Abstract
The mammalian dynamin-like protein DLP1/Drp1 has been shown to mediate both mitochondrial and peroxisomal fission. In this study, we have examined whether hFis1, a mammalian homologue of yeast Fis1, which has been shown to participate in mitochondrial fission by an interaction with DLP1/Drp1, is also involved in peroxisomal growth and division. We show that hFis1 localizes to peroxisomes in addition to mitochondria. Through differential tagging and deletion experiments, we demonstrate that the transmembrane domain and the short C-terminal tail of hFis1 is both necessary and sufficient for its targeting to peroxisomes and mitochondria, whereas the N-terminal region is required for organelle fission. hFis1 promotes peroxisome division upon ectopic expression, whereas silencing of Fis1 by small interfering RNA inhibited fission and caused tubulation of peroxisomes. These findings provide the first evidence for a role of Fis1 in peroxisomal fission and suggest that the fission machinery of mitochondria and peroxisomes shares common components.
INTRODUCTION
Peroxisomes are ubiquitous subcellular organelles that participate in a variety of important catabolic and anabolic functions, including hydrogen peroxide and lipid metabolism (van den Bosch et al., 1992). An interesting feature of peroxisomes is their ability to proliferate and multiply, or be degraded in response to nutritional and environmental stimuli. The prevailing model of peroxisome biogenesis (Lazarow and Fujiki, 1985) predicts that peroxisomes grow by uptake of newly synthesized proteins from the cytosol and multiply by division. The majority of the proteins controlling peroxisome biogenesis (collectively named peroxins) are linked to matrix protein import (Purdue and Lazarow, 2001), whereas proteins that are specifically involved in peroxisome proliferation are scarce. Members of the Pex11p family of peroxisomal membrane proteins (PMPs) have been proposed to function in the regulation of peroxisome size and number in a variety of species (Erdmann and Blobel, 1995; Marshall et al., 1995; Sakai et al., 1995; Lorenz et al., 1998; Passreiter et al., 1998; Schrader et al., 1998). Pex11p promotes peroxisome elongation and subsequent division upon ectopic expression, whereas loss of Pex11p results in reduced peroxisome number (Erdmann and Blobel, 1995; Marshall et al., 1995; Schrader et al., 1998; Li et al., 2002). A striking increase in elongated forms of peroxisomes on expression of Pex11p has been observed in all organisms studied, indicating that tubule formation of peroxisomes may be an important aspect of peroxisome division (Schrader et al., 1996, 1998).
Recently, we and others found that the dynamin-like protein DLP1 is involved in peroxisome fission in mammals (Koch et al., 2003, 2004; Li and Gould, 2003). Similarly, the dynamin-related protein Vps1p was found to mediate peroxisome division in Saccharomyces cerevisiae (Hoepfner et al., 2001). The dynamin family of large GTPases has been implicated in tubulation and fission events of cellular membranes (Praefcke and McMahon, 2004). Mammalian DLP1 and its homologues Dnm1p (S. cerevisiae) and Drp1 (Caenorhabditis elegans) are also known to mediate mitochondrial fission (Otsuga et al., 1998; Yoon et al., 1998; Labrousse et al., 1999; Bleazard et al., 1999; Sesaki and Jensen, 1999; Smirnova et al., 2001).
In a previous study, we presented evidence that peroxisome elongation and constriction can occur independently of DLP1, whereas the final fission step of peroxisomes requires DLP1 function (Koch et al., 2004). Furthermore, overexpression of DLP1 did not increase peroxisomal fission, indicating that other proteins/factors are required in the peroxisomal fission process. Studies on mitochondrial division in yeast indicate that mitochondrial fission is mediated by an interaction of three proteins, the dynamin-like enzyme Dnm1p, Mdv1p, and Fis1p through a multistep pathway (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000). Whereas a cognate homologue of Mdv1p has not yet been found in higher eukaryotes, homologues of Fis1p have been identified. It has recently been shown that hFis1, a human homologue of Fis1p, regulates mitochondrial fission in mammalian cells through an interaction with DLP1 (James et al., 2003; Yoon et al., 2003; Stojanovski et al., 2004). Fis1p is a 17-kDa transmembrane protein of the outer mitochondrial membrane. Its N-terminal region is predicted to be exposed to the cytosol and to adopt a novel tetratricopeptide repeat (TPR)-like helix bundle (Suzuki et al., 2003; Dohm et al., 2004), whereas a short C-terminal tail protrudes into the mitochondrial intermembrane space (Mozdy et al., 2000; Yoon et al., 2003). In regard of a similar mechanism of mitochondrial and peroxisomal membrane fission, we studied the role of Fis1 in peroxisome biogenesis. We report here that Fis1 is involved in peroxisomal fission and that it is one of the few transmembrane proteins described so far that is targeted to both mitochondria and peroxisomes.
MATERIALS AND METHODS
cDNAs and Antibodies
Untagged hFis1 (hFis1-UT), hFis1 fused to green fluorescent protein (GFP), or the Myc epitope tag (hFis1-GFP, hFis1-Myc, GFP-hFis1, Myc-hFis1), and the C-terminally (Myc-hFis1-ΔC, Myc-hFis1-ΔTM/C) and N-terminally truncated hFis1 constructs [Myc-hFis1(32-152), Myc-hFis1(61-152), and Myc-hFis1(92-152)] were described previously (Yoon et al., 2003). A construct encoding the C-terminal 26 amino acids of hFis1 tagged to yellow fluorescent protein (YFP) (hFis1-YFP-TM/C) was kindly provided by D. I. James (University of Geneva, Geneva, Switzerland). The dominant-negative mutant of DLP1 (GFP-DLP1-K38A) was described previously (Pitts et al., 1999). The C-terminally tagged version of Pex11pβ-Myc was described in Schrader et al. (1998). The construct pmitoGFP for the labeling of the mitochondrial matrix and rabbit anti-porin polyclonal antibodies were kindly provided by R. Lill (University of Marburg, Marburg, Germany). Rabbit anti-PMP70, rabbit anti-catalase, and rabbit antiacyl-CoA oxidase polyclonal antibodies were a gift from A. Völkl (University of Heidelberg, Heidelberg, Germany). A polyclonal antibody to the Myc epitope was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-hFis1 polyclonal antibody used for immunoblotting was described previously (Yoon et al., 2003). Rabbit anti-hFis1 polyclonal antibody used for morphological studies was obtained from Alexis Biochemicals (Grünberg, Germany). The following monoclonal antibodies were used: antimyc epitope 9E10 (kindly provided by M. Eilers, University of Marburg, Germany) and anti-tubulin DM1α (Sigma-Aldrich, St. Louis, MO). Species-specific anti-IgG antibodies conjugated to tetramethylrhodamine B isothiocyanate (TRITC) or fluorescein isothiocyanate (FITC) were obtained from Dianova (Hamburg, Germany).
Cell Culture, RNA Interference, and Transfection Experiments
COS-7, HepG2, and COS-7 cells stably expressing a GFP construct bearing a C-terminal peroxisomal targeting signal 1 (GFP-PTS1) were cultured in DMEM containing 10% fetal calf serum as described previously (Schrader et al., 2000). Cells were transfected with DNA constructs by incubation with polyethylenimine (Sigma-Aldrich) or by electroporation (Schrader et al., 1998; Koch et al., 2004). To knock down the expression of hFis1 (accession no. AF151893) by RNA interference, 21-nucleotide small interfering RNA (siRNA) (sense strand, 5′-CGAGCUGGUGUCUGUGGAGdTdT-3′) (Dharmacon, Lafayette, CO) was transfected into the cells using Oligofectamine (Invitrogen, Carlsbad, CA). As a control, cells were transfected with siRNA duplexes targeting luciferase (Dharmacon). Cells were transfected with siRNA duplexes 24 and 48 h after seeding and assayed for silencing and organelle morphology 72–96 h after seeding.
Immunofluorescence and Microscopy
Cells grown on glass coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline, pH 7.4, permeabilized with 0.2% Triton X-100 or 25 μg/ml digitonin, and incubated with primary and secondary antibodies as described (Schrader et al., 1998). For the detection of endogenous Fis1, cells were first fixed with 4% paraformaldehyde and afterwards treated with cold methanol for 5 min. Transfected cells were processed for immunofluorescence 24–48 h after transfection. Samples were examined using an Olympus BX-61 microscope (Olympus Optical, Hamburg, Germany) equipped with the appropriate filter combinations and a 100× objective (Olympus Plan-Neofluar; numerical aperture, 1.35). Fluorescence images were acquired with an F-view II charge-coupled device camera (Soft Imaging System, Münster, Germany) driven by Soft imaging software. Digital images were optimized for contrast and brightness using Adobe Photoshop software.
Isolation of Peroxisomes
Peroxisomes were isolated from the livers of adult male Wistar rats (Charles River, Sulzfeld, Germany) according to standard procedures (Völkl et al., 1996). Animals were handled according to the German law for the protection of animals, with the permit of the local authorities. Isolation of peroxisomes from rats that were fed with the peroxisome proliferator bezafibrate (Beier et al., 1988) (Roche Diagnostics, Mannheim, Germany) was performed in cooperation with A. Völkl (University of Heidelberg). Briefly, one liver was homogenized using a Potter S homogenizer (Braun, Melsungen, Germany) in ice-cold homogenization buffer (5 mM MOPS, pH 7.4, 250 mM sucrose, 1 mM EDTA, 0.1% ethanol, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM ε-aminocaproic acid, and 0.2 mM dithiothreitol). After subcellular fractionation, a crude peroxisomal fraction was sedimented into exponentially shaped OptiPrep (Axis-Shield, Oslo, Norway) gradients. The highly purified peroxisomal fraction with a density of 1.24 g/ml (>95% pure) was used for further experiments. Purified peroxisomes were lysed by freezing and thawing and separated into a matrix and membrane fraction by centrifugation at 100,000g for 1 h in a swinging-bucket rotor (Beckman SW50.1). For determination of catalase and cytochrome c oxidase activities, standard procedures were used (Völkl et al., 1996). Protein was determined using the bicinchoninic acid assay. All assays were run with a recording spectrophotometer (Uvikon 810, Kontron, Munich, Germany; or a Beckman model 24, Beckman Instruments, Munich, Germany).
Gel Electrophoresis and Immunoblotting
Protein samples were separated by SDS-PAGE, transferred to nitrocellulose using a semidry apparatus, and analyzed by immunoblotting. Immunoblots were processed using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence reagents (GE Healthcare, Piscataway, NJ). For quantification, immunoblots were scanned and processed using Pcbas software.
Quantitation and Statistical Analysis of Data
For quantitative evaluation of peroxisome morphology, 100–200 cells per coverslip were examined and categorized as cells with elongated, tubular (> 2 μm in length; Figure 6C) or spherical peroxisomes (0.3–1 μm; including rod-shaped peroxisomes; Figures 1B and 5C) as described previously (Schrader et al., 1996). Furthermore, cells exhibiting elongated peroxisomes with a segmented, “beads on a string”-like appearance (Figure 3, C and D) and cells with very small, punctiform peroxisomes (<0.3 μm) (Figures 3C and 7B) were counted. Usually, three to five coverslips per preparation were analyzed, and three to five independent experiments were performed. Significant differences between experimental groups were detected by analysis of variance for unpaired variables using Microsoft Excel. Data are presented as means ± SD, with an unpaired t test used to determine statistical differences. p values <0.05 are considered as significant, and p values <0.01 are considered as highly significant.
Figure 6.
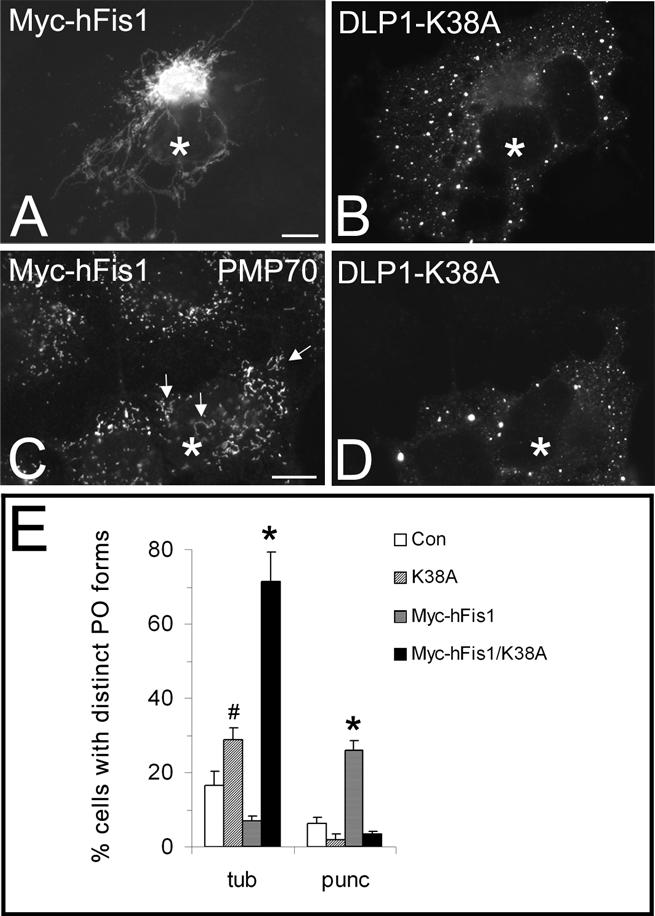
Inhibition of DLP1 function interferes with peroxisomal fission induced by hFis1 expression. COS-7 cells were cotransfected with Myc-hFis1 and GFP-DLP1-K38A (A–D) and immunostained with antibodies to the Myc tag of hFis1 (A) or to PMP70 (C). The corresponding GFP fluorescence of DLP1-K38A is shown in B and D. Note the pronounced elongation of peroxisomes in DLP1-K38A coexpressing cells (arrows). A quantitative analysis of peroxisome morphology is shown in E. Cells were categorized as cells with tubular (tub) or punctiform (punc) peroxisomes (percentage of total) (see Materials and Methods); *p < 0.01, # p < 0.05 compared with controls. Con, untransfected control. Asterisks, coexpressing cells. N, nucleus. Bars, 10 μm.
Figure 1.
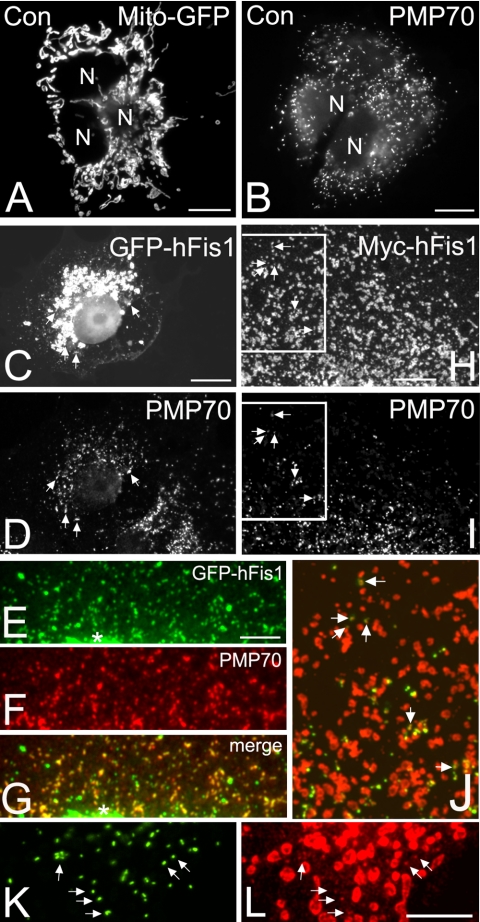
Fis1 localizes to peroxisomes and mitochondria in mammalian cells. Normal mitochondrial (A) and peroxisomal (B) morphology in COS-7 control cells processed for immunofluorescence microscopy. Mitochondria in A are visualized by expression of a Mito-GFP construct, whereas peroxisomes (B) are stained with antibodies to PMP70, a peroxisomal membrane protein. GFP-tagged (C–G) and Myc-tagged (H–J) hFis1 protein (GFP-hFis1, Myc-hFis1) colocalizes with peroxisomes. COS-7 cells were transfected with either GFP-hFis1 (C–G) or Myc-hFis1 (H–J) and processed for immunofluorescence microscopy using antibodies to PMP70 (D, F, and I) and the Myc epitope tag (H). Note that the expression of GFP-hFis1 causes aggregation of mitochondria around the nucleus (C), whereas expression of Myc-hFis1 induces their fragmentation (H). (E–G) Higher magnification view of an area close to the nucleus in a COS-7 cell transfected with GFP-hFis1. The asterisks in E and G mark the beginning of a mitochondrial aggregate. (G) Overlay (merge) of E and F. (J) Overlay and higher magnification view of boxed region in H and I. Myc-hFis1/TRITC (red), PMP70/FITC (green). (K and L) Detection of endogenous Fis1 (L) in COS-7 cells expressing a GFP-PTS1 construct (K). Arrows highlight some regions of colocalization. N, nucleus. Bars, 10 μm (A–D, H–L); 5 μm (E–G).
Figure 5.

Silencing of Fis1 induces elongation of peroxisomes. COS-7 cells were transfected with Fis1 siRNA duplexes (siRNA) and processed for immunofluorescence (A–F) and immunoblotting (H) with anti-hFis1 antibodies. (A and B) Mitochondrial morphology in control cells (Con) expressing Mito-GFP (A), and in cells silenced for Fis1 (B). (C–F) Peroxisome morphology in control cells (Con) expressing GFP-PTS1 (C), and in cells silenced for Fis1 (E). Note the elongated, segmented peroxisomes in E (arrows). (D and F) Staining of Fis1 in control cells (D) and after transfection with Fis1 siRNA duplexes (F). (G) Quantification of peroxisome morphology after silencing of Fis1. Cells were categorized as cells with spherical (sph) or elongated (elong) peroxisomes (percentage of total) (see Materials and Methods); p < 0.01 compared with controls. (H) Immunoblots of homogenates prepared from controls (Con) and transfected cells (siRNA) using anti-hFis1 and anti-tubulin (Tub) antibodies. Equal amounts of protein (hFis1, 50 μg/lane; Tub, 20 μg/lane) were loaded onto the gels. Anti-tubulin was used to check for equal loading and integrity of the cells after transfection. N, nucleus. Bars, 10 μm.
Figure 3.
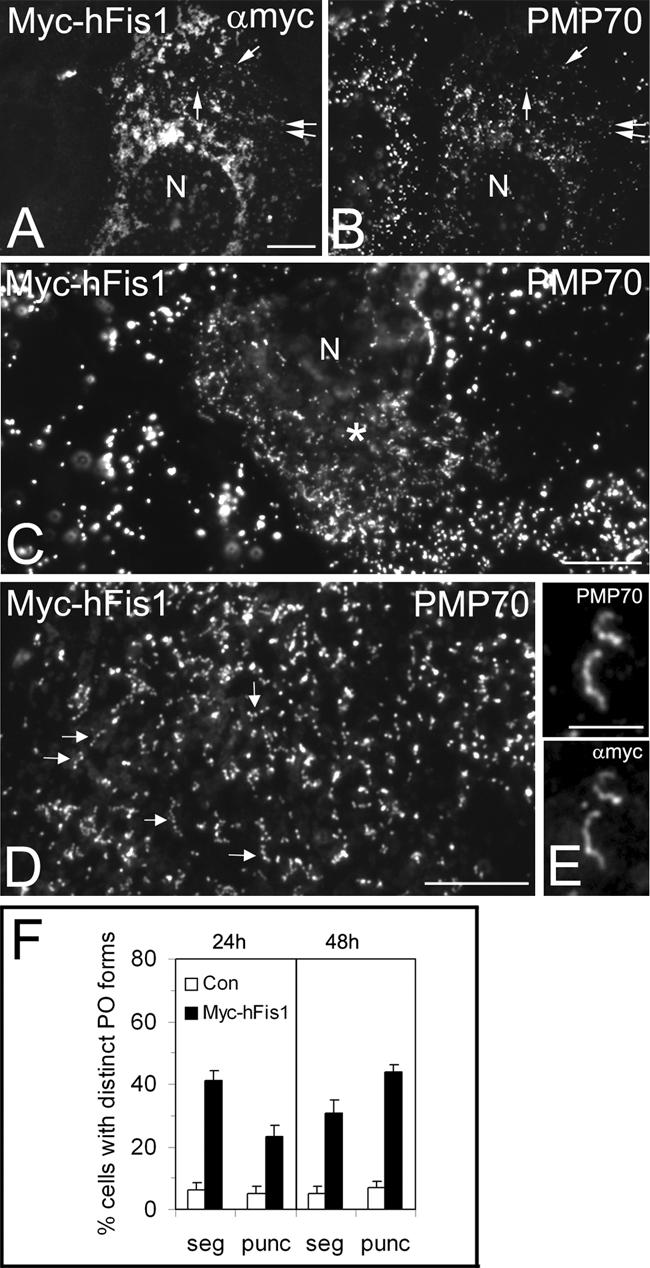
hFis1 promotes peroxisomal fission. COS-7 cells were transfected with Myc-hFis1 (A–E) and immunostained with antibodies to Myc (A and E) and PMP70 (B–E). Cells expressing Myc-hFis1 contained fragmented mitochondria (A). Note that in A, contrast and brightness have been optimized for the visualization of mitochondrial morphology and that therefore not all Myc-hFis1 positive peroxisomes are visible. Arrows in A and B point to some regions of colocalization. Peroxisomes were observed to segment and to separate into numerous punctiform organelles in Myc-hFis1expressing cells (C, D, and F). Note the normal-sized peroxisomes in the untransfected cell on the left (C). (D) Higher magnification view of separating peroxisomes (arrows). (E) Colocalization of Myc-hFis1 and PMP70 on tubular peroxisomes with beginning segmentation. (F) Quantitative evaluation of peroxisome morphology. Cells were categorized as cells with segmented (seg) and punctiform (punc) peroxisomes (% of total) (see Materials and Methods). Note the high frequency of small, punctiform peroxisomes in cells expressing Myc-hFis1. p < 0.01 compared with controls. Con, control (untransfected and vector only). Asterisks in C highlight a transfected cell. N, nucleus. Bars, 10 μm (A–D); 5 μm (E).
Figure 7.
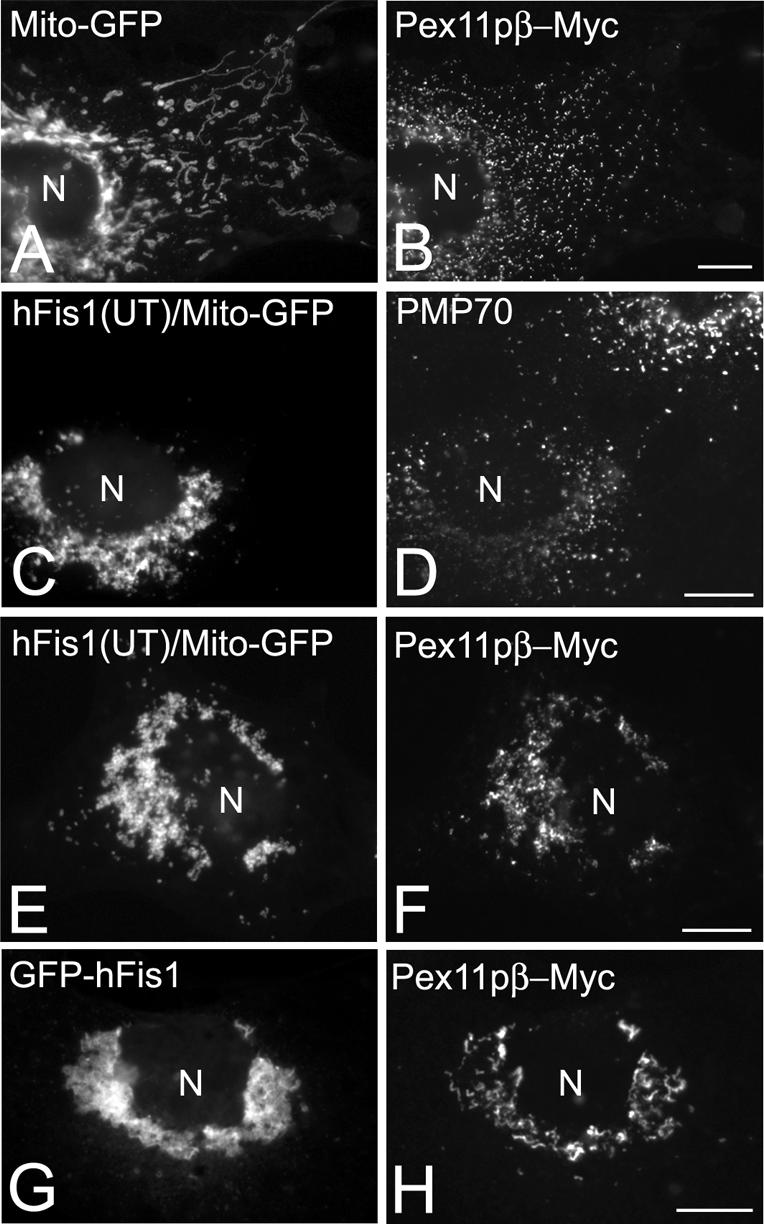
Coexpression of hFis1 and Pex11pβ influences the intracellular distribution of peroxisomes. COS-7 cells were cotransfected with Mito-GFP and Pex11pβ-Myc (A and B), with Mito-GFP and untagged hFis1(hFis1-UT) (C and D), with Mito-GFP, Pex11pβ-Myc and untagged hFis1(hFis1-UT) (E and F), or with GFP-hFis1 and Pex11pβ-Myc (G and H). Twenty-four hours after transfection, cells were immunostained with anti-Myc (B, F, and H) or anti-PMP70 (D) antibodies. Note the juxtanuclear clustering of peroxisomes and their association with mitochondria in cells coexpressing hFis1 and Pex11pβ (E–H). N, nucleus. Bars, 10 μm.
RESULTS
Fis1 Localizes to Mitochondria and Peroxisomes
To examine whether Fis1 localizes to peroxisomes, COS-7 cells were transfected with two hFis1 constructs tagged with GFP (GFP-hFis1) or Myc (Myc-hFis1) at the N terminus of hFis1. These constructs have recently been reported to be targeted to mitochondria and to alter mitochondrial morphology (Yoon et al., 2003). In control cells processed for immunofluorescence mitochondria exhibited a heterogeneous morphology with spherical, tubular, and bulbous structures (Figure 1A and supplemental data). Peroxisomes in controls were of spherical or rod-like shape and showed a typical uniform intracellular distribution (Figure 1B and supplemental data). In cells expressing GFP-hFis1, mitochondria had a banded or striped morphology and became aggregated around the nucleus (Figure 1C and 4E). This aggregation might be mediated by an interaction of the GFP tags on the mitochondrial surface (Yoon et al., 2003). Through the aggregation of mitochondria in the cell center, a fine, punctate staining pattern in the cytoplasm became visible. Remarkably, many of the punctate structures positive for GFP-hFis1 colocalized with peroxisomal marker proteins (e.g., PMP70 [Figure 1, C–G], catalase, acyl-CoA oxidase [our unpublished data]). Similar to mitochondrial aggregation, some cells also exhibited small aggregates of peroxisomes (our unpublished data; Figure 4, A and B). As reported previously (Yoon et al., 2003), expression of MychFis1 causes a pronounced fragmentation of mitochondria (Figure 1, H and J). Like GFP-hFis1, Myc-hFis1 was also found to colocalize with peroxisomes, although detection was rendered more difficult due to the strong labeling intensity of fragmented mitochondria that were scattered throughout the cytoplasm (Figures 1, H–J, and 4, E and F). Peroxisomes were much smaller in diameter than the fragmented mitochondria, and Myc-hFis1- as well as GFP-hFis1 labeling of peroxisomes was heterogeneous. Colocalization of GFP-hFis1 and Myc-hFis1 with peroxisomes was further confirmed by confocal laser scanning microscopy (supplemental data). Both GFP- and Myc-tagged hFis1 were labeled under differential permeabilization conditions (25 μg/ml digitonin or 0.2% Triton X-100 for 5 min) with anti-GFP or anti-Myc antibodies (our unpublished data). Localization under these two permeabilization conditions suggests that hFis1 is not intraperoxisomal, but it is inserted in the peroxisomal membrane with its N terminus exposed to the cytoplasm. To verify the observations made with the hFis1 constructs, endogenous Fis1 was detected in COS-7 cells expressing a GFP construct bearing the C-terminal peroxisomal targeting signal 1 (GFP-PTS1) by incubation with a hFis1 antibody suitable for morphological studies. Besides a prominent staining of mitochondria, a specific and heterogeneous staining of peroxisomes was observed (Figure 1, K and L).
Figure 4.

Localization of hFis1 to peroxisomes requires an intact C terminus. COS-7 cells were transfected with hFis1-Myc (A and B), with Myc-hFis1-ΔC (C and D), or with hFis1-YFP-TM/C (E and F), and immunostained with antibodies to the Myc tag of hFis1 (A and C) and to PMP70 (B, D, and F). The inset in E shows a mitochondrial aggregate induced by the expression of YFP-hFis1-TM/C. Arrows in F point to peroxisomal aggregates. N, nucleus. Bars, 10 μm.
Fis1 Associates with Highly Purified Peroxisomes
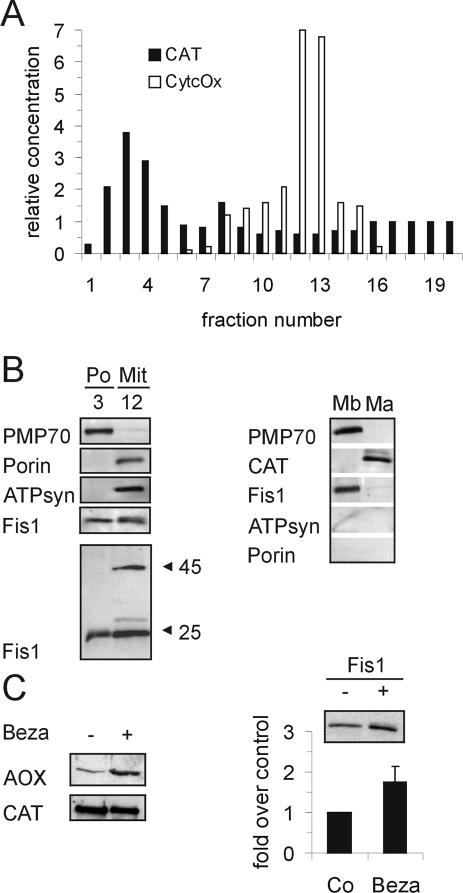
To determine whether Fis1 was associated with purified peroxisomes and peroxisomal membranes, we isolated a highly purified peroxisomal fraction from rat liver by gradient centrifugation (Figure 2A). Peroxisomes were mainly recovered in fractions 2 to 4 with the mean equilibrium density of 1.24 g/ml, whereas mitochondria mostly band in a density range of ∼1.15 g/ml (fractions 12 and 13). Determination of cytochrome c oxidase activity revealed no contamination of the peroxisomal fraction with mitochondria (relative specific activity of fraction 3, 0.01 ± 0.2; fraction 12, 1.4 ± 0.4). The peroxisomal and mitochondrial fractions were analyzed by immunoblotting with an anti-hFis1 antibody that has been shown to cross-react with both hFis1 and rat Fis1 (Yoon et al., 2003) (Figure 2B). The Fis1-specific antibody detected a single band of the expected size (17 kDa) in the peroxisomal fraction; in the mitochondrial fraction, an additional band of ∼45 kDa was detected under our experimental conditions. ATP synthase α, an abundant mitochondrial marker, and porin, an outer mitochondrial membrane protein, were absent from the peroxisomal fraction. When isolated peroxisomes were separated into a membrane and matrix fraction, Fis1p was absent from the matrix fraction but was found to be associated with the peroxisomal membrane. Furthermore, the amount of Fis1p in the rat peroxisomal fraction was found to be increased approximately twofold when peroxisome proliferation was induced by treatment of the animals with the potent peroxisome proliferator bezafibrate (Figure 2C). After treatment, a typical induction of acyl-CoA oxidase, a key enzyme of peroxisomal β-oxidation was observed, whereas catalase is usually not or only slightly induced (Figure 2C).
Figure 2.
Fis1 is associated with highly purified peroxisomes and is induced during peroxisome proliferation. (A) Highly purified peroxisomes were isolated from rat liver. Distribution of peroxisomal and mitochondrial marker enzyme activities after OptiPrep gradient centrifugation of a crude peroxisomal fraction. Relative concentration = (U ×∑ΔV) (∑U ×ΔV)–1. ΔV represents the volume of a single fraction, U its corresponding enzyme activity, ∑ΔV the total gradient volume, and ∑U the total units recovered from all gradient fractions. CAT, catalase; CytcOx, cytochrome c oxidase. (B) Left, localization of Fis1 in the peroxisomal (Po) and mitochondrial (Mit) fractions. Equal amounts of protein (5 μg/lane) from a single preparation were run on 12.5% acrylamide gels, blotted onto nitrocellulose membranes, and incubated with antibodies to hFis1, PMP70, porin, and ATP synthase α (ATPsyn). In the mitochondrial fraction, an additional band of ∼45 kDa was detected with the hFis1 antibody when higher amounts of protein were applied (20 μg/lane) (immunoblot obtained with another organelle preparation tested negative for mitochondrial contamination). Right, highly purified peroxisomes were separated into a matrix (Ma) and a membrane fraction (Mb). Proteins (PMP70, CAT, 4 μg/lane; hFis1, ATPsyn, porin, 10 μg/lane) were immunoblotted using antibodies to PMP70, catalase (CAT), hFis1, porin, or ATP synthase α. (C) Highly purified peroxisomes from controls (–) (Co) and rats treated with the peroxisome proliferator bezafibrate (+) (Beza) were isolated and immunoblotted using antibodies to peroxisomal acyl-CoA oxidase (AOX), catalase (CAT) or hFis1. Amounts of protein loaded onto the gel: AOX, CAT, 3 μg/lane; hFis1, 6 μg/lane. For hFis1, immunoblots loaded with different amounts of protein (5–35 μg/lane) were quantitated and are expressed as means ± SD. The positions of molecular mass markers (in kilodaltons) are indicated on the right. For acyl-CoA oxidase, only the B subunit (52 kDa) is shown.
Overexpression of hFis1 Promotes Peroxisomal Fission
To investigate an influence of hFis1 on peroxisomal fission and morphology, COS-7 cells were transfected with Myc-hFis1, which in contrast to GFP-hFis1 seemed to be fully functional (Figures 1 and 7; Yoon et al., 2003). Cells expressing Myc-hFis1 exhibited fragmented mitochondria, and Myc-hFis1 was localized to both mitochondria and peroxisomes (Figures 3, A and B, arrows; also see 1, H–J, and 4, E and F). A quantitation of peroxisomal forms revealed that peroxisomes in Myc-hFis1 expressors displayed significant changes in morphology (Figure 3F). About 40% of the cell population expressing Myc-hFis1 exhibited peroxisomes with a segmented, “beads on a string”-like appearance (compared with 6% in controls), which has been shown to be indicative for peroxisomal constriction and fission (Figure 3, C, D, and F) (Schrader et al., 1996; Koch et al., 2004). Myc-hFis1 was uniformly distributed along the membrane of the segmented peroxisomes and was not found to be concentrated at the constriction sites (Figure 3E). Most notably, a drastic, time-dependent increase of very small, punctiform peroxisomes was observed in Myc-hFis1-expressing cells (Figure 3, C, D, and F). Expression of PMP70-Myc, a peroxisomal ATP-binding cassette transporter, or expression of nonfunctional, N-terminally truncated versions of Myc-hFis1 were not observed to alter peroxisomal or mitochondrial morphology (supplemental data; Table 1). These findings indicate that the morphological changes observed are not just due to overproduction of membrane proteins. Interestingly, the morphological changes of peroxisomes observed after expression of hFis1 resemble those induced by expression of Pex11pβ, which has been implicated to function in peroxisomal division (Schrader et al., 1998). In controls, segmented peroxisomes as well as punctiform peroxisomes were less frequent, and large peroxisomal spheres represented the predominant form (Figure 3C). The formation of small, punctiform peroxisomes was also observed when an untagged hFis1 was coexpressed with a fluorescent marker protein (e.g., Mito-GFP) to detect transfected cells (Figure 7, C and D). These observations indicate that an elevated level of Fis1 induces peroxisome abundance by increasing peroxisomal fission.
Table 1.
Overview of cellular distribution and alterations in peroxisomal and mitochondrial morphology of different constructs used in this study
| Construct | Distribution | PO morphology | Mito morphology |
|---|---|---|---|
| hFis1 (UT) | Mito/PO | Segmentation/fission | Fragmentation/aggregation |
| GFP-hFis1 | Mito/PO | Normal/small aggregates | Aggregation |
| Myc-hFis1 | Mito/PO | Segmentation/fission | Fragmentation |
| hFis1-GFP | Diffuse | Normal | Normal |
| hFis1-Myc | Diffuse | Normal | Normal |
| Myc-hFis1-ΔC | Diffuse | Normal | Normal/collapsed tubules |
| Myc-hFis1-ΔTM/C | Diffuse | Normal | Normal/collapsed tubules |
| hFis1-YFP-TM/C | Mito/PO | Normal/small aggregates | Aggregation |
| Myc-hFis1(32-152) | Mito/PO | Normal | Normal/swollen |
| Myc-hFis1(61-152) | Mito/PO | Normal | Normal/swollen |
| Myc-hFis1(92-152) | Mito/PO | Normal | Normal |
Mito, mitochondrial; PO, peroxisomal; UT, untagged.
GFP, YFP, and Myc were used as an epitope tag at either the N or C terminus of hFis1.
Peroxisomal Localization of hFis1 Requires an Intact C Terminus
We now examined the influence of C-terminal modifications of hFis1 on peroxisomal targeting and morphology. When hFis1 tagged with Myc at its C-terminal end (hFis1-Myc) was expressed in COS-7 cells, a diffuse cytosolic distribution was observed (Figure 4A). hFis1-Myc was not found to localize to peroxisomes or to change peroxisomal morphology and to induce peroxisomal fission (Figure 4B). Similar observations were made when hFis1 tagged with GFP at its C terminus (hFis1-GFP) was expressed (Table 1). Next, we expressed an hFis1 construct lacking the five amino acid tail (Myc-hFis1-ΔC). The truncated protein also failed to localize to peroxisomes and showed a diffuse cytosolic distribution (Figure 4C). In addition, peroxisomes seemed normal in these transfected cells (Figure 4D). Similar observations were made when a hFis1 construct lacking the transmembrane domain and the tail (Myc-hFis1-ΔTM/C) was expressed (Table 1). The C-terminally tagged or truncated constructs of hFis1 were also found to disrupt mitochondrial localization and did not result in mitochondrial fragmentation (Table 1) (Yoon et al., 2003). These results suggest that the intact C-terminal structure is required for proper peroxisomal (and mitochondrial) distribution. To further confirm this prediction, the last C-terminal 26 amino acids of hFis1 (hFis1-YFP-TM/C) were expressed in COS-7 cells. Interestingly, hFis1-YFP-TM/C was successfully targeted to peroxisomes and mitochondria (Figure 4, E and F). The localization to both organelles was further confirmed by confocal microscopy (supplemental data). Similar to the expression of GFP-hFis1 (Figure 1C), mitochondria had a banded or striped morphology and became aggregated around the nucleus (Figure 4E, inset). Peroxisomes were also found to form small aggregates in some of the cells but increased fission or segmentation of peroxisomes was not observed. These findings indicate that the C-terminal transmembrane domain and the last five amino acids are both necessary and sufficient to target hFis1 to mitochondria and peroxisomes.
Whereas the C-terminal region of hFis1 is important for its localization, the N-terminal region has been demonstrated to be required for mitochondrial fission (Yoon et al., 2003). To examine the role of the N-terminal region in peroxisomal fission, we expressed three N-terminally truncated hFis1 constructs [Myc-hFis1(32-152), Myc-hFis1(61-152), Myc-hFis1(92-152)] in COS-7 cells. Like the hFis1-YFP-TM/C construct (Figure 4, E and F), all three truncated hFis1 constructs localized to peroxisomes and mitochondria (Table 1 and supplemental data). In contrast to hFis1-YFP-TM/C, aggregation of mitochondria or peroxisomes was not observed with the Myc-tagged constructs. Furthermore, mitochondrial and peroxisomal morphology was normal in the majority of the transfected cells, and increased mitochondrial fragmentation or peroxisomal fission was not observed (supplemental data). These observations indicate that the N terminus of hFis1 is required for peroxisomal fission but dispensable for localization. A summary of the constructs, their localization, and their effects on peroxisomal and mitochondrial morphology is shown in Table 1.
Silencing of Fis1 Induces Elongation of Peroxisomes
To examine the effects of reduced Fis1 mRNA and Fis1 protein levels on peroxisome morphology and division, we conducted RNA interference experiments. Fis1 siRNA duplexes specific for human Fis1 were transfected into COS-7 cells. Immunoblots of cell homogenates demonstrated that the Fis1 protein level was reduced to 20–30% of the control level (Figure 5H). The transfection efficiency is usually ∼70–80%, and the protein level is likely to be even lower in those cells that took up the cognate siRNA (Koch et al., 2004). A significant reduction of Fis1 protein level was not observed in controls transfected with a luciferase siRNA duplex (Figure 5H). In 40–50% of the treated cells, mitochondria were found to collapse around the nucleus and often exhibited an elongated phenotype (Figure 5, A and B). Such morphological changes of mitochondria have been recently described after disruption of Fis1 function (Yoon et al., 2003; Stojanovski et al., 2004). A prominent mitochondrial staining for Fis1 was observed in control cells, whereas in silenced cells only a nonspecific nuclear staining was detected (Figure 5, D and F). The peroxisomes in control cells had an overwhelmingly spherical appearance, whereas peroxisomes in Fis1silenced cells exhibited an elongated, tubular morphology (Figure 5, C, E, and G). Interestingly, the elongated peroxisomes induced by Fis1 silencing also had a segmented/constricted appearance but did not separate into spherical organelles. The peroxisomal phenotype observed after silencing of Fis1 was similar to that found in cells silenced for DLP1, although elongation of peroxisomes was more pronounced after silencing of DLP1 (Koch et al., 2003, 2004). However, the peroxisomal phenotypic changes described above are consistent with a function of Fis1 in peroxisomal fission.
Inhibition of DLP1 Function Interferes with Peroxisomal Fission Induced by hFis1 Overexpression
To provide further evidence for a role of Fis1 in DLP1-mediated peroxisomal fission, we coexpressed Myc-hFis1 with a dominant-negative DLP1 mutant (GFP-DLP1-K38A). COS-7 cells were immunostained 24 h after cotransfection with antibodies to the Myc epitope tag or to PMP70. Of the transfected cells, 86 ± 8% were found to express both transfected constructs under our experimental conditions. GFP-DLP1-K38A assembled into large cytoplasmic aggregates in addition to associating with punctate vesicular structures (Figure 6, B and D) (Pitts et al., 1999; Yoon et al., 2001). In agreement with recent findings (James et al., 2003; Yoon et al., 2003), we found that expression of DLP1-K38A reduced mitochondrial fragmentation promoted by Myc-hFis1 (Figure 6A). Cells with fragmented mitochondria were reduced from ∼85 to 60%, whereas cells containing tubular mitochondria increased accordingly. Remarkably, peroxisomes in the cotransfected cells also exhibited an elongated, tubular morphology, and were not observed to divide into punctiform organelles (Figure 6, C and E), which were detected when Myc-hFis1 was expressed alone (Figures 3F and 6E). Expression of DLP1-K38A in the absence of a proliferative stimulus had only a modest but significant effect on peroxisome morphology (Figure 6E) (Koch et al., 2003). Interestingly, peroxisomes with a tubular morphology were more prominent in Myc-hFis1/DLP1-K38A co-expressors than in cells expressing DLP1-K38A alone, indicating that hFis1 expression exerts a stimulatory effect on peroxisome elongation in addition to fission (Figure 6, C and E). These results show that peroxisomal fission induced by hFis1 expression is suppressed through inhibition of DLP1 function.
Coexpression of hFis1 and Pex11pβ Changes the Uniform Intracellular Distribution of Peroxisomes
Because the expression of hFis1 had similar effects on peroxisome morphology and division to that of Pex11pβ, we tested whether these two proteins act together during the processes of peroxisomal growth and division. When we coexpressed untagged hFis1 or GFP-hFis1 and Pex11pβ-Myc in COS-7 cells, peroxisomes were found to change their normal uniform intracellular distribution and to accumulate in a juxtanuclear region (Figure 7). These peroxisomes were closely associated with aggregated/fragmented mitochondria (Figure 7, E–H), whereas the expression of Pex11pβ-Myc alone had no effect on the distribution of peroxisomes or mitochondria (Figure 7, A and B). Furthermore, a pronounced clustering of peroxisomes or association with mitochondria was not observed in cells expressing untagged hFis1 or by GFP-hFis1 alone (Figures 1, C–G, and 7, C and D). Expression of Pex11pβ-Myc alone is known to induce a pronounced proliferation of peroxisomes through a multistep process involving peroxisome elongation and division into punctiform peroxisomal structures (Schrader et al., 1998; Koch et al., 2003) (Figure 7B). Whereas the peroxisomal aggregates in cells coexpressing untagged hFis1 and Pex11pβ-Myc were composed of spherical and punctiform peroxisomes (Figure 7F), the aggregates in cells coexpressing GFP-hFis1 and Pex11pβ-Myc had a tubulo-reticular appearance (Figure 7H). Colocalization of both proteins on peroxisomal tubulo-reticular aggregates was confirmed by confocal microscopy and by deconvolution microscopy (supplemental data). These observations indicate that peroxisomal fission induced by Pex11pβ is disturbed in GFP-hFis1-expressing cells and support the notion that GFP-hFis1 is not fully functional. Furthermore, elevated levels of both proteins seem to influence peroxisomal distribution.
To test whether Fis1 and Pex11pβ interact with each other, we performed coimmunoprecipitation experiments. However, we failed to detect Fis1 and DLP1 in immunoprecipitates obtained from Pex11pβ-Myc-expressing cells even in the presence of a membrane-permeant cross-linker. In contrast, immune complexes isolated from Myc-hFis1-expressing cells contained DLP1 (supplemental data). In agreement with these findings, an interaction of hFis1 and DLP1 has recently been demonstrated (Yoon et al., 2003), whereas evidence for a physical interaction between DLP1 and Pex11p was not provided (Li and Gould, 2003). Our findings now indicate that Fis1 and Pex11p are presumably not part of the same complex.
DISCUSSION
Fis1 on Peroxisomes and Mitochondria
Studies with yeast and mammalian Fis1p have provided evidence that Fis1p is a transmembrane protein of the outer mitochondrial membrane with its N-terminal region exposed to the cytosol and a short C-terminal tail protruding into the mitochondrial intermembrane space (Mozdy et al., 2000; Yoon et al., 2003). Our morphological and biochemical data demonstrate that Fis1 also localizes to peroxisomes and indicates that peroxisomal Fis1 is a transmembrane protein with the short five-amino acid C-terminal tail protruding into the peroxisomal matrix (Figures 1 and 2). We observed that the addition of a GFP- or Myc-tag at the C terminus, or the truncation of the five-amino acid C-terminal tail of hFis1 interfered with its proper peroxisomal (and mitochondrial) localization (Figure 4). The last C-terminal 26 amino acids of hFis1, however, were efficiently targeted to both peroxisomes and mitochondria. These results suggest that an intact C-terminal structure of hFis1 is required for proper targeting to both mitochondria and peroxisomes. Furthermore, the targeting signal of hFis1 can be restricted to the last C-terminal 26 amino acids (composed of the transmembrane domain and the short five-amino acid tail), which are both necessary and sufficient for sorting hFis1 to peroxisomes and mitochondria. The C-terminal tail contains two lysine residues at positions 149 and 151, and it is likely that the overall basic charge within the tail is crucial for targeting. In support of this, Stojanovski et al. (2004) have demonstrated that mutation of the lysine residues to alanine disturbed mitochondrial targeting. Remarkably, this targeting information can be recognized by both the peroxisomal and the mitochondrial import machinery. Although only few targeting signals of PMPs are known, a hydrophilic peptide containing a group of positively charged amino acids adjacent to at least one hydrophobic patch or transmembrane domain was observed several times (Purdue and Lazarow, 2001; Rottensteiner et al., 2004). Whereas some soluble matrix proteins that are targeted to both peroxisomes and mitochondria are known (Szewczyk et al., 2001; Danpure et al., 2003), Fis1 is one of the few transmembrane proteins described, which localize to both organelles in mammalian cells (Miyazawa et al., 1985). Because we are just beginning to understand the import of membrane proteins into peroxisomes, we can currently not answer to the interesting question how this dual targeting is achieved. We speculate that it is mediated by the different import machineries and not primarily by the information in the hFis1 sequence. This assumption is currently under investigation.
Tagging with either GFP or Myc at the N terminus or truncation of the N-terminal region of hFis1 did not disrupt peroxisomal or mitochondrial localization (Table 1; supplemental data) (Yoon et al., 2003), indicating that the N terminus is not required for proper targeting. The N-terminally tagged proteins influenced peroxisomal morphology in different ways: whereas Myc-hFis1 promoted peroxisomal (and mitochondrial) fission, GFP-hFis1 seemed to be less functional. Mitochondria in some GFP-hFis1-expressing cells had a banded or striped morphology and formed large aggregates, whereas the uniform intracellular distribution of peroxisomes was mostly unaffected. Similar observations were made after expression of a YFP-hFis1-TM/C construct indicating that aggregation might be mediated by an interaction of the GFP/YFP tags on the organelle surface. We observed that besides the addition of a large GFP/YFP-tag at the N terminus, truncations of the N-terminal region of hFis1 interfered with its proper function in organelle fission (Table 1; supplemental data). Recent structural data show that the cytosolic amino acids of hFis1 form six α-helices and build up TPR-like binding motifs that mediate protein–protein interaction and support the idea that hFis1 functions as an adaptor protein (Suzuki et al., 2003; Dohm et al., 2004). Interestingly, microinjection of an antibody to the residues 12–38 disrupted hFis1 function in mitochondrial fission, suggesting a steric hindrance of a possible protein–protein interaction at the core domain containing the TPR motifs (Yoon et al., 2003).
A Role for Fis1 in Peroxisomal Division
In this study, we have shown that overexpression of hFis1 induces not only mitochondrial but also peroxisomal fission, resulting in the formation of small, punctiform peroxisomes and fragmented mitochondria (Figures 1 and 3). Experimental evidence has been presented that yeast and mammalian Fis1p participate in the Dnm1p/DLP1-mediated mitochondrial fission pathway (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; James et al., 2003; Yoon et al., 2003; Stojanovski et al., 2004). Recent data on the solution structure of hFis1 support its function as a molecular adaptor in the process of organelle fission (Suzuki et al., 2003; Dohm et al., 2004). Yeast Fis1p recruits Dnm1p and Mdv1p, a WD-40-repeat-containing peripheral membrane protein, to fission sites on the mitochondrial outer membrane (Mozdy et al., 2000; Tieu et al., 2002), and hFis1 has recently been shown to interact with DLP1 (Yoon et al., 2003). This interaction, which has been confirmed in this study (supplemental data), requires the use of a cross-linker and is suggested to be unstable and transient (Yoon et al., 2003). Interestingly, DLP1 has recently been shown to be a key component in both peroxisomal and mitochondrial fission in mammalian cells (Koch et al., 2003, 2004; Li and Gould, 2003). Another dynamin-related protein, DRP3A, has been identified in higher plants, which is also involved in both peroxisomal and mitochondrial division (Mano et al., 2004), whereas peroxisomal division in S. cerevisiae depends on the dynaminrelated protein Vps1p, and not on Dnm1p (Hoepfner et al., 2001). These findings indicate that in higher eukaryotes, peroxisomes and mitochondria use a common fission machinery. It is therefore likely that Fis1 fulfills similar functions during peroxisomal and mitochondrial fission, e.g., that it facilitates targeting of cytosolic DLP1 to the peroxisomal membrane, or stimulates DLP1 function in the final fission step. In support of this, an increase in the expression of Fis1 induces division of peroxisomes, whereas loss of Fis1 results in highly elongated peroxisomes (Figures 3 and 5). Interestingly, further expression of DLP1, which is abundant in the cytosol, does not induce peroxisomal fission (Koch et al., 2004), indicating that a molecular adaptor/regulator is required. Furthermore, the expression of a DLP1 mutant, which blocks peroxisomal fission, interferes with peroxisomal division promoted by hFis1 overexpression (Figure 6). These observations indicate that Fis1 and DLP1 function together in peroxisomal fission. Interestingly, overexpression of hFis1 (either alone or in combination with DLP1-K38A) (Figures 3 and 6) had also a stimulatory effect on peroxisome elongation in addition to its stimulatory effect on division. Because elongation of spherical peroxisomes seems to be a prerequisite for division, we assume that Fis1 might exert a regulatory function/stimulating activity on “upstream” components of the fission machinery.
It cannot be ruled out that Fis1 cooperates with other components of the fission machinery including those to recruit or influence DLP1. These components might be specific lipid molecules or Mdv1p-like proteins. Our observation that elongation of peroxisomes after silencing of Fis1 is less pronounced than elongation induced by silencing of DLP1 might point in that direction. Another protein that has been implicated to function in peroxisomal division is Pex11p (Erdmann and Blobel, 1995; Marshall et al., 1995; Schrader et al., 1998; Li et al., 2002). It has been proposed recently that Pex11p overexpression, which promotes peroxisome proliferation and division in a multistep process, recruits DLP1 to the peroxisomal membrane (Koch et al., 2003; Li and Gould, 2003). However, a direct interaction between Pex11p and DLP1 was not detected, indicating an indirect mechanism (Li and Gould, 2003). We also failed to detect an interaction between Pex11p and DLP1 using a cross-link approach. Furthermore, Fis1 and Pex11p were not found to be part of a common complex (supplemental data). Remarkably, coexpression of both proteins changed the normal uniform intracellular distribution of peroxisomes (Figure 7). An explanation might be a disturbance of the binding to or movement along microtubules (Schrader et al., 2003), suggesting a possible link between peroxisome formation and the cytoskeleton/motor proteins.
In a previous study, we presented evidence that peroxisomal elongation, constriction, and fission require distinct sets of proteins and that elongation and constriction can occur independently of DLP1 (Koch et al., 2004). Whereas DLP1, which associates in spots along the membrane of elongated peroxisomes, is absolutely required for the final fission step, Pex11p is suggested to function primarily in peroxisome elongation (Koch et al., 2003, 2004). The proteins mediating constriction of the peroxisomal membrane to allow DLP1 function are currently unknown. Like Pex11p, Fis1 does not seem to be required for peroxisomal constriction because constricted peroxisomes were also observed after silencing of Fis1 (Figure 5). It is likely that Fis1 acts downstream of Pex11p and in close association with DLP1 in peroxisome division. The missing link between these proteins might be the yet unidentified components of the peroxisomal constriction machinery. In summary, these results demonstrate that Fis1 and DLP1 are part of a general fission machinery used by mitochondria and peroxisomes. Although both mitochondria and peroxisomes use some organelle-specific components for membrane division and maintenance of morphology, we speculate that they might have other components of the fission machinery in common. Further studies of DLP1-mediated organelle fission should therefore be of considerable value for the understanding of mitochondrial and peroxisomal biogenesis.
Supplementary Material
Acknowledgments
We thank D. I. James, J.-C. Martinou (University of Geneva), D. Stojanovski, M. Ryan (La Trobe University, Melbourne, Australia), R. Lill (University of Marburg), and A. Völkl (University of Heidelberg) for providing antibodies and cDNA constructs; and G. Schneider, W. Ackermann, and V. Kramer (Marburg, Germany) for excellent technical assistance.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0159) on August 17, 2005.
Abbreviations used: GFP, green fluorescent protein; PMP, peroxisomal membrane protein; TPR, tetratricopeptide repeat; YFP, yellow fluorescent protein.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Beier, K., Völkl, A., Hashimoto, T., and Fahimi, H. D. (1988). Selective induction of peroxisomal enzymes by the hypolipidemic drug bezafibrate. Detection of modulations by automatic image analysis in conjunction with immunoelectron microscopy and immunoblotting. Eur J. Cell Biol. 46, 383–393. [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J. M., King, E. J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J. M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danpure, C. J., Lumb, M. J., Birdsey, G. M., and Zhang, X. (2003). Alanine: glyoxylate aminotransferase peroxisome-to-mitochondrion mistargeting in human hereditary kidney stone disease. Biochim. Biophys. Acta 1647, 70–75. [DOI] [PubMed] [Google Scholar]
- Dohm, J. A., Lee, S. J., Hardwick, J. M., Hill, R. B., and Gittis, A. G. (2004). Cytosolic domain of the human mitochondrial fission protein fis1 adopts a TPR fold. Proteins 54, 153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, R., and Blobel, G. (1995). Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes, P., Shepard, K. A., and Yaffe, M. P. (2000). Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H. F., and Hettema, E. H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, D. I., Parone, P. A., Mattenberger, Y., and Martinou, J. C. (2003). hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 278, 36373–36379. [DOI] [PubMed] [Google Scholar]
- Koch, A., Schneider, G., Lüers, G. H., and Schrader, M. (2004). Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117, 3995–4006. [DOI] [PubMed] [Google Scholar]
- Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M. A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605. [DOI] [PubMed] [Google Scholar]
- Labrousse, A. M., Zappaterra, M. D., Rube, D. A., and van der Bliek, A. M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- Lazarow, P. B., and Fujiki, Y. (1985). Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530. [DOI] [PubMed] [Google Scholar]
- Li, X., Baumgart, E., Morrell, J. C., Jimenez-Sanchez, G., Valle, D., and Gould, S. J. (2002). PEX11 beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol. Cell. Biol. 22, 4358–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and Gould, S. J. (2003). The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278, 17012–17020. [DOI] [PubMed] [Google Scholar]
- Lorenz, P., Maier, A. G., Baumgart, E., Erdmann, R., and Clayton, C. (1998). Elongation and clustering of glycosomes in Trypanosoma brucei overexpressing the glycosomal Pex11p. EMBO J. 17, 3542–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano, S., Nakamori, C., Kondo, M., Hayashi, M., and Nishimura, M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38, 487–498. [DOI] [PubMed] [Google Scholar]
- Marshall, P. A., Krimkevich, Y. I., Lark, R. H., Dyer, J. M., Veenhuis, M., and Goodman, J. M. (1995). Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129, 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa, S., Hashimoto, T., and Yokota, S. (1985). Identity of long-chain acyl-coenzyme A synthetase of microsomes, mitochondria, and peroxisomes in rat liver. J. Biochem. 98, 723–733. [DOI] [PubMed] [Google Scholar]
- Mozdy, A. D., McCaffery, J. M., and Shaw, J. M. (2000). Dnm1p GTPasemediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga, D., Keegan, B. R., Brisch, E., Thatcher, J. W., Hermann, G. J., Bleazard, W., and Shaw, J. M. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passreiter, M., Anton, M., Lay, D., Frank, R., Harter, C., Wieland, F. T., Gorgas, K., and Just, W. W. (1998). Peroxisome biogenesis: involvement of ARF and coatomer. J. Cell Biol. 141, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts, K. R., Yoon, Y., Krueger, E. W., and McNiven, M. A. (1999). The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell 10, 4403–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke, G. J., and McMahon, H. T. (2004). The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell. Biol. 5, 133–147. [DOI] [PubMed] [Google Scholar]
- Purdue, P. E., and Lazarow, P. B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17, 701–752. [DOI] [PubMed] [Google Scholar]
- Rottensteiner, H., Kramer, A., Lorenzen, S., Stein, K., Landgraf, C., VolkmerEngert, R., and Erdmann, R. (2004). Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals (mPTS). Mol. Biol. Cell 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, Y., Marshall, P. A., Saiganji, A., Takabe, K., Saiki, H., Kato, N., and Goodman, J. M. (1995). The Candida boidinii peroxisomal membrane protein Pmp30 has a role in peroxisomal proliferation and is functionally homologous to Pmp27 from Saccharomyces cerevisiae. J. Bacteriol. 177, 6773–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, M., Burkhardt, J. K., Baumgart, E., Luers, G., Spring, H., Volkl, A., and Fahimi, H. D. (1996). Interaction of microtubules with peroxisomes. Tubular and spherical peroxisomes in HepG2 cells and their alterations induced by microtubule-active drugs. Eur. J. Cell Biol. 69, 24–35. [PubMed] [Google Scholar]
- Schrader, M., King, S. J., Stroh, T. A., and Schroer, T. A. (2000). Real time imaging reveals a peroxisomal reticulum in living cells. J. Cell Sci. 113, 3663–3671. [DOI] [PubMed] [Google Scholar]
- Schrader, M., Reuber, B. E., Morrell, J. C., Jimenez-Sanchez, G., Obie, C., Stroh, T. A., Valle, D., Schroer, T. A., and Gould, S. J. (1998). Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273, 29607–29614. [DOI] [PubMed] [Google Scholar]
- Schrader, M., Thiemann, M., and Fahimi, H. D. (2003). Peroxisomal motility and interaction with microtubules. Microsc. Res. Tech. 61, 171–178. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R. E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Griparic, L., Shurland, D. L., and van der Bliek, A. M. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski, D., Koutsopoulos, O. S., Okamoto, K., and Ryan, M. T. (2004). Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 117, 1201–1210. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Jeong, S. Y., Karbowski, M., Youle, R. J., and Tjandra, N. (2003). The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J. Mol. Biol. 334, 445–458. [DOI] [PubMed] [Google Scholar]
- Szewczyk, E., Andrianopoulos, A., Davis, M. A., and Hynes, M. J. (2001). A single gene produces mitochondrial, cytoplasmic, and peroxisomal NADPdependent isocitrate dehydrogenase in Aspergillus nidulans. J. Biol. Chem. 276, 37722–37729. [DOI] [PubMed] [Google Scholar]
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak, V., Naylor, K., and Nunnari, J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch, H., Schutgens, R. B., Wanders, R. J., and Tager, J. M. (1992). Biochemistry of peroxisomes. Annu. Rev. Biochem. 61, 157–197. [DOI] [PubMed] [Google Scholar]
- Völkl, A., Baumgart, E., and Fahimi, H. D. (1996). Isolation and characterization of peroxisomes. In: Subcellular Fractionation: A Practical Approach, ed. J. Graham and D. Rickwood, Oxford, United Kingdom: Oxford University Press, 143–167.
- Yoon, Y., Krueger, E. W., Oswald, B. J., and McNiven, M. A. (2003). The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 23, 5409–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, Y., Pitts, K. R., Dahan, S., and McNiven, M. A. (1998). A novel dynaminlike protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J. Cell Biol. 140, 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, Y., Pitts, K. R., and McNiven, M. A. (2001). Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12, 2894–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.