferred into a PCR tube, and the PCR was performed under the condition described above. Amplification of the mutant and the wild-type allele were performed with primer pairs used for genotyping of heterozygous and wild-type offspring. The PCR products were transferred to a nylon membrane and hybridized with a probe spanning the amplified mutant and wild-type region. The blots were exposed to a Fuji x-ray film.
Western Blotting.
Tissue preparation, processing for Western blot analysis and immunohistochemistry was performed as described (30). Primary antibodies included the anti-Smn antibody IgG1, 250 μg/ml (Transduction Laboratories, Lexington, KY), a rabbit antiserum against human Gemin2 (16), a SmD3 rabbit antiserum, and the monoclonal anti-actin antibody (1 mg/ml) (Roche) were diluted 1:1,000 in 5% instant milk in Tris-buffered saline (TBS). Goat anti-mouse and anti-rabbit horseradish peroxidase-conjugated antibodies (Roche) were used at a dilution of 1:10,000 with 5% instant milk in TBS. The quantification analysis of the Western blots was performed by using the aida software as described (12).
Immunohistochemistry.
For immunohistochemistry, the mouse monoclonal antibody Y12 (32) was used at a 1:1,000 dilution in TBS/10% BSA. Anti-mouse Cy3 (Rockland, Gilbertsville, PA) was applied as a secondary antibody, diluted 1:300 in TBS/10% BSA.
Nissl Staining of Spinal and Facial Motoneurons.
Mice were deeply anaesthetized and transcardially perfused with fixative as described (33). Paraffin serial sections of brainstem and spinal cord were prepared, and motoneurons were counted in every 5th section of the brainstem and every 10th section of the lumbar spinal cord (L1–L6). Raw counts were corrected for double counting of split nucleoli as described (33). Differences between groups were evaluated with ANOVA test (significance level P < 0.05), applying GraphPad prism software (San Diego).
Results
Gene Targeting of the Murine Gemin2 Gene Leads to an Embryonic Lethal Phenotype.
To assess the consequence of Gemin2 deficiency for motoneuron degeneration and U snRNP assembly, we inactivated Gemin2 by a gene targeting approach. For this purpose, a genomic bacterial artificial chromosome clone containing the entire coding region for murine Gemin2, including its 5′ and 3′ flanking regions, was selected. Exon 1 and part of intron 1 were deleted in the targeting construct (Fig. 1a). The construct was electroporated into E14Tg2a-IV embryonic stem cells and clones were selected in the presence of G418. Homologous integration of the targeting construct into the Gemin2 locus was analyzed by PCR and Southern blot analysis (Fig. 1b).
Gemin2-targeted stem cell clones were injected into C57BL/6 blastocysts and gave rise to germ line transmission of the mutated allele. Chimeras were mated with C57BL/6 females and the heterozygous offspring was backcrossed with C57BL/6 mice. Gemin2 heterozygous intercrosses segregated Gemin2+/− and Gemin2+/+ in a 2:1 ratio. Gemin2+/− mice appeared healthy and were backcrossed with normal C57BL/6 mice for at least three generations to achieve a C57BL/6 background.
No homozygous Gemin2−/− mice with C57BL/6 background were born, indicating that Gemin2 is essential for embryonic viability. To determine the time point when homozygous deficient Gemin2 mice die, preimplantation embryos from Gemin2+/− intercrosses were isolated as uncompacted 5–7 cell morulae at 56 h post coitum (Fig. 1c Upper). After incubation for 3 days, the embryos had progressed to the blastocyst stage (Fig. 1c Lower), which allowed the determination of their genotype by PCR (Fig. 1d). Gemin2−/− and Gemin2+/− embryos were indistinguishable from wild-type controls, indicating that the development to the blastocyst stage progressed normally (Fig. 1c). Analysis of E12 embryos derived from heterozygous intercrosses did not show any Gemin2−/− homozygous deficient progenies or any signs of uterine resorption. Together, these data suggest that Gemin2−/− embryos die during or shortly after implantation.
Gemin2 Protein Levels in Gemin2+/− Mice Are Not Reduced in Spinal Cord Tissue.
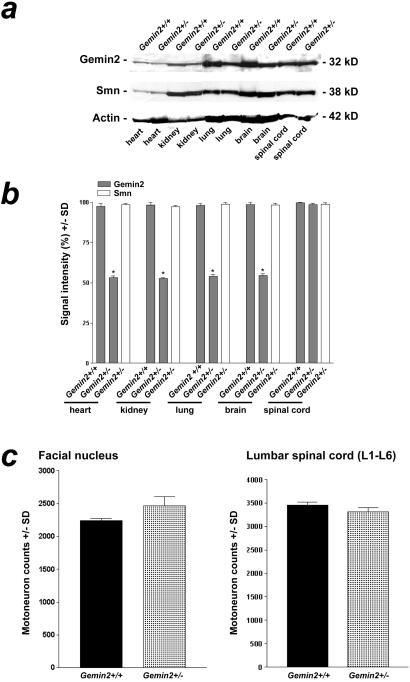
Previous studies have shown that Smn and Gemin2 protein levels are co-regulated in SMA patients and mice with reduced expression of Smn (30). Thus, Smn+/− mice exhibit reduced Gemin2 in addition to reduced Smn protein levels. To investigate whether Gemin2 gene-dose reduction results in altered Gemin2 and Smn protein levels, various tissues from 7-month-old wild-type and heterozygous deficient Gemin2 mice were investigated by Western blot analysis (Fig. 2 a and b). As expected, Gemin2 levels in Gemin2+/− mice were reduced by about 50% in heart, kidney, lung, and brain. However, a reduction of Smn protein was not observed in any of these tissues, indicating that Smn protein levels are not affected by alterations in Gemin2 expression. Remarkably, despite of the lack of one Gemin2 allele, Gemin2 protein levels in spinal cord were unaltered in comparison to wild-type mice. Thus, Gemin2 is either stabilized or its expression is up-regulated in the spinal cord of Gemin2+/− mice.
Figure 2.
Quantification of Gemin2 and Smn protein levels in tissues of 7-month-old heterozygous and wild-type Gemin2 mice. (a) Gemin2 protein was detected by Western blotting using a Gemin2-specific antiserum (Top). The signal for Gemin2 was reduced in heart, lung, kidney, and brain of Gemin2+/− mice. In contrast, Gemin2 protein levels were unchanged in spinal cord of Gemin2+/− mice. Western blotting of the same extract with monoclonal anti-Smn antibody (Middle) revealed similar signal intensities in tissues of wild-type and Gemin2+/− mice. (Bottom) Reprobing of the Western blot with a monoclonal anti-actin antibody as a control for equal loading of the gel. (b) Gemin2 and Smn protein content in various tissues of 7-month-old wild-type and Gemin2+/− mice was determined by scanning and quantification of the intensity of the Gemin2 and Smn immunoreactive bands from Western blots. Gemin2+/− mice showed similar Gemin2 protein levels in spinal cord tissue. Signal intensities for Gemin2 in heart, kidney, lung, and brain of Gemin2+/− mice (gray bars) were significantly reduced of about 50% (*). Smn signal intensity in each tissue was similar in Gemin2+/− and wild-type mice (white bars). (c) Quantification of motoneurons in the facial nucleus (Left) and in the spinal cord (Right) of heterozygous deficient and wild-type Gemin2 mice. Motoneuron counts in the facial nucleus and in the spinal cord of heterozygous deficient and wild-type Gemin2 mice revealed no differences.
Mice Double Heterozygous Deficient for Smn and Gemin2 Exhibit Enhanced Motoneuron Loss in Spinal Cord.
The Gemin2-deficient mice were used to investigate the role of Smn and Gemin2 in motoneuron cell death. Previous studies have shown that a significant number of lumbar motoneurons is lost in 1-year-old Smn+/− mice, thus resembling the SMA type III phenotype in humans (12). As these mice exhibit both reduced levels of Smn and Gemin2 proteins, the question remained whether the loss of motoneurons is attributable to a reduction of either Smn or Gemin2, or both. To address this issue, we compared facial and lumbar spinal motoneuron loss in wild-type, Gemin2+/−, and Smn+/− mice and in mice double heterozygous deficient for Smn and Gemin2.
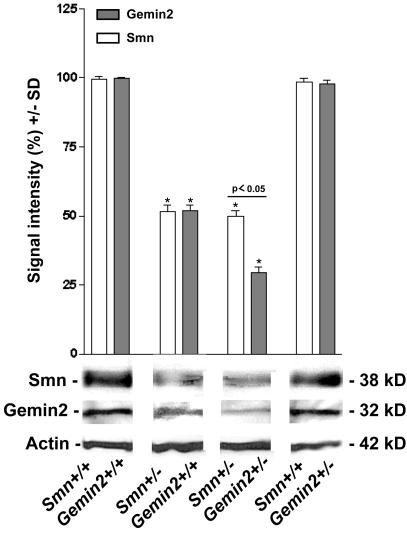
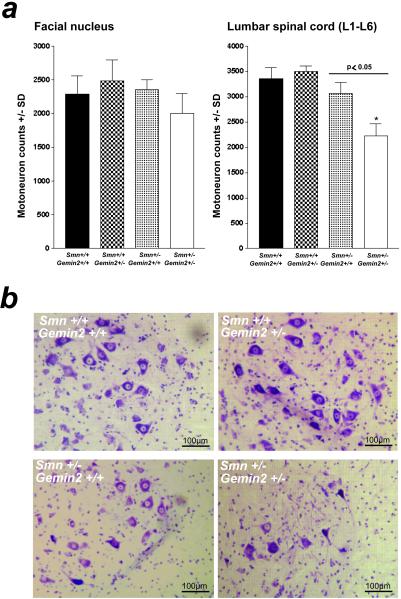
The number of lumbar and facial motoneurons is not reduced in 7-month-old Gemin2 heterozygous deficient mice (Fig. 2c). This finding fits well with our observation that neither Smn nor Gemin2 levels were altered in these cells on inactivation of one Gemin2 allele (see above). To obtain mice with reduced levels of Gemin2 in motoneurons, Smn and Gemin2 heterozygous deficient mice were crossbred. As expected, double homozygous deficient mice were embryonic lethal (data not shown). However, 5-month-old Gemin2 and Smn double heterozygous deficient mice were viable. As determined by Western blotting, these mice showed a 75% reduction of Gemin2 and a 50% reduction of Smn protein levels in spinal cord compared with wild-type and Gemin2+/− animals. Importantly, the observed Gemin2 protein level in double heterozygous deficient mice was significantly lower than in Smn+/− mice, which showed only a 50% reduction (Fig. 3). Strikingly, as shown in Fig. 4, the number of lumbar motoneurons in 5-month-old double heterozygous deficient mice was significantly decreased by 34% in comparison to a reduction by 9% in Smn+/− mice (Fig. 4a Right). In contrast, the population of facial motoneurons, which is less affected in patients with SMA, showed only a marginally enhanced degeneration that did not reach the level of statistical significance (Fig. 4a Left). As motoneurons of Gemin2+/−/Smn+/− mice differ only in their level of Gemin2 in comparison to Smn+/− mice, the enhanced motoneuron degeneration in spinal cord can only be attributable to the reduction of Gemin2 protein expression. Thus, these results indicate that reduced Gemin2 protein levels enhance lumbar spinal motoneuron degeneration.
Figure 3.
Quantification of the Gemin2 and Smn protein levels in spinal cord of 5-month-old Gemin2 and Smn heterozygous deficient mice. Gemin2 and Smn protein levels in spinal cord of wild-type, Smn+/−, Smn+/−/Gemin2+/−, and Gemin2+/− mice were compared by quantification of the intensities of Gemin2 and Smn immunoreactive bands from Western blots using a Gemin2-specific antiserum and a monoclonal anti-Smn antibody. Similar protein expression of Gemin2 and Smn was observed in spinal cord tissue of wild-type and Gemin2+/− mice. Only 50% of Smn (*, white bar) and Gemin2 (*, gray bar) protein was expressed in spinal cord of Smn+/− mice. Immunodetection of Gemin2 in spinal cord of Smn+/−/Gemin2+/− mice showed a stronger reduction (−75%) compared with Smn (−50%) (P < 0.05).
Figure 4.
Quantitative analysis of facial and lumbar spinal motoneurons of 5-month-old Gemin2+/−, Smn+/−, Gemin2+/−/Smn+/−, and wild-type mice. (a) Counting of facial motoneurons in 5-month-old Smn+/−, Gemin2+/−, Smn+/−/Gemin2+/−, and wild-type littermates revealed no differences (Left). In contrast, the number of lumbar spinal motoneurons in Smn+/−/Gemin2+/− mice (*) was significantly reduced (−34%) in comparison to wild-type, Gemin2+/−, and Smn+/− mice (P < 0.05; Right). (b) Nissl staining of lumbar spinal motoneurons in Gemin2+/−, Smn+/−, Smn+/−/Gemin2+/−, and wild-type mice.
Impaired Nuclear Accumulation of Sm Proteins in Gemin2- and Smn-Deficient Spinal Motoneurons.
Several lines of evidence suggested that Smn and Gemin2 play an essential role in the assembly of spliceosomal U snRNPs in the cytoplasm. However, it is not known so far whether the U snRNP assembly is defective in motoneurons of SMA patients or in mouse models of SMA. We therefore investigated spliceosomal U snRNP assembly in motoneurons of Smn+/−/Gemin2+/− mice. It appeared particularly interesting to test whether U snRNP assembly is disturbed at a time period when motoneuron degeneration occurs in the Smn and Gemin2 double heterozygous deficient mice (i.e., the fifth month).
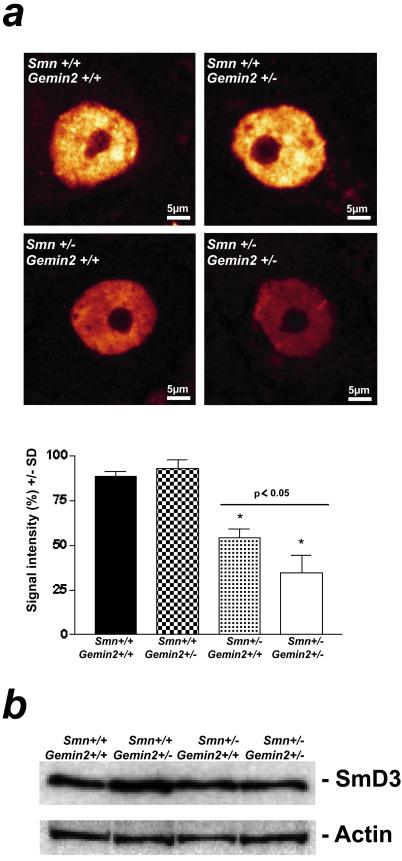
As the low number of motoneurons in the spinal cord precluded isolation and biochemical analysis, U snRNP assembly was assessed indirectly via semiquantitative analysis of nuclear Sm proteins in situ. Frozen spinal cord sections of 5-month-old wild-type, Gemin2+/−, Smn+/− and double heterozygous deficient mice were stained with the anti-Sm monoclonal antibody Y12. As shown in Fig. 5a Upper, Sm proteins are highly concentrated in the nucleus of motoneurons in wild-type and Gemin2+/− mice, both of which show normal levels of Smn and Gemin2. In contrast, Sm protein levels in the nucleus of Smn+/− mice were significantly reduced by 39%. Strikingly, the loss of Sm proteins was further enhanced to 61% in double heterozygous deficient mice (Fig. 5a Lower). Of note, only the total amount of Sm proteins were altered in the nucleus of spinal motoneurons of these mice but not the subnuclear distribution (Fig. 5a). The impaired nuclear accumulation of Sm proteins may be caused by the down-regulation of these proteins in response to impaired expression of Gemin2 and Smn. Alternatively, production of Sm proteins may be normal, but because of a failure to assemble into U snRNPs, these proteins are retained in cytoplasmatic storage pools, and hence cannot be detected by immunofluorescence. To distinguish between both possibilities, we analyzed the spinal cord in targeted and normal mice by Western blotting. As shown in Fig. 5b, spinal cord of either genotype showed similar amounts of SmD3 protein. Together, these data support the hypothesis that U snRNP assembly is reduced in lumbar spinal motoneurons of mice with reduced levels of Smn and Gemin2.
Figure 5.
Sm protein immunoreactivity in the nucleus of spinal motoneurons of 5-month-old Gemin2+/−, Smn+/−, Gemin2+/−/Smn+/−, and wild-type mice. (a) The monoclonal anti-Sm protein antibody Y12 was used for immunohistochemical analysis of the Sm protein distribution in the nucleus of spinal motoneurons of 5-month-old Gemin2+/−, Smn+/−, Gemin2+/−/Smn+/−, and wild-type mice. A significant reduction of Sm proteins in the nucleus of spinal motoneurons was detectable in Smn/Gemin2 double heterozygous deficient mice (*) in comparison to wild type, Gemin2+/−, and Smn+/− motoneurons. Distribution of the Sm proteins within the cell body of each genotype was not altered. (Lower) The semiquantitative analysis of Sm protein levels in spinal motoneuron nuclei. Reduced Sm protein signal intensity was measured in spinal motoneurons of Smn+/− (−39%, *) and even more (P < 0.05) in Smn+/−/Gemin2+/− mice in comparison to wild-type and Gemin2+/− mice. (b) Similar signal intensities for SmD3 proteins in spinal cord of wild-type, Gemin2+/−, Smn+/−, and Smn/Gemin2 double heterozygous-deficient mice were observed. The anti-actin antibody was used as a control showing equal protein concentration in each lane.
Discussion
Although genetic studies with patients and mouse models have provided clear evidence that mutations in the SMN1 gene are the primary cause of SMA, only very little is known about the molecular events that lead to motoneuron degeneration. Here we have applied a genetic approach to analyze cell death of spinal motoneurons in correlation with the cellular level of the Smn complex and the U snRNP assembly. Our data indicate that reduced expression of components of the Smn complex and subsequent impaired (or disturbed) U snRNP biogenesis go in parallel with motoneuron cell death.
Biochemical studies indicated an essential role for Gemin2 in the assembly of spliceosomal U snRNPs (26). As this function is relevant for most cells of an organism, targeting of the Gemin2 gene was expected to cause a severe phenotype in mice. Consistently, we observed that Gemin2-deficient mice die early during development. However, in contrast to Smn-deficient mice, which do not enter the blastocyst stage (8), Gemin2−/− mice exhibit a slightly milder phenotype and thus die later during embryonic development. This delay might be caused by higher stability of the maternal Gemin2 protein (possibly by binding to Smn) and hence to a longer persistence in the developing embryo.
In contrast to Smn+/− mice, which develop motoneuron degeneration at an age of 6 to 7 months, Gemin2+/− mice appear normal and do not show any signs of motoneuron loss. Interestingly, the analysis of protein levels revealed that despite of the lack of one Gemin2 allele, Gemin2 protein is present at wild-type level in spinal cord. In contrast, other tissues exhibit the expected 50% reduction of Gemin2 protein levels. The lack of any disease symptoms correlates with normal levels of Gemin2 and Smn protein in the spinal cord of Gemin2+/− mice. These data suggest that the Gemin2 protein levels are specifically regulated in spinal cord. The mechanism that underlies this phenomenon is unclear but may involve the stabilization of Gemin2 protein.
By crossbreeding Smn+/− and Gemin2+/− mice, we obtained Smn+/−/Gemin2+/− double heterozygous animals. These mice express approximately 50% of normal Smn protein but, unexpectedly, only 25% of the amount of Gemin2 found in wild-type animals, and develop a stronger motoneuron degeneration in comparison to Smn+/− mice. Indeed, at 5 months of age, when motoneuron degeneration in Smn+/− mice has just started, already 34% of the lumbar motoneurons were lost in Smn and Gemin2 double heterozygous deficient mice. This finding shows that motoneuron cell death cannot only be induced by impaired expression of Smn (12, 13) but also by reduced levels of Gemin2. In fact, based on these data, it is tempting to speculate that any condition that leads to the impaired production of the Smn complex (for example mutations in other components of the complex) likewise causes this phenotype.
A wealth of data points to a role of the Smn complex in the cytoplasmic assembly of spliceosomal U snRNPs (21, 24, 26, 27). To investigate whether motoneuron degeneration can artificially be induced by the reduction of functional Smn complex, we have examined mice that are heterozygous deficient for Smn or double heterozygous deficient for Smn and Gemin2. As the capacity to assemble U snRNPs cannot be assessed directly (i.e., by biochemical means), we have determined the intracellular localization of Sm proteins in motoneurons at an age of 5 months when degeneration is prominent in Smn+/−/Gemin2+/− mice. Indeed, Sm protein levels in the nucleus of motoneurons are reduced by 39% in Smn+/− mice in comparison to matched wild-type control mice and Gemin2 heterozygous deficient mice, which show normal Smn and Gemin2 levels. Importantly, motoneurons of double heterozygous deficient mice show an even stronger reduction of nuclear Sm proteins (on average −61%) at an age of 5 months. Based on these results, we favor the idea that the observed reduction of the nuclear Sm protein pool is primarily caused by a defect in U snRNP assembly. However, we cannot rigorously exclude the possibility that also other events, such as impaired regeneration of U snRNPs or enhanced degradation of U snRNAs contribute to the loss of Sm proteins from the nuclear compartment. In vivo studies that directly monitor the assembly of U snRNPs in Gemin2/Smn heterozygous cells are necessary to further address this question.
In conclusion, our data reveal a correlation between the capacity of the cell to assemble, regenerate and/or maintain spliceosomal U snRNPs, and motoneuron degeneration in a Gemin2 and Smn double heterozygous deficient mouse model. Our data further suggest that a similar biochemical defect occurs in SMA patients and contributes to motoneuron loss. Thus, strategies that lead to enhanced efficacy of the cell to assemble snRNP complexes might be a target to develop therapies for patients with SMA.
Acknowledgments
We thank J. Steitz and I. Mattaj for donation of the Y12 antibody, S. Klüpfel, J. Kara, and G. Sowa for excellent technical work, and K. Grohmann for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grants SFB 581 B1 and Fi-573/2-2.
Abbreviations
- SMA
spinal muscular atrophy
- SMN
survival motor neuron
- snRNP
small nuclear ribonucleoproteins
References
- 1.Crawford T O. Neurology. 1996;46:335–340. doi: 10.1212/wnl.46.2.335. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Endrizzi M, Huang S, Scharf J M, Kelter A R, Wirth B, Kunkel L M, Miller W, Dietrich W F. Genomics. 1999;60:137–151. doi: 10.1006/geno.1999.5910. [DOI] [PubMed] [Google Scholar]
- 4.Lorson C L, Hahnen E, Androphy E J, Wirth B. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wirth B. Hum Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Rochette C F, Gilbert N, Simard L R. Hum Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 7.Hannus S, Buhler D, Romano M, Seraphin B, Fischer U. Hum Mol Genet. 2000;9:663–674. doi: 10.1093/hmg/9.5.663. [DOI] [PubMed] [Google Scholar]
- 8.Schrank B, Gotz R, Gunnersen J M, Ure J M, Toyka K V, Smith A G, Sendtner M. Proc Natl Acad Sci USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miguel-Aliaga I, Chan Y B, Davies K E, van den Heuvel M. FEBS Lett. 2000;486:99–102. doi: 10.1016/s0014-5793(00)02243-2. [DOI] [PubMed] [Google Scholar]
- 10.Miguel-Aliaga I, Culetto E, Walker D S, Baylis H A, Sattelle D B, Davies K E. Hum Mol Genet. 1999;8:2133–2143. doi: 10.1093/hmg/8.12.2133. [DOI] [PubMed] [Google Scholar]
- 11.Paushkin S, Charroux B, Abel L, Perkinson R A, Pellizzoni L, Dreyfuss G. J Biol Chem. 2000;275:23841–23846. doi: 10.1074/jbc.M001441200. [DOI] [PubMed] [Google Scholar]
- 12.Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M. Hum Mol Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 13.Monani U R, Sendtner M, Coovert D D, Parsons D W, Andreassi C, Le T T, Jablonka S, Schrank B, Rossol W, Prior T W, et al. Hum Mol Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Fischer U, Wang F, Dreyfuss G. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Dreyfuss G. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 16.Meister G, Buhler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U. Hum Mol Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 17.Campbell L, Hunter K M, Mohaghegh P, Tinsley J M, Brasch M A, Davies K E. Hum Mol Genet. 2000;9:1093–1100. doi: 10.1093/hmg/9.7.1093. [DOI] [PubMed] [Google Scholar]
- 18.Charroux B, Pellizzoni L, Perkinson R A, Shevchenko A, Mann M, Dreyfuss G. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss M D, Hille A, Barth S, Spurk A, Hennrich F, Holzer D, Mueller-Lantzsch N, Kremmer E, Grasser F A. J Virol. 2001;75:11781–11790. doi: 10.1128/JVI.75.23.11781-11790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charroux B, Pellizzoni L, Perkinson R A, Yong J, Shevchenko A, Mann M, Dreyfuss G. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 22.Gubitz A K, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. J Biol Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 23.Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. J Biol Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 24.Buhler D, Raker V, Luhrmann R, Fischer U. Hum Mol Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 25.Pellizzoni L, Charroux B, Dreyfuss G. Proc Natl Acad Sci USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer U, Liu Q, Dreyfuss G. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 27.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 28.Will C L, Luhrmann R. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 29.Helmken C, Wetter A, Rudnik-Schoneborn S, Liehr T, Zerres K, Wirth B. Eur J Hum Genet. 2000;8:493–499. doi: 10.1038/sj.ejhg.5200479. [DOI] [PubMed] [Google Scholar]
- 30.Jablonka S, Bandilla M, Wiese S, Buhler D, Wirth B, Sendtner M, Fischer U. Hum Mol Genet. 2001;10:497–505. doi: 10.1093/hmg/10.5.497. [DOI] [PubMed] [Google Scholar]
- 31.Vernet M, Bonnerot C, Briand P, Nicolas J F. Methods Enzymol. 1993;225:434–451. doi: 10.1016/0076-6879(93)25030-6. [DOI] [PubMed] [Google Scholar]
- 32.Lerner E A, Lerner M R, Hardin J A, Janeway C A, Jr, Steitz J A. Arthritis Rheum. 1982;25:761–766. doi: 10.1002/art.1780250709. [DOI] [PubMed] [Google Scholar]
- 33.Masu Y, Wolf E, Holtmann B, Sendtner M, Brem G, Thoenen H. Nature (London) 1993;365:27–32. doi: 10.1038/365027a0. [DOI] [PubMed] [Google Scholar]