Abstract
Although the Arp2/3 complex localizes to the leading edge of motile cells, endocytic structures, and mitochondria in budding yeast, the mechanism for targeting the Arp2/3 complex to different regions in the cell is not well understood. We find that Jsn1p, a member of the PUF family of proteins, facilitates association of Arp2/3 complex to yeast mitochondria. Jsn1p localizes to punctate structures that align along mitochondria, cofractionates with a mitochondrial marker protein during subcellular fractionation, and is both protease sensitive and carbonate extractable in isolated mitochondria. Thus, Jsn1p is a peripheral membrane protein that is associated with the outer leaflet of the mitochondrial outer membrane. Jsn1p colocalized and coimmunoprecipitated with mitochondria-associated Arc18p-GFP, and purified Arp2/3 complex bound to isolated TAP-tagged Jsn1p. Moreover, deletion of JSN1 reduces the amount of Arc18p-GFP that colocalizes and is recovered with mitochondria twofold, and jsn1Δ cells exhibited defects in mitochondrial morphology and motility similar to those observed in Arp2/3 complex mutants. Thus, Jsn1p has physical interactions with mitochondria-associated Arp2/3 complex and contributes to physical and functional association of the Arp2/3 complex with mitochondria.
INTRODUCTION
The Arp2/3 complex stimulates actin nucleation and force generation on bacterial pathogens in infected host cells and at endogenous cellular membranes, including the leading edge of motile cells, endosomes in all cell types studied, and mitochondria in budding yeast (Merrifield et al., 1999; Cossart, 2000; Taunton et al., 2000; Boldogh et al., 2001; Insall et al., 2001; Zhang et al., 2002; Chang et al., 2003; Pollard and Borisy, 2003; Southwick et al., 2003). Targeting of the Arp2/3 complex to endogenous membranes requires protein kinase C and Cdc42 in Xenopus extracts, and phosphatidylinositol-4-phosphate 5-kinase, the SH3-SH2 adapter protein Nck, and the WASp interacting protein WIP in cultured fibroblasts (Rozelle et al., 2000; Taunton et al., 2000; Benesch et al., 2002). However, membrane-specific proteins that mediate binding of the Arp2/3 complex or its activators to intracellular membranes remain to be determined. Here, we provide evidence that Jsn1p, a PUF family protein, is a peripheral mitochondrial outer membrane protein that contributes to recruitment of Arp2/3 complex to mitochondria in budding yeast.
In budding yeast, the Arp2/3 complex and its activators are present in actin patches, endosomes that are coated with F-actin and form at sites of polarized cell surface growth (Moreau et al., 1996; Winter et al., 1997; Chang et al., 2003; Kaksonen et al., 2003; Huckaba et al., 2004). Arp2/3 complex is not required for linear, long-distance endosome movement (Huckaba et al., 2004). However, it may contribute to membrane invagination, membrane internalization, and/or short-distance endosome movement at the cell cortex. Arp2/3 complex also contributes to later steps in endocytosis. Arp2/3 complex and F-actin localizes to the sites of vacuole fusion. Moreover, actin remodeling has been implicated in vacuole docking before fusion, late stages of vacuole fusion, and recruitment of 3-phosphoinostides to sites of vacuole fusion (Eitzen et al., 2002; Fratti et al., 2004).
In addition to its role in the endomembrane system, Arp2/3 complex is required for mitochondrial movement and inheritance in budding yeast (Boldogh et al., 2001). During S, G2, and M phases of the yeast cell cycle, mitochondria display a pattern of movement that resembles chromosome movement during cell division. That is, they undergo poleward movement and retention at the poles, which results in equal distribution between mother and daughter cells. During poleward movement, mitochondria undergo linear movement in the anterograde direction (toward the bud tip) or in the retrograde direction (toward the distal tip of the mother cell). After mitochondria have reached the bud tip or mother cell tip, they are immobilized at these cellular poles. Finally, at the end of the cell division cycle, mitochondria are released from the retention zones, and are redistributed in the dividing cells (Simon et al., 1997; Yang et al., 1999; Boldogh et al., 2004; Fehrenbacher et al., 2004).
Actin cables, bundles of actin that assemble in the bud tip and bud neck and extend along the mother-bud axis, are required for poleward mitochondrial movement. Retrograde mitochondrial movement is driven by retrograde actin flow, assembly-driven movement of actin cables from the mother cell into the bud (Yang and Pon, 2002). Binding of mitochondria to actin cables undergoing retrograde flow is both necessary and sufficient to drive retrograde movement of the organelle toward the mother cell tip (Fehrenbacher et al., 2004). In contrast, anterograde movement requires actin cables and Arp2/3 complex-driven actin polymerization (Boldogh et al., 2001, 2005; Fehrenbacher et al., 2004). That is, Arp2/3 complex subunits and activity are detected in isolated mitochondria and in mitochondria in intact yeast cells. Moreover, mutations that dampen actin dynamics or compromise Arp2/3 complex inhibit anterograde mitochondrial movements but have no effect on retrograde movement of the organelle.
Here, we provide evidence for a role of Jsn1p in recruiting the Arp2/3 complex to mitochondria. Jsn1p is a member of the PUF family of proteins, which show sequence similarity to the abdominal pattern formation protein Pumilio of Drosophila (Barker et al., 1992; MacDonald, 1992; Murata and Wharton, 1995). PUF proteins contain the Pumilio homology domain, which consists of up to eight tandem repeats that can confer RNA binding (Zamore et al., 1997; Wang et al., 2002). The six PUF proteins of budding yeast are encoded by PUF1/JSN1, PUF2, PUF3, PUF4, PUF5/MPT5/UTH4, and PUF6. Although all yeast PUF proteins copurify with ribonucleoprotein complexes (Gerber et al., 2004), direct RNA binding activity has only been demonstrated for Puf3p, Puf4p, and Puf5p. Puf3p shows transcript-specific binding and promotes decay of bound mRNA by enhancing mRNA deadenylation (Olivas and Parker, 2000). In contrast, Puf5p and Puf6p bind to the 3′ untranslated region of mRNAs and mediate translational repression (Tadauchi et al., 2001; Gu et al., 2004). Thus, although these PUF proteins can bind to RNA, PUF protein activity subsequent to RNA binding is variable.
Jsn1p was initially identified in yeast because JSN1 overexpression suppresses the temperature sensitivity of a microtubule-stabilizing tubulin mutation (Machin et al., 1995). A link between Jsn1p and mitochondrial motility in budding yeast emerged from genome-wide two-hybrid screens (Uetz et al., 2000; Ito et al., 2001). These studies revealed that Jsn1p can bind to Arc18p, a subunit of the Arp2/3 complex, and to integral mitochondrial outer membrane proteins, including Fis1p, a protein that recruits the yeast dynamin-like fission mediator Dnm1p to mitochondrial membranes and Tom20p/Mas20p, a mitochondrial protein import receptor (Sollner et al., 1989; Mozdy et al., 2000). We find that Jsn1p is associated with mitochondria and mitochondria-associated Arp2/3 complex. In addition, we show that Jsn1p contributes to recruiting the Arp2/3 complex to yeast mitochondria. These studies provide additional evidence for a role for the Arp2/3 complex in control of actin-based mitochondrial movement in budding yeast and offer insight into the mechanism for intracellular targeting of the Arp2/3 complex.
MATERIALS AND METHODS
Yeast Media, Plasmids, and Strains
Table 1 lists yeast strains used for this study. Yeast cell growth and manipulations were carried out according to Sherman (2002).
Table 1.
Yeast strains used in this study
| Strain name | Genotype |
|---|---|
| 2GS4-B-4 | (BY4741) MATahis3Δ1 leu2Δ0 ura3Δ0 met15Δ0 JSN1-TAP:HIS3JSN1-TAP:HIS3 |
| BY4741 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 |
| BY4743 | MATa/α his3Δ1/his3D1 leu2Δ0/leu2D0 ura3Δ0/ura3D0 met15Δ0/MET15 lys2Δ0/LYS2 |
| KFY103 | MATaade2 ura3-1 leu2-3, 112 TUB1-GFP:LEU2 |
| KFY119 | (BY4743) arc18:GFP-kanMX6/ARC18 |
| KFY118 | (BY4743) jsn1Δ::kanMX4/jsn1Δ::kanMX4, arc18: GFP-HIS3MX6/ARC18 |
| KFY200 | (BY4743) jsn1Δ::kanMX4/jsn1Δ::kanMX4 |
| KFY206 | (BY4743) [pCIT1-GFP:URA3] |
| KFY202 | (BY4743) jsn1Δ::kanMX4/jsn1Δ::kanMX4 [pCIT1-GFP:URA3] |
| KFY208 | MATatrp1-1 his3D200 lys2-801 ura3-52 leu2-3 arc15Δ::TRP1 [pCIT1-GFP:URA3] |
| KFY209 | MATatrp1-1 his3D200 leu2-3 arp2Δ::TRP1 [pCIT1-GFP:URA3] |
| KFY212 | MATatrp1-1 his3D200 lys2-801 ura3-52 leu2-3 [pCIT1-GFP:URA3] |
| KFY214 | MATa/α his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2 arc18::kanMX6 [pCIT1-GFP:URA3] |
| KFY218 | MATaarp2-H330L ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 [pCIT1-GFP:URA] |
| KFY219 | MATaade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 [pCIT1-GFP:URA] |
| KFY301 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 jsn1Δ::kanMX4 TUB1-GFP:LEU2 |
| KFY404 | (BY4743) jsn1Δ::kanMX4/jsn1Δ::kanMX4, arc18: GFP-HIS3MX6/ARC18 [pF0ATPase-subunit 9-RFP:URA3] |
| KFY407 | (BY4743) arc18:GFP-kanMX6/ARC18 [pF0ATPase-subunit 9-RFP:URA3] |
Most strains used in this study were from the yeast deletion set (Open Biosystems, Huntsville, AL), except KFY208, KFY209, KFY212, and KFY214, which derive from RLY570, RLY574, RLY514 (Winter et al., 1999), and ABY1803 (Evangelista et al., 2002), respectively. KFY219 and KFY218 derive from YPH499 and YMW82, respectively (Moreau et al., 1996).
Tagging of ARC18
The COOH terminus of Arc18p was tagged with green fluorescent protein (GFP) using PCR-based insertion into the chromosomal copy of the ARC18 gene (Longtine et al., 1998; Nowakowski et al., 2001). The primers used for tagging were forward, 5′-GTTGGCGTTCACAAGAAGAAGATTCATGAACAAATCTCTACGGATCCCCGGGTTAATTAA-3′ and reverse, 5′-TAAATTCTAGTTTTTATTATTTTCCTTGCACCGTATACCTGAATTCGAGCTCGTTTAAAC-3′. Underlined sequences correspond to the tagging plasmid sequence. Yeast cells were transformed with PCR products using the lithium acetate method (Gietz et al., 1995). Transformants, positive for integration at the target locus, were validated by PCR. Expression and localization of GFP-tagged proteins were analyzed using Western blots and fluorescence microscopy. Arc18p-GFP localized to cortical patch-like structures that stained with Arp3p antibodies and displayed movement similar to those reported for actin patches. Finally, expression of Arc18p-GFP had no effect on cell growth, actin organization or function, or mitochondrial structure, respiration, or motility.
Visualization of Mitochondria
Mitochondria were visualized using a fusion protein consisting of the mitochondrial signal sequence of citrate synthase 1 fused to GFP (pCIT1-GFP; Okamoto et al., 2001). Images were collected with a Nikon E600 microscope (Nikon USA, Melville, NY) using a Plan-Apochromat 100×, 1.4 numerical aperture objective lens, and a cooled charge-coupled device camera (Orca-ER; Hamamatsu, Bridgewater, NJ). Illumination with a 100-W mercury arc lamp was controlled with a MAC5000 shutter controller and Ludl filter wheel (Ludl Electronic Products, Hawthorne, NY). Ludl filter wheels or a Dual View image splitter (Optical Insights, Santa Fe, NM) were used for two-color imaging. Hardware control and image enhancement were performed using Openlab software (Improvision, Coventry, United Kingdom).
For three-dimensional (3-D) imaging, 25 z-sections were obtained at 0.2-μm intervals through the entire cell using a piezo-electric focus motor mounted on the objective lens of the microscope (Polytech PI, Auburn, MA). Out-of-focus light was removed by iterative deconvolution of each image section, and each series of deconvolved images was projected and rendered using Volocity software (Improvision).
Quantitation of Mitochondrial Movement In Vivo
Mitochondria were defined as motile if they displayed linear movement for three consecutive still frames. In all cases, the only portion of the organelle that was evaluated for movement was the tip of the organelle in the mother cell in a single focal plane. Polarized movement was defined as that which achieves a net displacement toward or away from the bud and is expressed as the percentage of all observable mitochondrial tips over the time-lapse course (60 s).
Immunoprecipitation
Mitochondria were isolated according to Lazzarino et al. (1994). Further purification of the crude mitochondrial fraction was carried out by isopycnic centrifugation using Nycodenz gradients [5-(N-2,3-dihydroxypropylacetamido-2,4,6-triiodo-N,N′-bis (2,3-dihydroxy-propyl) isophthalamide; Sigma-Aldrich, St. Louis, MO) (Glick and Pon, 1995). Isolated mitochondria were solubilized to 1 mg/ml with digitonin (0.5%) and HEPES, pH 7.4, according to Kerscher et al. (1997). Immunoprecipitated proteins were analyzed by Western blots using antibodies raised against Jsn1p, GFP, and the mitochondrial marker protein porin (gifts from G. Barnes [University of California at Berkeley, Berkeley, CA], P. Silver [Harvard Medical School, Boston, MA], and G. Schatz [Swiss Science and Technology Council, Bern, Switzerland]).
Conjugation of Jsn1p-TAP with IgG Sepharose Beads
The TAP-tagged Jsn1p strain (2GS4-B-4) was obtained from Open Biosystems and consists of a calmodulin binding peptide, TEV cleavage site, two IgG binding domains of Staphylococcus aureus protein A, and a selectable HIS1 marker integrated into the C terminus of Jsn1p. Correct genomic integration of each tag was verified by PCR and by immunoblot analysis of cell extracts (our unpublished data).
Wild-type (BY4743) and Jsn1p-TAP tagged strains (2GS4-B-4) were grown to mid-log phase in lactate liquid media at 30°C in a shaking incubator. Cells were collected by centrifugation (2000 × g). Ten milliliters of wet cells were resuspended in 25 ml of buffer containing 50 mM HEPES, pH 7.5, 100 mM KCl, 30 mM MgCl2 10 mM EGTA, 0.2 mM ATP, 1 mM dithiothreitol, and protease inhibitor cocktail (Lazzarino et al., 1994), and lysed with bead beating (6 × 30-s pulse 30-s rest) on ice. The lysates were centrifuged at 100,000 × g. The supernatants were filtered through a 0.2-μm filter and incubated with 600 μl of IgG-conjugated Sepharose beads (GE Healthcare, Uppsala, Sweden) for overnight at 4°C. IgG-conjugated Sepharose beads were prepared according to the manufacturer's instructions and were washed in 5 volumes of lysing buffer before incubation with the supernatants. After incubation, the beads were collected on a small column and washed with 10 volumes of lysing buffer.
Jsn1p-TAP Binding Assay with Purified Arp2/3 Complex
Ten microliters of IgG-Sepharose beads, incubated with yeast lysate from untagged cells or cells expressing TAP tagged-Jsn1p, were washed two times with 5 volumes of binding buffer (50 mM HEPES, pH 7.4, 100 mM NaCl containing protease inhibitor cocktail) and incubated with purified Arp2/3 complex (0.2 mg/ml; 2.5 volume) (gift from B. Goode, Brandeis University, Waltham, MA; Goode et al., 2001) for 2 h at 4°C. Beads (containing bound material) were centrifuged (1000 × g) and washed twice with 4 volumes of binding buffer to remove unbound Arp2/3 complex. Bead pellets washed four times with 5 volumes of binding buffer with no protease inhibitors were incubated with 20 U of TEV protease (Invitrogen, Carlsbad, CA) in 100-μl volume for 1.5 h at 23°C. Supernatant consisting of material released by TEV protease treatment was collected, as described above. Equal volumes of samples were analyzed by SDS-PAGE and Western blot using a polyclonal rabbit IgG against both Arp2p (Boldogh et al., 2001) and a region of the TAP tag (amino acid sequence: SSGALDYDIPTTASENLYFQ) that is N-terminal to the TEV protease cleavage site (Open Biosystems).
Carbonate Extraction and Protease Treatment
For protease protection assays, Nycodenz-purified mitochondria were washed and resuspended to a final concentration of 0.5 mg/ml in protease inhibitor-free breaking buffer, and mitochondria were incubated with trypsin and chymotrypsin (150 μg/ml final concentration) for 30 min at 23°C. Protease treatment was stopped by addition of the protease inhibitor mixture described above and soybean trypsin inhibitor (0.15 mg/ml final concentration). For protease-free controls, protease inhibitors were added alone. The reaction mixtures were subjected to centrifugation at 12,500 × g for 5 min at 4°C, and mitochondrial pellets were analyzed by PAGE and Western blots.
For carbonate extraction and salt treatment, mitochondria were resuspended to a final concentration of 0.25 mg/ml in breaking buffer containing protease inhibitors. Na2CO3 or KCl were added to final concentrations of 0.1 and 1 M, respectively, and samples were incubated at 4°C for 30 min. Reaction mixtures were then subjected to centrifugation at 356,000 × g for 20 min at 4°C. Trichloroacetic acid (TCA) was added to a final concentration of 20% to the supernatants. After incubation at 4°C for 15 min, TCA-precipitated material was collected by centrifugation at 12,500 × g for 15 min at 4°C. Proteins in the carbonate- or salt-extracted pellets and in the TCA-precipitated supernatants were analyzed by PAGE and Western blots.
Other Methods
Protein concentrations were determined using the bicinchoninic acid assay (Pierce Chemical, Rockford, IL). Gel electrophoresis and Western blot analysis were performed as described previously (Boldogh et al., 1998). Immunofluorescence and visualization of the actin cytoskeleton with Alexa-594 phalloidin were carried out essentially as described previously (Daum et al., 1982; Burke et al., 2000; Fehrenbacher et al., 2004). Subcellular fractionation was carried out according to Daum et al. (1982). S1 and P1, supernatant and pellet recovered after centrifugation of homogenized spheroplasts at 2000 × g; P2, crude mitochondria recovered after centrifugation of S1 at 12,500 × g; NM, Nycodenz-purified P2; P3 and S3, pellet and supernatant recovered after centrifugation of postmitochondrial supernatant at 125,000 × g.
RESULTS
Jsn1p Is a Peripheral Mitochondrial Outer Membrane Protein
The findings that Jsn1p can bind to an Arp2/3 complex subunit and to integral mitochondrial outer membrane proteins support a role for Jsn1p in recruiting Arp2/3 complex to mitochondria. If this is true, then Jsn1p should localize to mitochondria. To localize Jsn1p, we used an affinity-purified Jsn1p antibody generously provided by Dr. G. Barnes. This antibody recognized a doublet band using Western blot analysis that was enriched in mitochondria isolated from wild-type cells, absent in mitochondria isolated from jsn1Δ mutants, and had electrophoretic mobilities consistent with predicted molecular weight for Jsn1p (Figure 1A).
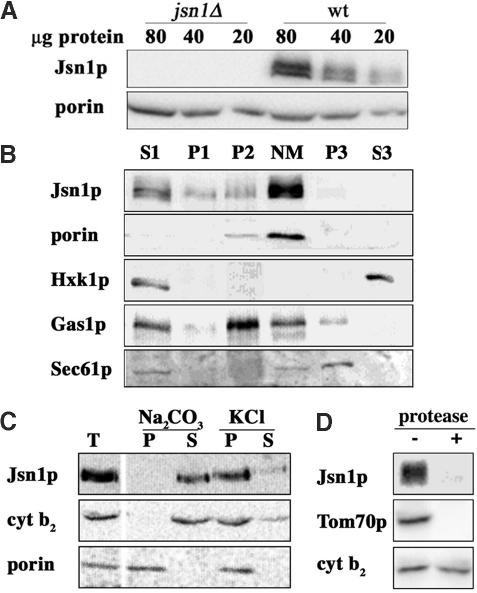
Figure 1.
Jsn1p is a peripheral mitochondrial outer membrane protein. (A) Mitochondria were isolated from a jsn1Δ mutant (KFY200) and the corresponding wild-type cells (BY4743) by differential and Nycodenz gradient centrifugation. Recovery of Jsn1p and porin with isolated yeast mitochondria was analyzed using Western blots. (B) Wild-type cells (BY4743) were grown to mid-log phase in lactate medium and separated into subcellular fractions as described in Materials and Methods. Equal amounts of protein from S1, P1, crude mitochondrial pellet (P2), Nycodenz-purified mitochondria (NM), P3, and S3 were analyzed using Western blots and antibodies that recognize the mitochondrial marker porin, the cytosolic marker hexokinase (Hxk1p), the plasma membrane marker Gas1p, and the endoplasmic reticulum marker Sec61p. (C) Nycodenz-purified mitochondria prepared from a wild-type strain (BY4743) treated with sodium carbonate or KCl as described in Materials and Methods. Carbonate or salt-extractable and -inextractable material was analyzed using Western blots and antibodies raised against Jsn1p, porin (an integral mitochondrial membrane protein), and cytochrome b2 (a peripheral mitochondrial inner membrane protein). T, total mitochondrial extract; P, pellet; and S, supernatant. (D) Nycodenz-purified mitochondria were washed with protease inhibitor-free breaking buffer and incubated with a protease inhibitor cocktail or trypsin and chymotrypsin (150 μg/ml) for 30 min at 23°C. After addition of protease inhibitors to the proteasetreated sample, mitochondria were separated from the reaction mixture. Proteins in the mitochondrial pellets were analyzed using Western blots and antibodies raised against Jsn1p, Tom70 (a mitochondrial outer membrane protein), and cytochrome b2(a peripheral membrane protein on the outer leaflet of the inner mitochondrial membrane).
Jsn1p cofractionated with porin, a mitochondrial marker protein, upon fractionation by differential and Nycodenz gradient centrifugation (Figure 1B). Jsn1p was enriched in crude (P2) and Nycodenz-purified mitochondria (NM), and depleted in the postmitochondrial pellet (P3) and cytosolic fractions (C) to the same extent as porin. In addition, Jsn1p could be extracted from mitochondrial membranes with sodium carbonate but remained associated with mitochondria after treatment with high salt (Figure 1C). Finally, Jsn1p was degraded upon treatment of Nycodenz-purified mitochondria with trypsin and chymotrypsin (Figure 1D). This protease treatment degraded the mitochondrial outer membrane protein Tom70p without affecting the recovery of the inner membrane protein cytochrome b2 with mitochondria. Therefore, it resulted in degradation of mitochondrial surface proteins without affecting the integrity of the organelle. Together, these results indicate that Jsn1p is a peripheral mitochondrial membrane protein that is associated with the cytosolic face of the mitochondrial outer membrane.
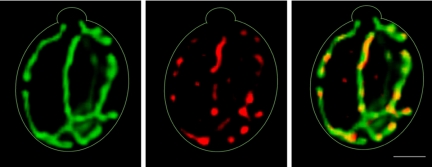
Previous immunofluorescence studies showed that Jsn1p localized to punctate, cortical structures when overexpressed in budding yeast (Machin et al., 1995). Here, we studied the localization of endogenous Jsn1p with respect to mitochondria. Consistent with previous work, we observed that Jsn1p-containing particles localize to foci near the cortex of the cell. With the improved spatial resolution of deconvolution microscopy and 3-D reconstruction, and visualization of Jsn1p and mitochondria in the same cells, we found that Jsn1p-containing particles colocalized with mitochondria in cells bearing small buds (Figure 2). In typical cells, >85% of the Jsn1-containing particles align along mitochondria. Jsn1p particles that did not colocalize with mitochondria were weakly fluorescent, and likely due to background staining by the polyclonal antibody. Although single projections of the 3-D volumes are shown, colocalization of Jsn1p with mitochondria was confirmed by examining the projections at multiple angles.
Figure 2.
Jsn1 localizes to mitochondria in intact yeast. Jsn1p localizes to punctate structures that colocalize with mitochondria. Yeast (KFY206) expressing a fusion protein consisting of the mitochondrial signal sequence of citrate synthase fused to GFP (pCIT1-GFP) were grown to mid-log phase, fixed, and converted to spheroplasts. Jsn1p was visualized by indirect immunofluorescence using an affinity-purified polyclonal anti-Jsn1p antibody. Mitochondria were visualized using fluorescence imaging of pCIT1-GFP. Z-sections through the cell were collected, deconvolved, and reconstructed into a 3-D volume. The images shown are two-dimensional projections of reconstructed 3-D volumes. Mitochondria were resolved as long, tubular structures that align along the mother-bud axis (green). Jsn1p was detected in punctate cytoplasmic structures (red). The overlay of Jsn1p (red) with mitochondria (green) reveals colocalization of Jsn1p-containing particles with mitochondria (right). Gray outlines show cell boundaries detected by phase contrast imaging. Bar, 1 μm.
Jsn1p Colocalizes with Mitochondria-associated Arc18p and Contributes to Association of Arc18p to Mitochondria
Colocalization and interactions of Jsn1p and Arp2/3 complex were tested using Arc18p, a subunit of the Arp2/3 complex, that was tagged at its chromosomal locus with GFP. The GFP tag had no obvious effect on yeast cell growth or on the organization or function of the actin cytoskeleton (our unpublished data). In addition, Arc18p-GFP localization was similar to that reported for other tagged or untagged Arp2/3 complex subunits (Moreau et al., 1996; Winter et al., 1997). That is, it localized to cortical patch-like structures (Figure 3) that stained with Arp3p antibodies and therefore consist of fully assembled Arp2/3 complex (our unpublished data). In the bud, Arc18p-GFP patches contained Abp1p, an actin patch marker, and displayed patterns and rates of movement similar to those reported for actin patches (our unpublished data). In the mother cell, we detect an average of 10 and 12 (n = 35 and 54; SEM = 0.45 and 0.63) Arc18p-GFP patches in fixed and live cells, respectively. Eighty-one percent of the Arc18p-GFP patches in the mother cell colocalized with mitochondria (Figure 3B) and do not contain Abp1p (our unpublished data).
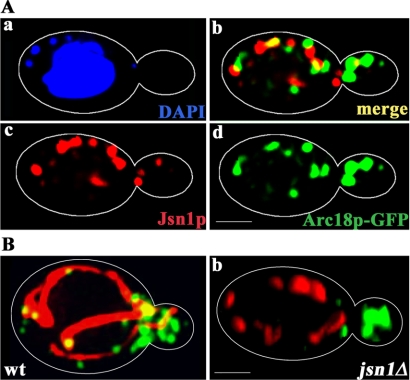
Figure 3.
Jsn1p colocalizes with mitochondria-associated Arc18p-GFP and is required for localization of Arc18p-GFP to mitochondria. (A) Yeast expressing ARC18 tagged at its chromosomal locus with GFP (KFY119) were grown to mid-log phase, fixed, and stained for Jsn1p as for Figure 1, and for DNA using DAPI. Z-sections through the cell were collected, deconvolved, and reconstructed into a 3-D volume. The images shown are three projections. (a) DAPI-stained material. Nuclear DNA was resolved as a single large spherical structure in the mother cell. mtDNA was resolved as punctate cytoplasmic material. b, c, and d show the localization of Arc18p-GFP (green) and Jsn1p (red) either alone (c and d) or merged (b). Arc18p-GFP was resolved as punctate structures that were enriched in the bud. Jsn1p was resolved as punctate structures that were enriched in the mother cell. Visualization of 3-D reconstructions at different rotation planes revealed that Arc18p-GFP in mother cells were associated with Jsn1p. In rotation angle shown, Jsn1p structures colocalized or overlapped with Arc18p-GFP-labeled structures. White outlines show cell boundaries detected by phase contrast imaging. Bar, 1 μm. (B) Wild-type yeast cells (KFY401, a), jsn1Δ cells (KFY404, b) expressing Arc18p-GFP and a fusion protein consisting of the mitochondrial signal sequence of subunit 9 of F1 F0-ATPase fused to RFP were grown to mid-log phase and visualized by epifluorescence microscopy. Arc18p-GFP is shown in green and mitochondria labeled with the targeted RFP are shown in red. Z-sections through the cell were collected, deconvolved, and reconstructed into a 3-D volume. The images shown are two-dimensional projections of reconstructed 3-D volumes. Colocalization was defined as instances where two fluorescent structures either completely or partially overlapped at multiple viewing angles in rendered 3-D volumes. Deletion of JSN1 resulted in fragmentation of mitochondria and a decrease in the amount of Arc18p-GFP that colocalized with mitochondria. White outlines show cell boundaries detected by phase contrast imaging. Bar, 1 μm.
Because actin patches contain the Arp2/3 complex and have recently been identified as endosomes that can associate with elongating actin cables (Huckaba et al., 2004), it is possible that the localization of the Arp2/3 complex to mitochondria is a consequence of actin cable association with mitochondria. However, we find that destabilization of actin cables, by short-term incubation of the bni1-11 bnr1Δ formin mutant at restrictive temperature (Evangelista et al., 2002), had no significant effect on the amount of Arc18p-GFP that associates with mitochondria (our unpublished data).
In contrast, we found that Jsn1p colocalizes with Arc18p-GFP and contributes to recruitment of Arc18p-GFP to the organelle. For localization studies, yeast expressing Arc18p-GFP were fixed and stained for mitochondrial DNA (mtDNA) using the DNA binding dye 4′,6-diamidino-2-phenylindole (DAPI) and for Jsn1p by indirect immunofluorescence. In wild-type cells, mtDNA localizes to nucleoids, punctate DNA-containing structures that are present in multiple copies per organelle. Visualization of 3-D reconstructions of yeast labeled for mitochondria, Jsn1p, and Arp2/3 complex at different rotation planes revealed that all of the Arc18p-GFP structures that localized to mitochondria in mother cells were associated with Jsn1p-containing structures (Figure 3A).
To determine whether Jsn1p contributes to targeting of the Arp2/3 complex to mitochondria, we studied the localization of Arc18p-GFP and mitochondria in a JSN1 deletion mutant and the corresponding wild-type cell. We found that deletion of JSN1 resulted in a 50% decrease in the amount of Arc18p-GFP that colocalized with mitochondria in the mother cell (Figure 3B; p < 0.001). In complementary studies, we studied the effect of deletion of JSN1 on recovery of Arp2/3 complex from isolated yeast mitochondria (Figure 4). Deletion of JSN1 had no obvious effect on 1) the overall cellular levels of Arp2p, Arc18p, or the mitochondrial marker protein, cytochrome b2; 2) the recovery of cytochrome b2 with mitochondria; or 3) the level of plasma membrane contaminants in the mitochondrial fraction. In contrast, the amount of Arp2p and Arc18p-GFP recovered with Nycodenz-purified mitochondria was reduced by 50% in jsn1Δ mutants compared with wild type (p < 0.05; Figure 4, A and B). Thus, Jsn1p contributes to recruitment of Arp2/3 complex to the organelle.
Figure 4.
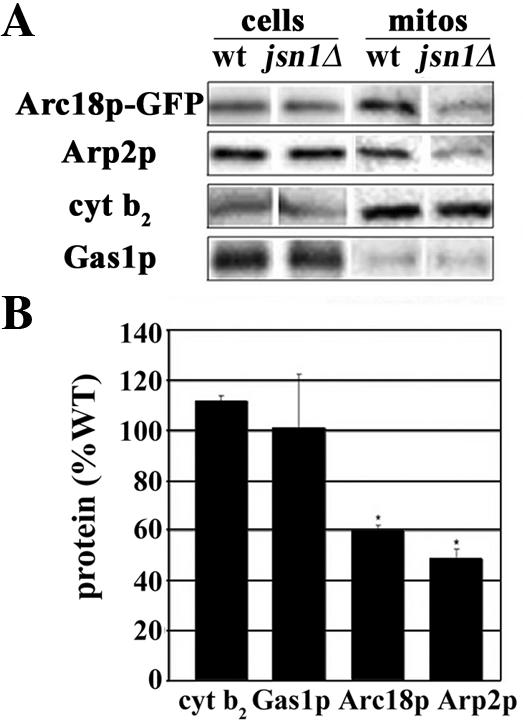
Deletion of JSN1 results in a reduction in the amount of Arp2/3 complex subunits that are recovered with mitochondria after subcellular fractionation. (A) Wild-type yeast (KFY119) and jsn1Δ cells (KFY118) that express ARC18 tagged at its chromosomal locus with GFP were grown to mid-log phase. Whole cell extracts (20 μg) and isolated mitochondria (20 μg) from these yeast cells were analyzed using Western blots and antibodies raised against GFP, Arp2p, cytochrome b2, and Gas1p. (B) Quantitation of the amount of Arc18p-GFP, Arp2p, cytochrome b2, and Gas1p revealed equal recovery of markers for mitochondria and plasma membrane in mitochondria isolated from wild-type and jsn1Δ cells, and a 50% decrease in the recovery of both Arc18p-GFP and Arp2p in the jsn1Δ mitochondria compared with wild-type mitochondria.
Deletion of JSN1 Produces Mitochondrial Morphology and Motility Defects Similar to Those Observed in Arp2/3 Complex Mutants
In wild-type cells, mitochondria were resolved as long, tubular structures that align along the mother-bud axis and accumulate in the bud tip and in the tip of the mother cell distal to the bud. In contrast, jsn1Δ cells displayed defects in mitochondrial morphology (Figure 5A). To quantitate mitochondrial morphology defects in jsn1Δ cells, we scored the number of cells that displayed defects in continuity (fragmentation) or distribution (aggregation). Cells that showed a defect in either category were scored as abnormal. Although only 17% of wild type cells (n = 100) exhibited abnormal mitochondrial morphology, 83% of jsn1Δ mutant cells displayed defects in mitochondrial morphology. The defects in mitochondrial morphology observed in the jsn1Δ mutant were similar to those observed in cells carrying a temperature-sensitive mutation in ARP2 (arp2-1) or an insertion mutation in ARC15 (Boldogh et al., 2001).
Figure 5.
Deletion of JSN1 results in defects in mitochondrial morphology but has no effect on cytoskeletal organization or association of mitochondria with actin cables. (A) Mitochondria in midlog phase wild-type yeast (KFY206, a and c) and jsn1Δ cells (KFY202, b and d) were visualized using pCIT1-GFP and fluorescence microscopy. Images shown are two-dimensional projections of 3-D reconstructions obtained as for Figure 2. Gray outlines show cell boundaries detected by phase contrast imaging. Bar, 1 μm. (B) Microtubules were visualized in wild-type cells (KFY103, a) and jsn1Δ cells (KFY301, b) using a tubulin-GFP fusion protein. Bar, 1 μm. (C) Wild-type yeast (KFY206) and jsn1Δ cells (KFY202) expressing pCIT1-GFP were grown to mid-log phase, fixed, and stained for actin using Alexa-phalloidin. Mitochondria (a and d), actin (b and e), and an overlay showing mitochondria (green) and actin (red) (c and f) for wild-type cells (a–c) and jsn1Δ cells (d–f) are shown. Bar, 1 μm.
Previous studies revealed that actin organization and association of mitochondria with actin cables were not affected by mutations in ARP2 or ARC15 (Boldogh et al., 2001). However, because overexpression of the JSN1 gene suppressed the temperature sensitivity of a microtubule-stabilizing tubulin mutation (Machin et al., 1995), it is possible that the defects in mitochondrial morphology and motility observed in the jsn1Δ cells were due to defects in cytoskeletal organization. To address this issue, we examined the microtubules and actin in the jsn1Δ mutant.
Yeast bearing deletions in JSN1 progressed normally through the cell cycle. Moreover, microtubule architecture was indistinguishable in wild-type and jsn1Δ cells (Figure 5B). Finally, although actin cables were less robust in jsn1Δ cells, deletion of JSN1 had no obvious effect on actin cable polarity or on association of mitochondria with the actin cytoskeleton (Figure 5C). Numerous actin cables traversed the length of the mother cell in wild-type cells and jsn1Δ cells. In addition, mitochondria coaligned with actin cables in wild-type and jsn1Δ cells. Specifically, 83% of mitochondria visible in the average jsn1Δ mutant mother cell coaligned with actin cables, whereas an average of 82% of mitochondria coaligned with actin cables in the wild-type cell (n = 100). Thus, our observed mitochondrial morphology defects in jsn1Δ cells were not due to indirect effects on the cytoskeleton.
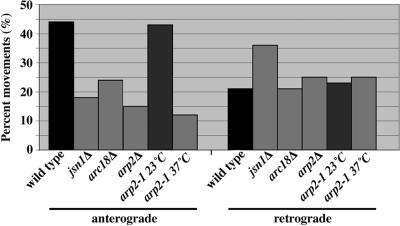
Extending this analysis with live-cell imaging, we found that deletion of JSN1 conferred defects in mitochondrial motility that were also similar to those observed in Arp2/3 complex mutants (Figure 6). In wild-type cells, we observed that 44% of the mitochondria detected in a single focal plane over a 60-s period moved in the anterograde direction, whereas 21% of mitochondria moved in a retrograde direction. The number of retrograde mitochondrial movements was either elevated or not reduced in yeast bearing mutations in JSN1 or Arp2/3 complex subunits (jsn1Δ, arc18Δ, arp2Δ, or arp2-1). In contrast, we found that the number of anterograde mitochondrial movements in yeast bearing deletions in JSN1 was reduced to 41% of that observed in wild-type cells, and that mutations in the Arp2/3 complex produced a similar reduction in anterograde movement of that in wild-type cells (n = 100). Reduction in anterograde mitochondrial motility observed in these mutants was not a consequence of defects in mitochondrial morphology. In wild-type cells, mitochondria, which were similar in size and shape to those seen in jsn1Δ, arp2Δ, or arp2-1 mutants, exhibited normal motility. Overall, these findings support a role for the Arp2/3 complex and Jsn1p in anterograde mitochondrial movement.
Figure 6.
Deletion of JSN1 affects anterograde but not retrograde mitochondrial movement. Mitochondrial motility in yeast bearing deletions in JSN1 (KFY202), ARC18 (KFY214), ARP2 (KFY209), or temperature-sensitive mutation in ARP2 (KFY218), and the corresponding wild-type cells was evaluated using pCIT1-GFP to label mitochondria and time-lapse fluorescence microscopy. Mitochondria undergoing anterograde and retrograde movement were scored in a single focal plane, recorded as described in Materials and Methods, and graphed as a percentage of total movements (n >100). Images were acquired at 1-s intervals.
Jsn1p Interacts with Mitochondria-associated Arp2/3 Complex
Genome-wide two-hybrid screens revealed that Jsn1p has the capacity to bind to Arp2/3 complex subunits. In light of this, it is possible that Jsn1p contributes to targeting Arp2/3 complex to mitochondria through interactions between mitochondria-associated Jsn1p and Arp2/3 complex. Here, we tested whether Jsn1p has physical interactions with Arp2/3 complex and whether this interaction is physiologically relevant.
First, we isolated TAP-tagged Jsn1p from whole cell extracts using affinity chromatography and tested whether purified Arp2/3 complex would bind to immobilized, TAP-tagged Jsn1p (Figure 7A). Arp2/3 complex did not show significant binding to or release from control beads. In contrast, we found that Arp2/3 complex bound to TAP-tagged Jsn1p and that tagged Jsn1p and Arp2/3 complex could be released from beads after TEV treatment.
Figure 7.
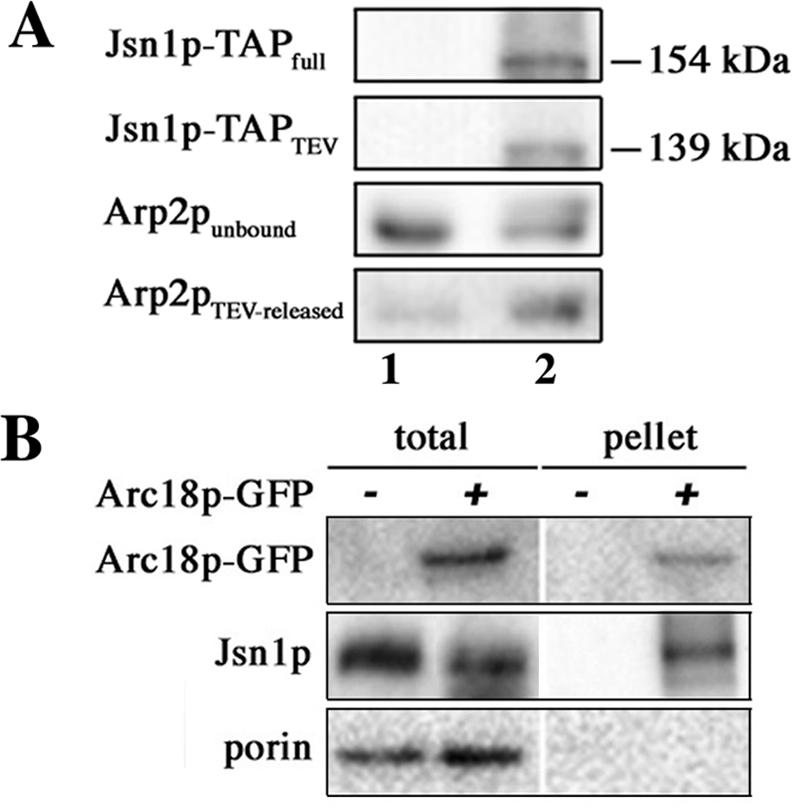
Jsn1p interacts with mitochondria-associated Arc18p-GFP. (A) Whole cells extracts from untagged controls (BY4743; lane 1) or yeast expressing TAP-tagged Jsn1p (2GS-B-4; lane 2) were purified using IgG-coupled Sepharose beads, as described in Materials and Methods. IgG-Sepharose beads were incubated with purified Arp2/3 complex (0.5 μg) for 2 h at 4°C. The beads were concentrated by centrifugation, and unbound Arp2/3 complex in the supernatant was removed. Beads were washed, incubated with TEV protease, and material released by protease treatment was isolated from the beads, as described above. All samples described below were analyzed using Western blots. Jsn1p-TAPfull: IgGSepharose beads after incubation with whole cell extracts from untagged or tagged cells, probed with an antibody raised against the TAP tag. Jsn1p-TAPTEV, material released from the beads after treatment with TEV protease, probed with an antibody raised against the TAP tag. Arp2punbound, Arp2/3 complex that did not bind to the beads, probed with anti-Arp2p antibody. Arp2pTEV-released, material released from the beads after treatment with TEV protease, probed with anti-Arp2p antibody. (B) Mitochondria were isolated from mid-log phase yeast cells expressing GFP-tagged Arc18p (KFY119) or untagged ARC18 (BY4743). After solubilization of mitochondria in digitonincontaining buffers, Arc18p-GFP was immunoprecipitated using protein G-Sepharose beads conjugated to polyclonal rabbit anti-GFP IgGs. Aliquots of extracted mitochondria (total) and proteins immunoprecipitated from mitochondrial extracts (pellet) were analyzed using Western blots and antibodies raised against GFP, Jsn1p, and porin.
Second, we tested whether Jsn1p coimmunoprecipitates with Arp2/3 complex. To do so, we isolated mitochondria from cells expressing Arc18p-GFP and immunoprecipitated the tagged Arp2/3 complex subunit from mitochondrial extracts using anti-GFP antibody. We found that Arc18p-GFP coimmunoprecipitated with Jsn1p and not with an abundant, unrelated mitochondrial outer membrane protein (porin) (Figure 7B). Neither Jsn1p nor Arc18p-GFP were recovered with IgG-free beads after incubation with mitochondria isolated from cells expressing GFP-tagged or untagged Arc18p (our unpublished data). Thus, coimmunoprecipitation of Jsn1p with Arc18p-GFP seems to be specific. Together, these findings provide additional support that Jsn1p interacts with Arp2/3 complex. Moreover, they indicate that mitochondria are the site of Jsn1p–Arp2/3 complex interactions and that these interactions are physiologically relevant.
DISCUSSION
Several signal transduction proteins have been implicated in the recruitment of the Arp2/3 complex activator N-WASp to endosomes. In addition, the mechanisms of targeting the Arp2/3 complex or its activators to pathogen surfaces are well understood (Frischknecht and Way, 2001). However, until now, the organelle-specific proteins that contribute to recruiting the Arp2/3 complex or its activator to endogenous membranes were not identified.
Our studies support a role for Jsn1p in recruiting the Arp2/3 complex to mitochondria in budding yeast. First, the localization of Jsn1p is consistent with a role in recruiting the Arp2/3 complex to mitochondria. Using affinity-purified anti-Jsn1p antibodies, we found that Jsn1p localized to punctate structures that colocalized with mitochondria. Consistent with this, we found that Jsn1p 1) cofractionated with mitochondria during subcellular fractionation, 2) was extracted from mitochondria using sodium carbonate, and 3) was protease sensitive in intact mitochondria. Thus, Jsn1p behaves like a peripheral mitochondrial membrane protein that is associated with the outer leaflet of the mitochondrial outer membrane.
Second, we find that Jsn1p colocalizes with mitochondriaassociated Arp2/3 complex and contributes to recruiting Arp2/3 complex protein to yeast mitochondria. Using Arc18p-GFP as a marker for Arp2/3 complex, we found that Jsn1p and Arp2/3 complex localize to punctate structures that align along mitochondria in the mother cell. All detectable mitochondria-associated Arp2/3 complex colocalized with Jsn1p. In addition, deletion of JSN1 results in a 50% reduction in the amount of Arp2p and Arc18p-GFP that is recovered with isolated mitochondria and a 50% reduction in the number of Arc18p-GFP puncta that associate with mitochondria in vivo. Finally, deletion of JSN1 produces mitochondrial abnormalities similar to those observed in Arp2/3 complex mutants. That is, mitochondria are fragmented or aggregated in jsn1Δ cells and in arp2 or arc15 mutants (Boldogh et al., 2001). Moreover, deletion of JSN1, ARP2, ARC18, or ARC15 inhibited the amount of anterograde mitochondrial movement to similar extents and had either no effect or increased the amount of retrograde mitochondrial movement. Because microtubule and microfilament architecture and association of mitochondria with actin cables were indistinguishable between jsn1Δ cells and wild-type cells, the defects in mitochondrial morphology and motility were not consequences of defects in cytoskeletal organization or organelle–cytoskeleton interactions. Rather, our data support the model that Jsn1p contributes to targeting of the Arp2/3 complex to mitochondria.
Finally, our preliminary results indicate that Jsn1p may recruit Arp2/3 complex to mitochondria through direct or indirect interactions between mitochondria-associated Jsn1p and Arp2/3 complex. As described above, genome-wide two-hybrid screens indicate that Jsn1p can bind to Arp2/3 complex subunits. We obtained three lines of evidence that this Jsn1p has physiologically relevant interactions with Arp2/3 complex. First, Arp2/3 complex, which is associated with mitochondria and not actin patches, colocalizes with Jsn1p. Second, Arp2/3 complex subunits bind to TAP-tagged Jsn1p. Third, Arc18p-GFP and Jsn1p coimmunoprecipitate from mitochondrial extracts. These findings raise the possibility that Jsn1p may be part of the Arp2/3 complex receptor on yeast mitochondria.
As described above, Jsn1p was originally discovered because JSN1 overexpression suppresses the temperature sensitivity of a microtubule-stabilizing tubulin mutation (Machin et al., 1995). We do not detect any obvious defects in microtubule or microfilament organization in jsn1Δ cells. However, it is possible that Jsn1p function in microtubule dynamics and mitochondrial motility may reflect cross-talk between actin cables and microtubules. Because Jsn1p recruits the Arp2/3 complex to mitochondria, it could contribute to actin polymerization and assembly at the mitochondria–actin cable interface in ways that are not detectable with light microscopy. This, in turn, could affect actin cable integrity and function in localization of spindle alignment elements and other proteins that affect microtubule dynamics, producing the suppression observed in previous overexpression studies.
Finally, although Jsn1p is necessary for efficient targeting of the Arp2/3 complex to mitochondria, we suspect that it is not sufficient for this targeting event. First, Jsn1p behaves like a peripheral mitochondrial outer membrane protein and does not contain any obvious mitochondrial targeting sequences or hydrophobic stretches typical of integral membrane proteins. Therefore, there must be other protein and/or membrane associations that target Jsn1p to mitochondria. Second, deletion of JSN1 does not completely abolish the association of the Arp2/3 complex with mitochondria. Thus, there may be other proteins that contribute to recruiting Jsn1p and the Arp2/3 complex to yeast mitochondria.
Ongoing studies are directed toward identifying other components of the Arp2/3 complex-recruiting machinery on mitochondria. Potentially redundant Arp2/3 complex recruiters may include other yeast PUF family proteins. Because Jsn1p, like all PUF family proteins, contains a putative RNA binding domain, Jsn1p function in Arp2/3 complex recruitment may require RNA. Indeed, Jsn1p may be present within a ribonucleoprotein particle (Gerber et al., 2004) or use RNA for its Arp2/3 complex recruiting activity.
Acknowledgments
We thank Thomas Huckaba for database searches that revealed Jsn1p-related two-hybrid interactions; Medha Goyal for assistance in strain construction; members of the Pon laboratory for critical evaluation of the manuscript; Drs. G. Barnes, P. Silver, and G. Schatz for antibodies; Dr. P. Crews for latrunculin-A (whose work is supported by a grant from the National Institutes of Health Grant CA4715); Drs. T. Stearns and J. Shaw for plasmids; Drs. P. Thorsness, R. Li, B. Winsor, and A. Bretscher for yeast strains; Drs. M. Longtine and J. Pringle for tagging cassette DNA; Dr. T. Takeda for TAP purification reagents; and Dr. B. Goode for providing purified Arp2/3 complex. This work was supported by research grants to L. P. from the National Institutes of Health (GM-45735 and GM-66037), and training grant awards to K. F. from the National Institutes of Health (1 T32 DK07786 and 5 T32 NS07062).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–06–0590) on August 17, 2005.
Abbreviations used: DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; mtDNA, mitochondrial DNA.
References
- Barker, D. D., Wang, C., Moore, J., Dickinson, L. K., and Lehman, R. (1992). Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 6, 2312–2326. [DOI] [PubMed] [Google Scholar]
- Benesch, S., Lommel, S., Steffen, A., Stradal, T. E., Scaplehorn, N., Way, M., Wehland, J., and Rottner, K. (2002). Phosphatidylinositol 4,5-biphosphate (PIP2)-induced vesicle movement depends on N-WASp and involves Nck, WIP, and Grb2. J. Biol. Chem. 277, 37771–37776. [DOI] [PubMed] [Google Scholar]
- Boldogh, I. R., Fehrenbacher, K. L., Yang, H. C., and Pon, L. A. (2005). Mitochondrial movement and inheritance in budding yeast. Gene 354, 28–36. [DOI] [PubMed] [Google Scholar]
- Boldogh, I. R., Ramcharan, S., and Pon, L. A. (2004). A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol. Biol. Cell. 15, 3994–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I., Vojtov, N., Karmon, S., and Pon, L. A. (1998). Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J. Cell Biol. 141, 1371–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I. R., Yang, H. C. Nowakowski, W. D., Karmon, S. L., Hays, L. G., Yates, J. R., 3rd, and Pon, L. A. 2001. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc. Natl. Acad. Sci. USA. 98, 3162–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., Dawson, D., and Stearns, T. (2000). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 133–135.
- Chang, F. S., Stefan, C. J., and Blumer, K. J. (2003). A WASp homolog powers actin polymerization-dependent motility of endosomes in vivo. Curr. Biol. 13, 455–463. [DOI] [PubMed] [Google Scholar]
- Cossart, P. (2000). Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2, 195–205. [DOI] [PubMed] [Google Scholar]
- Daum, G., Bohni, P. C., and Schatz, G. (1982). Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028–13033. [PubMed] [Google Scholar]
- Eitzen, G., Wang, L., Thorngren, N., and Wickner, W. (2002). Remodeling of organelle-bound actin is required for yeast vacuole fusion. J. Cell Biol. 158, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D. C., Boone, C., and A. Bretscher. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 260–269. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher, K. L., Yang, H. C., Gay, A. C., Huckaba, T., and Pon, L. A. (2004). Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 14, 1996–2004. [DOI] [PubMed] [Google Scholar]
- Fratti, R. A., Jun, Y., Merz, A. J., Margolis, N., and Wickner, W. (2004). Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 6, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht, F., and Way, M. (2001). Surfing pathogens and the lessons learned for actin polymerization. Trends Cell Biol. 11, 30–38. [DOI] [PubMed] [Google Scholar]
- Gerber, A. P., Herschlag, D., and Brown, P. O. (2004). Extensive association of functionally and cytotopically related mRNAs with Puf Family RNA-binding proteins in yeast. PloS Biol. 2, 342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Glick, B. S., and Pon, L. A. (1995). Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213–223. [DOI] [PubMed] [Google Scholar]
- Goode, B. L., Rodal, A. A., Barnes, G., and Drubin, D. G. (2001). Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 153, 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, W., Deng, Y., Zenklusen, D., and Singer, R. H. (2004). A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 18, 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba, T. M., Gay, A. C., Pantalena, L. F., Yang, H. C., and Pon, L. A. (2004). Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 167, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall, R., Muller-Taubenberger, A., Machesky, L., Kohler, J., Simmeth, E., Atkinson, S. J., Weber, I., and Gerisch, G. (2001). Dynamics of the Dictyostelium Arp 2/3 complex in endocytosis, cytokinesis, and chemotaxis. Cell Motil. Cytoskeleton 50, 115–128. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D. G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475–487. [DOI] [PubMed] [Google Scholar]
- Kerscher, O., Holder, J., Srinivasan, M., Leung, R. S., and Jensen, R. E. (1997). The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139, 1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarino, D. A., Boldogh, I., Smith, M. G., Rosand, J., and Pon, L. A. (1994). Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol. Biol. Cell. 5, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Macdonald, P. M. (1992). The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development 114, 221–232. [DOI] [PubMed] [Google Scholar]
- Machin, N. A., Lee, J. M., and Barnes, G. (1995). Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol. Biol. Cell. 6, 1241–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield, C. J., Moss, S. E., Ballestrem, C., Imhof, B. A., Giese, G., Wunderlich, I., and Almers, W. (1999). Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nat. Cell Biol. 1, 72–74. [DOI] [PubMed] [Google Scholar]
- Moreau, V., Madania, A., Martin, R. P., and Winsor, B. (1996). The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J. Cell Biol. 134, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy, A. D., McCaffery, J. M., and Shaw, J. M. (2000). Dnm1p GTPasemediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, Y., and Wharton, R. P. (1995). Binding of Pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80, 747–756. [DOI] [PubMed] [Google Scholar]
- Nowakowski, D. W., Swayne, T. C., and Pon, L. A. (2001). Epitope tagging and visualization of nuclear-encoded mitochondrial proteins in yeast. Methods Cell Biol. 65, 257–276. [DOI] [PubMed] [Google Scholar]
- Okamoto, K., Perlman, P. S., and Butow, R. A. (2001). Targeting of green fluorescent protein to mitochondria. Methods Cell Biol. 65, 277–283. [DOI] [PubMed] [Google Scholar]
- Olivas, W., and Parker, R. (2000). The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Rozelle, A. L., Machesky, I. M., Yamamoto, M., Driessens, M. H., Insall, R. H., Roth, M. G., Luby-Phelps, K., Marriott, G., Hall, A., and Yin, H. L. (2000). Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raftenriched vesicles through WASP-Arp2/3. Curr. Biol. 10, 311–320. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (2002). Getting started with yeast. Methods Enzymol. 350, 3–41. [DOI] [PubMed] [Google Scholar]
- Simon, V. R., Karmon, S. L., and Pon, L. A. (1997). Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton 37, 199–210. [DOI] [PubMed] [Google Scholar]
- Sollner, T., Griffiths, G., Pfaller, R., Pfanner, N., and Neupert, W. (1989). MOM19, an import receptor for mitochondrial precursor proteins. Cell 59, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Southwick, F. S., Li, W., Zhang, F., Zeile, W. L., and Purich, D. L. (2003). Actin-based endosome and phagosome rocketing in macrophages: activation by the secretagogue antagonists lanthanum and zinc. Cell Motil. Cytoskeleton 54, 41–55. [DOI] [PubMed] [Google Scholar]
- Tadauchi, T., Matsumoto, K., Herskowitz, I., and Irie, K. (2001). Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 1, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton, J., Rowning, B. A., Coughlin, M. L., Wu, M., Moon, R. T., Mitchison, T. J., and Larabell, C. A. (2000). Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Wang, X., McLachlan, J., Zamore, P. D., and Hall, T. M. (2002). Modular recognition of RNA by a human pumilio-homology domain. Cell 110, 501–512. [DOI] [PubMed] [Google Scholar]
- Winter, D., Podtelejnikov, A. V., Mann, M., and Li, R. (1997). The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 7, 519–529. [DOI] [PubMed] [Google Scholar]
- Winter, D. C., Choe, E. Y., and Li, R. (1999). Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA 96, 7288–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. C., Palazzo, A., Swayne, T. C., and Pon, L. A. (1999). A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr. Biol. 9, 1111–1114. [DOI] [PubMed] [Google Scholar]
- Yang, H. C., and Pon, L. A. (2002). Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. USA 99, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P. D., Williamson, J. R., and Lehmann, R. (1997). The Pumilio protein binds RNA through a conserved domain that defines a new class of RNAbinding proteins. RNA. 3, 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Southwick, F. S., and Purich, D. L. (2002). Actin-based phagosome motility. Cell Motil. Cytoskeleton 53, 81–88. [DOI] [PubMed] [Google Scholar]