Abstract
The regulators of G protein signaling (RGS) proteins modulate heterotrimeric G protein signaling. RGS8 is a brain-specific RGS protein of 180 aa. Here we identified a short isoform of RGS8, RGS8S, that arises by alternative splicing. RGS8S cDNA encodes a N terminus of 7 aa instead of amino acids 1–9 of RGS8 and 10–180 of RGS8. The subcellular distribution of RGS8 and RGS8S did not differ significantly in transfected cells. RGS8S accelerated, not as efficiently as RGS8, the turning on and off of Gi/o-mediated modulation of G protein-gated inwardly rectifying K+ channels in Xenopus oocytes. We next examined the effects of RGS8 and RGS8S on Gq-mediated signaling. RGS8 decreased the amplitude of the response upon activation of m1 muscarinic or substance P receptors, but did not remarkably inhibit signaling from m3 muscarinic receptors. In contrast, RGS8S showed much less inhibition of the response of either of these Gq-coupled receptors. By quantitative analysis of the inhibitory effect and the protein expression level, we confirmed that the difference of inhibitory effect is caused by both the qualitative difference between RGS8 and RGS8S and the quantitative difference of the protein expression level. We also confirmed that the receptor-type specificity of inhibition is not caused by the difference of the expression level of the receptors. In summary, we showed that 9 aa in the N terminus of RGS8 contribute to the function to inhibit Gq-coupled signaling in a receptor type-specific manner and that the regulatory function of RGS8S is especially diminished on Gq-coupled responses.
Regulators of G protein signaling (RGS) proteins comprise a large family of more than 20 members, which modulate heterotrimeric G protein signaling. They share a homologous domain, the RGS domain, which is flanked by diverse N and C termini (1–3). The RGS domain alone is sufficient for activating the GTPase of Gα, whereas the flanking domains confer various regulatory properties (3). RGS8 was identified in rat brain and is a small RGS protein along with RGS4, RGS5, and RGS16 (4, 5). We recently showed that RGS8 protein was concentrated in nuclei of cells transfected with cDNA for RGS8 expression and that coexpression of a constitutively active Gαo resulted in the translocation of RGS8 protein to the plasma membrane. The deletion of the N-terminal region (35 aa) of RGS8 abolished its nuclear localization and active Gαo-induced redistribution. This truncated mutant of RGS8, however, is still functional in inhibiting pheromone signaling in yeast to some extent. When coexpressed with G protein-gated inwardly rectifying K+ (GIRK) channels, the truncated RGS8 accelerated both turning on and off similar to RGS8. Acute desensitization of GIRK current observed in the presence of RGS8, however, was not induced. Thus, we clarified that RGS8 requires its N terminus for subcellular localization and full regulatory function (6). On the other hand, Zeng et al. (7) reported that the N-terminal domain (1–33 aa) of RGS4 confers receptor-selective inhibition of Gq signaling. There is significant sequence conservation in the N-terminal region of RGS4 and RGS8 (6).
It was recently reported that RGS3 is expressed as two isoforms transcribed from alternate promoter sites within the RGS3 gene and that the RGS3T form lacks a large portion of the N-terminal domain of RGS3 (8, 9). RGS3 attenuates Gαq/11-mediated signaling and shows agonist-induced translocation from cytosol to the plasma membrane. Deletion of the N terminus of RGS3 prevents its translocation (10). On the other hand, a short form of RGS3, RGS3T, was reported to be localized to the nucleus and induce apoptosis (11). In this study, we examined the possibility of additional forms of RGS8 and identified a splice variant of RGS8, RGS8S, in which 9 aa at the N terminus are replaced with 7 aa. By comparing regulatory effects of RGS8 and RGS8S on Gi or Gq-coupled responses, functional significance of the N terminus was investigated.
Materials and Methods
Reverse Transcriptase–PCR (RT-PCR).
To identify RGS8 and related RGS proteins expressed in rat brain, RT-PCR using specific primers was performed. Total RNA was isolated from adult and developing rat brain, and the first-strand cDNA was synthesized as a template for RT-PCR. To obtain the entire coding sequence, primers specific for the 5′ and 3′ noncoding regions (5′ noncoding: ATGCATGCGTGAGCCTATGTGTCC; 3′ noncoding: TTCACGTTAGAATGTGGTCTCGGC) were used. The amplified DNAs were cloned into pGEM-T vector (Promega), and their sequences were determined. For the detection of expression levels, other primers corresponding to common sequences of RGS8 and RGS8S (CTCTGACCATCCACTTGGCAAA, TTTTCCCTGGGCTTGATCAAAACA) were used.
Yeast Pheromone Response Assay.
A bioassay was used to measure the sensitivity of the pheromone response in yeast that expresses RGS proteins as described (6, 12). As the expression level of RGS8 protein was not high enough in yeast, a sequence based on the Kozak consensus sequence and a N-terminus myc tag (MEQKLISEEDLSRGS) were fused to RGS8 or RGS8S cDNA to increase the expression level. These constructs were introduced into the pTS210 yeast expression vector under the control of a galactose-inducible promoter. By PCR amplification, cDNA fragments containing the coding sequence of RGS8 or RGS8S were isolated. After confirmation by sequencing analysis, they were fused in-frame immediately downstream of the myc tag in pTS210. The sst2 deletion mutant yeast SNY86 (13) was transformed with each cDNA in pTS210 and selected on ura− dropout plates. Independent colonies of each yeast transformant were grown and a halo bioassay was performed with soft agar plates containing galactose. We first determined a suitable dose of an inducer for quantitative comparison of the inhibitory function of RGS proteins by examining the relationship between the reduction in halo size and the galactose concentration (0.1–2%) and decided to use 0.5%. Expression levels of myc-tagged RGS proteins were then examined by Western blotting as described (6, 12).
Immunofluorescence Staining of Cultured Cells.
A Syrian hamster leiomyosarcoma cell line, DDT1MF2, was grown in DMEM supplemented with 10% FBS. Rat RGS8 or RGS8S cDNA was cloned into the pCXN2 expression vector (14). Gαq cDNA (GenBank accession no. U40038) was amplified by PCR using a human fetal brain cDNA library (CLONTECH) as template. The GαqQ209L mutant (GαqQL) was constructed by the overlapping PCR method. The PCR product was inserted into the EcoRI site of the mammalian expression vector pCMV. The cDNA for the Gαo Q205L mutant (GαoQL) was provided by J. D. Jordan, Mount Sinai School of Medicine, New York. The resultant plasmid DNA was transfected into cultured cells by using Fugene 6 (Roche Molecular Biochemicals). Immunofluorescence staining was performed with anti-RGS8 antibody, which was raised against a peptide corresponding to residues 27–42 of rat RGS8. Details of antibody specificity and microscopy methods have been described (15). Briefly, cells were cultured on glass coverslips and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. They were then permeabilized with 0.5% Triton X-100 in PBS for 5 min and blocked with 5% skim milk in PBS for 90 min. The cultures were reacted with anti-RGS8 antibody raised in rabbit and the reaction was visualized with Cy3-anti-rabbit IgG antibody (Jackson ImmunoResearch). After immunoreaction, cells were mounted with Fluoro Guard antifade reagent (BioRad). The specimens were observed with a Axioskop microscope (Zeiss) equipped with phase-contrast and epifluorescence optics. Images were recorded with a Kodak MegaPlus camera.
Two-Electrode Voltage Clamp.
Functional expression in Xenopus oocytes and electrophysiological analysis under a two-electrode voltage clamp were done as described (5). Gαi-coupled responses were recorded by coexpressing the m2 muscarinic receptor and GIRK1/2 channel (see Fig. 4). The bath solution contained 90 mM KCl, 3 mM MgCl2, and 10 mM Hepes. The change of GIRK1/2 current amplitude induced by ligand application was recorded at −80 mV. The substance P (SP), m1, or m3 receptor was coexpressed, and Gαq/11-coupled responses were monitored as an increase in the current amplitude through the endogenous Ca2+-activated Cl− channel in Xenopus oocytes (see Figs. 5–7). The bath solution was a standard frog Ringer's solution. Depolarizing step pulses to +60 mV were applied repeatedly from the holding potential of −80 mV every 2 s. The time course of changes in the current amplitude on application of ligand was plotted (see Fig. 5a), and the value of the peak amplitude was obtained. Oocytes were frozen after electrophysiological analysis and sonicated in PBS containing 0.1 mM phenylmethylsulfonyl fluoride (see Fig. 6). The homogenate was centrifuged at 3,000 rpm at 4°C for 10 min, and the resultant supernatant was used as a whole protein extract of oocytes. To compare expression levels of RGS proteins in oocytes, Western blotting of whole protein extracts was performed by using anti-RGS8 antibody. Both SP receptor and m3 receptor were coexpressed in the same oocyte, and the amplitudes of responses to SP, and then to acetylcholine (ACh) was measured (see Fig. 7).
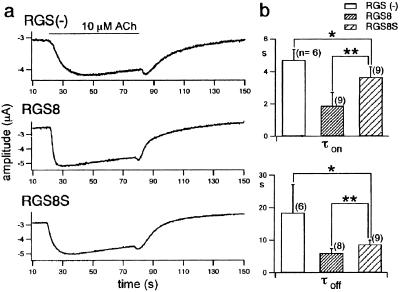
Figure 4.
Effects of RGS8 and RGS8S on turning-on and turning-off kinetics upon stimulation of the m2 muscarinic receptor. (a) GIRK1/2 and m2 muscarinic receptors without (Top) or with RGS8 (Middle) or RGS8S (Bottom) were coexpressed in Xenopus oocytes. Current traces at a holding potential of −80 mV are shown. ACh (10 μM) was applied at the time indicated by the bars. (b) Comparison of the time constants τon (Upper) and τoff (Lower) of the GIRK 1/2 current upon stimulation of m2 muscarinic receptors in the absence (Left) or presence of RGS8 (Center) or RGS8S (Right). From the traces in a, the increasing and decreasing phases were fitted with a single exponential function, τon and τoff were obtained, and the mean and standard deviation of each group were plotted. The n values are indicated in the graph. The values of RGS8S were significantly smaller than those without RGS protein (*, P < 0.05), and larger than those with RGS8 (**, P < 0.01) by Student's unpaired t test.
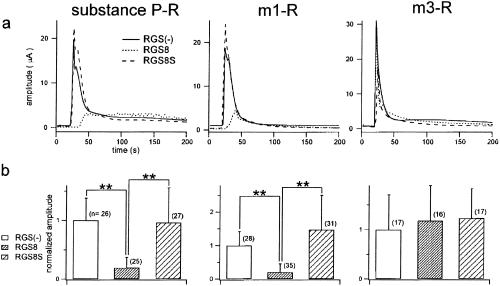
Figure 5.
Effects of RGS8 and RGS8S on the Gαq-coupled responses. The SP receptor (Left), m1 muscarinic receptor (Center), or m3 muscarinic receptor (Right) was expressed alone, with RGS8, or with RGS8S. The responses were monitored as an increase in the current amplitude of Ca2+–Cl− current. (a) Typical examples of the responses to the application of 10 nM SP (Left), 10 μM ACh (Center and Right). (b) Comparison of the peak amplitudes of the responses. To normalize the variation in the expression level among batches of oocytes, each recording was normalized by dividing by the average of the responses of RGS (−) oocytes of the same batch. Normalized data obtained from three (SP-R, m3-R) or five (m1-R) batches were pooled and the mean and standard deviation of each group were plotted. The n values are indicated in the graph. Differences judged to be significant (P < 0.01) by Student's unpaired t test are marked by **.
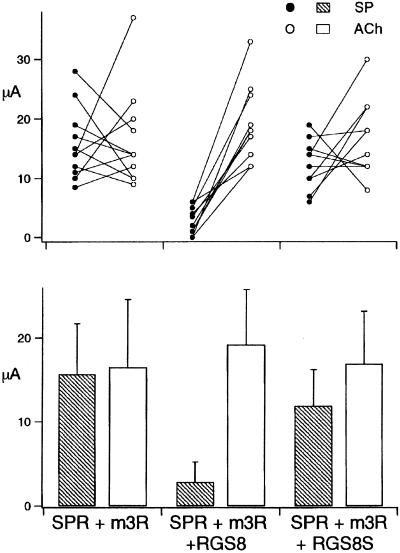
Figure 7.
Comparison of the inhibition of SP receptor and m3 receptor expressed in the same oocytes by RGS8 or RGS8S. SP receptor and m3 receptor were coexpressed in the same oocytes alone, or with RGS8 or RGS8S. SP (10 nM) was applied, and then it was washed out. Three minutes later, 10 μM ACh was applied to the same oocyte. The raw data (n = 10 or 11) are shown (Upper). Sets of two data connected by a line were obtained from the same oocyte. The mean and SD of the current amplitudes of the responses of each group were plotted (Lower).
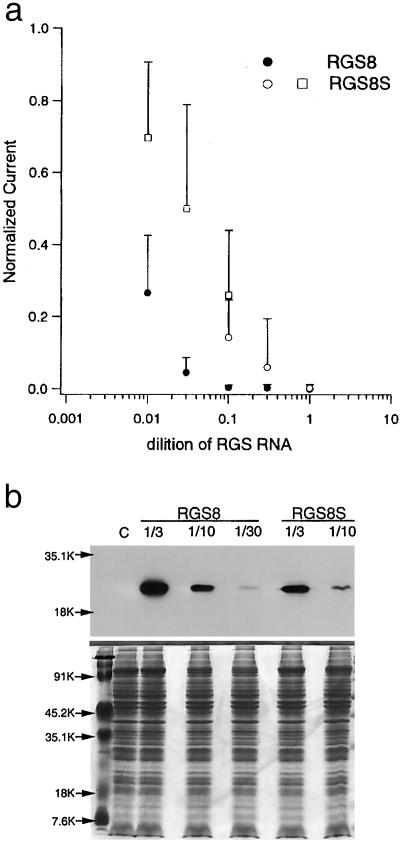
Figure 6.
Inhibition of m1 receptor-coupled response by various doses of RGS8 and RGS8S subcloned into a high expression vector and comparison of the amount of the expressed proteins. (a) RGS8 and RGS8S were subcloned into pGEMHE vector. It yields cRNA with 5′ and 3′ untranslated regions of Xenopus β-globin gene that expresses at a very high level in Xenopus oocytes. The amplitudes of Ca2+–Cl− current upon application of 10 μM ACh were normalized by the mean of the amplitudes in the absence of RGS proteins and plotted against the dilution of RGS cRNA. Dilution ×1 means full concentration, ≈1 μg/μl. ●, RGS8; ○, the results of RGS8S in the same set of experiments. □, data of RGS8S injected to the same batch of oocytes on the next day. The bars indicate SD (n = 10 or 13). (b) Oocytes of ● and ○ in a were frozen after electrophysiological analysis, and the whole protein was extracted. The amount of RGS proteins was compared by Western blotting using RGS8 antibody (Upper). The lanes correspond to noninjected control, 1/3, 1/10, 1/30 dilutions of RGS8, and 1/3, 1/10 dilutions of RGS8S, from left to right. The intensities of the bands quantified by Densitograph (Atto, Tokyo) were 3,841, 1,345, 96, 2,048, and 442 from left to right. Staining pattern with Coomassie brilliant blue of the whole protein extracts is also shown (Lower).
Results
Identification of a Form of RGS8, RGS8S.
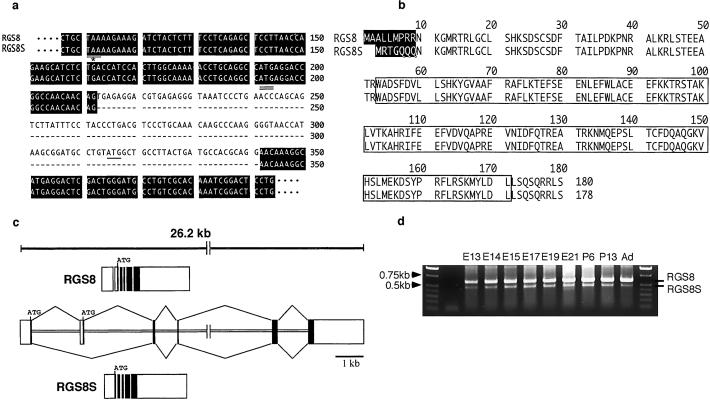
To examine whether two isoforms with different functional properties exist physiologically in the case of RGS8, we performed RT-PCR with rat brain cDNA and specific primers for 5′ and 3′ noncoding sequences of rat RGS8 cDNA. We detected a slightly smaller transcript in addition to a main product of 900 bp. By sequencing the products, we found that the main 900-bp DNA was an original rat RGS8 cDNA (5) and that the smaller one had an internal deletion of 129 bp. This 129-bp sequence includes the initiation ATG of the coding sequence of rat RGS8 (Fig. 1a, underlined). The single long ORF of the short RGS8 started from an initiation codon at 192 bp of rat RGS8 cDNA (GenBank accession no. AB006013), which is double-underlined in Fig. 1a. As a result, the N-terminal end of the additional RGS8 (RGS8S) is MRTGQQQ instead of MAALLMPRR, but the rest is the same as RGS8 (Fig. 1b). To elucidate the molecular mechanisms that generate the two forms of RGS8 and RGS8S, we searched for the RGS8 gene in the human genome. We found one locus (GenBank accession no. AL353778) on chromosome 1. By detailed comparison with coding sequences of rat RGS8 and RGS8S, this locus was found to contain a human RGS8 gene (>26.2 kb) consisting of at least six exons. The generation of RGS8 and RGS8S was explained by an alternative splicing of the second exon containing the initiation codon of RGS8 (Fig. 1c).
Figure 1.
Identification of a form of RGS8, RGS8S. (a) RT-PCR products obtained from rat brain with primers specific for 5′ and 3′ noncoding sequences of rat RGS8 cDNA were cloned and sequenced. In addition to the original rat RGS8 cDNA (5), a cDNA with an internal deletion of 129 bp was found. The nucleotide sequence of the short cDNA was aligned with that of rat RGS8 cDNA (GenBank accession no. AB006013). Nucleotide sequences between base pairs 100 and 400 containing the deletion are shown. The initiation codon of RGS8 is indicated by a single underline. In the case of RGS8S, the initiation codon is double-underlined, and the upstream stop codon in-frame is marked by an underline and star. Identical nucleotides are boxed in black. (b) Predicted amino acid sequences of RGS8 and RGS8S were aligned. Unique sequences are boxed in black. The remaining sequences are identical. The RGS domain is indicated by a white box. (c) The RGS8 gene was found in the human genome by homology search with sequences of RGS8 and RGS8S of rats (GenBank accession no. AL353778 on human chromosome 1). The predicted exon-intron organization is indicated. (d) Expression of RGS8S mRNA in developing brain. Total RNA was isolated from developing rat brain and RT-PCR analysis was performed by using primers corresponding to common sequences of RGS8 and RGS8S. E13: heads of 13-day embryo; E14, E15, E17, E19, E21: brains of 14-, 15-, 17-, 19-, and 21-day embryos, respectively; P6, P13: 6- and 13-day neonates, respectively; Ad: adults.
We next examined expression levels of the mRNAs of RGS8 and RGS8S in the developing brain. Total RNA was isolated from rat brain, and RT-PCR was performed. RGS8S mRNA was detected in 13-day embryos up to adults, but at levels lower than those of RGS8 mRNA at all stages examined (Fig. 1d).
Cellular Distribution of RGS8 and RGS8S.
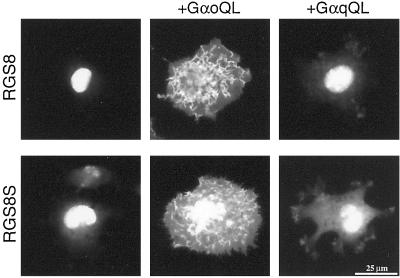
We recently demonstrated that the N terminus of RGS8 contributes to the subcellular localization in cultured cells (6). As the sequence of the N-terminal end differs between RGS8 and RGS8S, we compared the subcellular distribution and regulation of distribution patterns in transfected DDT1MF2 cells immunocytochemically. RGS8 protein was localized most abundantly in the nucleus in most of the transfected cells, and cotransfection of RGS8 and constitutively active Gαo (GαoQL) cDNA resulted in the translocation of RGS8 protein to the plasma membrane as described (6, 15). RGS8S protein showed similar nuclear localization and translocation to the plasma membrane induced by GαoQL (Fig. 2). In the case of coexpression of active Gαq (GαqQL), the membrane translocation of neither RGS8 nor RGS8S protein was observed (Fig. 2). Taken together, we could not detect any significant difference in the subcellular distribution and regulation of RGS8 and RGS8S proteins.
Figure 2.
Cellular distribution of RGS8S protein in cultured cells. DDT1MF2 cells were transfected with RGS8 or RGS8S cDNA alone, constitutively active Gαo (GαoQL), or constitutively active Gαq (GαqQL). After 48 h, cells were immunostained with anti-RGS8 antibody.
Effects of RGS8 and RGS8S on Pheromone Signaling in Yeast.
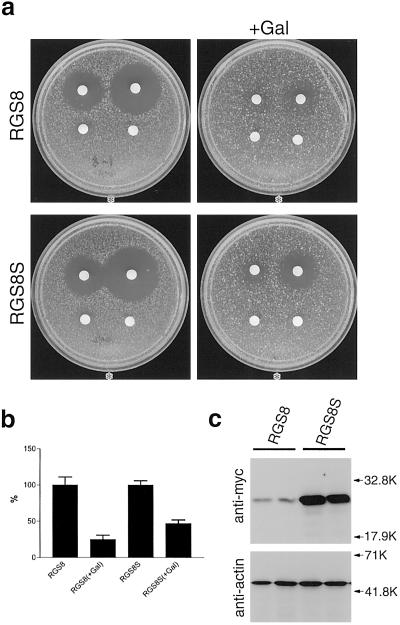
We next compared the functional difference between RGS8 and RGS8S in inhibiting the Gi-coupled system by using the yeast pheromone response pathway. Yeast cells stimulated with mating pheromone activate a heterotrimeric Gi-type protein-linked, mitogen-activated protein kinase pathway that induces G1 arrest and differentiation (16). We previously showed that expression of RGS8 reduces the size of a halo by inhibiting the growth arrest response to mating pheromone (6). We observed that RGS8S could also attenuate the pheromone signaling, but that its effect was slightly weaker than that of RGS8 (Fig. 3 a and b). As the expression level of RGS8S was apparently higher than that of RGS8 as detected by Western blotting (Fig. 3c), it was concluded that RGS8S regulates yeast pheromone signaling less effectively than RGS8.
Figure 3.
Effect of RGS8S on the response of yeasts to mating pheromone. (a) Cells of the sst2 strain (SNY86) carrying RGS8-pTS210 and RGS8S-pTS210 were plated on soft agar without or with 0.5% galactose (+Gal). Sterile filter disks were placed on the nascent lawn, and synthesized α-pheromone was applied to the disks. Plates were incubated at 30°C for 36 h. The amount of α-pheromone added to each disk was: 0 ng (Lower Right), 0.2 ng (Lower Left), 2 ng (Upper Left), and 20 ng (Upper Right). (b) The size of the halo of growth inhibition of cell lawns grown on agar plates (20 ng of α-pheromone) was calculated by measuring its diameter. Results were expressed as a percentage of the halo formed without galactose. The mean and standard deviation were as follows: RGS8 (0.5% Gal), 25.0 ± 5.9%; RGS8S (0.5% Gal), 46.8 ± 5.1%. (c) Two colonies isolated from each yeast (SNY86) transformed with RGS8-pTS210 and RGS8S-pTS210 were cultured in galactose-containing medium. The expression of RGS8 and RGS8S was detected by Western blotting with anti-myc antibody. The expression of actin was also examined.
Effects of RGS8 and RGS8S on On and Off Kinetics of the Gi/o-Coupled Response.
GIRK channels are known to be activated directly by Gβγ subunits released upon stimulation of Gi/o protein-coupled receptors such as the m2 muscarinic receptor. Doupnik et al. (17) and us (5) reported in 1997 that some RGS proteins including RGS8 accelerate both the activation and deactivation kinetics of GIRK upon receptor stimulation (for review see ref. 18). We compared here the effects of RGS8 and RGS8S on the on-off kinetics of GIRK current by using Xenopus oocytes coexpressing the GIRK1/2 heteromultimer and m2 receptor under a two-electrode voltage clamp (Fig. 4a). RGS8S accelerated both the turning on and off kinetics, but the effect was shown to be weaker than that of RGS8 by comparing the time constants for the fitted single exponential function of the activation and deactivation phases, τon and τoff (Fig. 4b). From the yeast pheromone response assays and the GIRK kinetics analysis, we concluded that RGS8S has an essentially similar but attenuated function in modulating Gi/o-mediated signaling in comparison with RGS8.
Effects of RGS8 and RGS8S on the Gαq-Coupled Responses.
Zeng et al. (7) reported that the N-terminal domain (1–33 aa) of RGS4 confers receptor-selective inhibition of Gq signaling. As there is significant sequence conservation in the N-terminal region of RGS4 and RGS8 (6), we speculated that RGS8 and/or RGS8S might also have a regulatory effect on Gq signaling, although our in vitro binding assay showed that RGS8 binds to Gαo and Gαi3 but only very weakly to Gαq (5). We monitored Gq signaling by using Xenopus oocytes as an increase in Ca2+-activated Cl− current caused by an increase in the intracellular Ca2+ (19, 20) upon stimulation of coexpressed Gq-coupled receptors, such as the m1 muscarinic, m3 muscarinic, or SP receptor. Expression of RGS8 suppressed the Gq signaling, from the m1 muscarinic or SP receptor, but the suppression of Gq signaling from the m3 muscarinic receptor was much less (Fig. 5). These results show that RGS8 suppresses Gq signaling to a different extent depending on the type of the receptor. On the other hand, RGS8S had reduced inhibitory effect on Gq signaling, although the unique region of RGS8S is only 7 aa long in the N terminus (Fig. 5).
There is a possibility that the remarkable difference of the inhibitory effect of RGS8 and RGS8S is not caused by the qualitative difference but by the difference of the expression level; i.e., it is possible that the expression level of RGS8S is low in the above experiment, and that RGS8S can also inhibit Gq-coupled responses when the expression level is high enough. To examine this possibility, we subcloned RGS8 and RGS8S to pGEMHE, which includes 5′ and 3′ noncoding sequences of Xenopus β-globin gene, enabling a high expression in oocytes (21). We observed that coinjection of full concentration (≈1 μg/μl) of RGS8S cRNA transcribed from pGEMHE vector clearly inhibited Gq-coupled responses by m1 receptor activation. As a next step, we injected cRNA of RGS8 or RGS8S of various dilution to compare the dose-inhibition relationship quantitatively (Fig. 6a). It was clearly observed that the inhibitory effect of RGS8S is much weaker compared with RGS8 when the RNA dose is the same (Fig. 6a). As a next step, we compared the amount of the expressed protein by Western blotting. The results showed that the amount of RGS8S protein was less than that of RGS8 even when the same amount of RNA was injected (Fig. 6b). From these results, it was shown that the remarkable difference of the dose-inhibition relationships of Fig. 6a was, at least partly, caused by the difference of the amount of the expressed protein. However, although the amount of RGS8S protein (1/3 RNA dilution) was more than that of RGS8 (1/10) (Fig. 6b), the effect was less (Fig. 6a). Also, although the amount of RGS8S protein (1/10) was four times more than that of RGS8 protein (1/30) (Fig. 6b), the inhibitory effect was significantly less (P < 0.02 by Student's unpaired t test) (Fig. 6a). Thus, we concluded that RGS8S has a qualitatively less inhibitory effect on the Gq-coupled response than RGS8, besides the lower expression level when the same amount of cRNA was injected.
It is also possible that the apparent receptor-type specificity of inhibition by RGS8 is caused by the difference of the expression level of receptors. To clarify this point, we carried out the experiment in Fig. 7. We coexpressed SP receptor and m3 receptor in the same oocytes, alone or with RGS8 or RGS8S, and measured the amplitudes of responses by application of SP and then ACh. In this experiment, the amplitudes of responses to SP and ACh were at a similar level in the absence of RGS (Fig. 7 Lower), confirming that the expression level of SP receptor and m3 receptor are comparable. By paired t test, it was shown that the amplitudes of responses to SP and ACh did not differ significantly (P > 0.05) in the absence of RGS and the presence of RGS8S. In contrast, there was a significant difference (P < 0.01) in the presence of RGS8 (Fig. 7 Upper). We, therefore, concluded that the receptor-type specificity of the inhibition by RGS8 is not caused by the difference of the expression level of the receptors.
Taken together, we uncovered a function of RGS8 to suppress Gq signaling in a receptor type-specific manner despite its weak in vitro binding to Gq protein, and that RGS8S, a splice variant of the N terminus of 9 aa, unexpectedly has a diminished function.
Discussion
In the present work, we report isolation and characterization of a splice variant of RGS8, RGS8S, in which 9 aa at the N terminus of RGS8 were replaced with 7 aa. It has been shown that the N-terminal 35 aa of RGS8 contribute to the subcellular localization in cultured cells. Therefore, we compared the subcellular distribution and regulation of distribution patterns of RGS8 and RGS8S in cultured cells. No significant difference was detected. The sequence of amino acids 10–35 of RGS8 was considered to play important roles in controlling its cellular distribution.
In yeast, RGS8S protein was expressed more abundantly than RGS8 protein, although RGS8S showed less inhibitory effect on pheromone signaling. When the N-terminal 7 aa of RGS8S was deleted, expression level of this truncated RGS8S decreased in yeast (data not shown). Therefore, the presence of the N terminus of RGS8S was considered to increase its protein stability in yeast. Transfection experiments using mammalian cells, however, did not indicate significant difference in the expression level of RGS8 and RGS8S. The lack of difference might be caused by an extremely high transcription level of the pCXN2 vector we used for transfection, which obscured the difference of protein stability.
We further compared regulatory effects of RGS8 and RGS8S on Gi-coupled responses. GIRK channels are activated by stimulation of Gi/o protein-coupled receptors. When coexpressed with GIRK channels, RGS8S accelerated the turning on and off of Gi/o-mediated GIRK response. This acceleration of RGS8S was not as efficient as RGS8. Taken together with the less contribution of RGS8S than RGS8 in yeast pheromone signaling, these results indicate that the regulatory function of RGS8S on Gi-coupled responses is weaker than that of RGS8. On the other hand, RGS8S induced acute desensitization of GIRK similar to RGS8. Acute desensitization of GIRK was demonstrated to be caused by the nucleotide exchange and hydrolysis cycle of G proteins (22). We previously showed that RGS8 requires its N-terminal 35 aa for the subcellular localization and the acute desensitization of GIRK (6). Thus, the N-terminal 9 aa of RGS8 were shown not to be involved in determining cellular distribution and inducing acute desensitization of GIRK.
We next examined the effects of RGS8 and RGS8S on Gq-mediated signaling. RGS8 suppressed Gq signaling in a receptor type-specific manner despite its weak in vitro binding to Gq protein, but RGS8S showed a diminished effect. What is the mechanism underlining the suppression of Gq signaling through the m1 or SP receptor by RGS8 and weakly by RGS8S? Because inhibition was observed to be receptor type specific, it is not likely that downstream pathways in Gq signaling are affected by RGS8. The first possibility is that RGS8 functions as a GTPase-activating protein for only a certain type of Gαq coupled to these receptors. If this is the case, it is speculated that the specific N terminus and RGS domain cooperatively determine the interaction of RGS8 with Gα, as the RGS domain is well known to bind to Gα subunits and is responsible for GTPase-activating protein function. The second possibility is that RGS8 recognizes a certain class of receptors by using its N-terminal region and that it suppresses signaling by directly interacting with these receptors. Considering that the binding activity of RGS8 for Gαq was weak (5), we speculate that the second possibility is the more likely. Supporting this possibility indirectly, we observed that RGS8 did not translocate from the nucleus to the plasma membrane remarkably when active Gαq mutant was coexpressed, in contrast to the case of Gαo. This result shows that Gq activation is not sufficient to induce significant membrane translocation of RGS8 and suggests that a certain type of Gq-coupled receptor may recruit RGS8 to the plasma membrane by direct interaction.
Further detailed study is required to understand the molecular mechanisms behind the receptor type-specific inhibition of the Gq response by RGS8 and the functional difference in the regulation of Gq signaling between RGS8 and RGS8S. We demonstrate, however, that alternative splicing for replacement of short N terminus of small RGS proteins, such as RGS8, changes its inhibitory function on G protein signaling and that these two forms are physiologically expressed in the brain.
Acknowledgments
We are grateful to Drs. T. Kubo for m1, m2, and m3 muscarinic receptor cDNA, M. Lazdunski for GIRK2 cDNA, S. Nakanishi for SP receptor cDNA, J. D. Jordan for Gαo Q205L cDNA, J. Miyazaki for pcXN2 expression vector, and J. Tytgat for pGEMHE vector. We thank Drs. H. Nakata and H. Abe for helpful discussion. This work was supported by research grants from the Ministry of Education, Science, Sports, and Culture of Japan (to O.S. and Y.K.), the Kato Memorial Bioscience Foundation (to O.S.), the Mitsubishi Foundation, Uehara Foundation, and Kowa Foundation (to Y.K.). Y.K. is also supported by the Core Research for Evolutional Science and Technology of the Japan Science and Technology Corporation.
Abbreviations
- RGS
regulators of G protein signaling
- GIRK
G protein-gated inwardly rectifying K+ channel
- SP
substance P
- RT-PCR
reverse transcriptase–PCR
- ACh
acetylcholine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dohlman H G, Thorner J. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 2.Berman D M, Gilman A G. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 3.Burchett S A. J Neurochem. 2000;75:1335–1351. doi: 10.1046/j.1471-4159.2000.0751335.x. [DOI] [PubMed] [Google Scholar]
- 4.Koelle M R, Horvitz H R. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 5.Saitoh O, Kubo Y, Miyatani Y, Asano T, Nakata H. Nature (London) 1997;390:525–529. doi: 10.1038/37385. [DOI] [PubMed] [Google Scholar]
- 6.Saitoh O, Masuho I, Terakawa I, Nomoto S, Asano T, Kubo Y. J Biol Chem. 2001;276:5052–5058. doi: 10.1074/jbc.M006917200. [DOI] [PubMed] [Google Scholar]
- 7.Zeng W, Xu X, Popov S, Mukhopadhyay S, Chidiac P, Swistok J, Danho W, Yagaloff K A, Fisher S L, Ross E M, et al. J Biol Chem. 1998;273:34687–34690. doi: 10.1074/jbc.273.52.34687. [DOI] [PubMed] [Google Scholar]
- 8.Druey K M, Blumer K J, Kang V H, Kehrl J H. Nature (London) 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee T K, Eapen A K, Fisher R A. J Biol Chem. 1997;272:15481–15487. doi: 10.1074/jbc.272.24.15481. [DOI] [PubMed] [Google Scholar]
- 10.Dulin N O, Sorokin A, Reed E, Elliott S, Kehrl J H, Dunn M J. Mol Cell Biol. 1999;19:714–723. doi: 10.1128/mcb.19.1.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dulin N O, Pratt P, Triruppathi C, Niu J, Voyno-Yasenetskaya T, Dunn M J. J Biol Chem. 2000;275:21317–21323. doi: 10.1074/jbc.M910079199. [DOI] [PubMed] [Google Scholar]
- 12.Saitoh O, Odagiri M, Masuho I, Nomoto S, Kinoshita N. Biochem Biophys Res Commun. 2000;270:34–39. doi: 10.1006/bbrc.2000.2379. [DOI] [PubMed] [Google Scholar]
- 13.Nomoto S, Adachi K, Yang L-X, Hirata Y, Muraguchi S, Kiuchi K. Biochem Biophys Res Commun. 1997;241:281–287. doi: 10.1006/bbrc.1997.7802. [DOI] [PubMed] [Google Scholar]
- 14.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 15.Itoh M, Odagiri M, Abe H, Saitoh O. Biochem Biophys Res Commun. 2001;287:223–228. doi: 10.1006/bbrc.2001.5489. [DOI] [PubMed] [Google Scholar]
- 16.Kurjan J. Annu Rev Genet. 1993;27:147–179. doi: 10.1146/annurev.ge.27.120193.001051. [DOI] [PubMed] [Google Scholar]
- 17.Doupnik C A, Davidson N, Lester H A, Kofuji P. Proc Natl Acad Sci USA. 1997;94:10461–10466. doi: 10.1073/pnas.94.19.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerangue N, Jan L Y. Curr Biol. 1998;8:R313–R316. doi: 10.1016/s0960-9822(98)70196-4. [DOI] [PubMed] [Google Scholar]
- 19.Rhee S G, Bae Y S. J Biol Chem. 1997;272:15045–15048. doi: 10.1074/jbc.272.24.15045. [DOI] [PubMed] [Google Scholar]
- 20.Berridge M J. Nature (London) 1993;361:315–332. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 21.Tytgat J, Vereecke J, Carmeliet E. J Physiol (London) 1994;481:7–13. doi: 10.1113/jphysiol.1994.sp020414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang H-H, Yu M, Jan Y N, Jan L Y. Proc Natl Acad Sci USA. 1998;95:11727–11732. doi: 10.1073/pnas.95.20.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]