Abstract
Long-distance transport is crucial for polar-growing cells, such as neurons and fungal hyphae. Kinesins and myosins participate in this process, but their functional interplay is poorly understood. Here, we investigate the role of kinesin motors in hyphal growth of the plant pathogen Ustilago maydis. Although the microtubule plus-ends are directed to the hyphal tip, of all 10 kinesins analyzed, only conventional kinesin (Kinesin-1) and Unc104/Kif1A-like kinesin (Kinesin-3) were up-regulated in hyphae and they are essential for extended hyphal growth. Δkin1 and Δkin3 mutant hyphae grew irregular and remained short, but they were still able to grow polarized. No additional phenotype was detected in Δkin1rkin3 double mutants, but polarity was lost in Δmyo5rkin1 and Δmyo5rkin3 mutant cells, suggesting that kinesins and class V myosin cooperate in hyphal growth. Consistent with such a role in secretion, fusion proteins of green fluorescent protein and Kinesin-1, Myosin-V, and Kinesin-3 accumulate in the apex of hyphae, a region where secretory vesicles cluster to form the fungal Spitzenkörper. Quantitative assays revealed a role of Kin3 in secretion of acid phosphatase, whereas Kin1 was not involved. Our data demonstrate that just two kinesins and at least one myosin support hyphal growth.
INTRODUCTION
Polarized growth of tip-growing plant cells, filamentous fungi, and neuronal cells is based on long-distance membrane transport along the fibrous elements of the cytoskeleton (Brady, 1995; Geitmann and Emons, 2000). Vesicle and organelle traffic is mediated by molecular motors that hydrolyze ATP and move their cargo along filamentous actin (F-actin) and/or microtubules (MTs; Hirokawa et al., 1998; Mermall et al., 1998). Among the 18 classes of actin-based myosin motors, class V myosins belong to an ancient group that is present in animals and fungi (Hodge and Cope, 2000). These myosins participate in transport of membranous vesicles and organelles (reviewed in Langford, 2002). The second major transport system involves MTs that usually extend their growing plus-ends toward the cell periphery or into the apical terminus in case of the axon (Heidemann et al., 1981). A large number of plus-end-directed kinesins use MTs to support anterograde (tip-directed) axonal transport (Hirokawa and Takemura, 2004). Among the numerous membrane motors in higher eukaryotes, only conventional kinesin (Brady, 1985; Scholey et al., 1985; Vale et al., 1985) and Unc104/Kif1A-like motors (Hall and Hedgecock, 1991; Aizawa et al., 1992) were found in the genome of Dictyostelium discoideum (Klopfenstein et al., 2002) and filamentous fungi (Schoch et al., 2003), where they support roles in or- ganelle transport (Pollock et al., 1999; Wedlich-Söldner et al., 2002). This indicates that Kinesin-1 (=conventional kinesin/KHC) and Kinesin-3 (=Unc104/Kif1A; nomenclature according to Lawrence et al., 2004) are organelle transporters that occurred early in evolution.
Recently, it emerged that microtubules and F-actin functionally cooperate in polar exocytosis. Axonal vesicles can switch from long-distance MT-based transport to short-distance F-actin-based movement (Kuznetsov et al., 1992). It was found that Myosin-V and Kinesin-1 physically interact and colocalize in mouse melanocytes (Huang et al., 1999) and squid neurons (Stafford et al., 2000), indicating that kinesins and Myosin-V cooperate in anterograde membrane traffic. This notion is confirmed by studies on melanophores, where bidirectional long-range pigment motility depends on a Kinesin-2 and dynein, whereas F-actin and Myosin-V are involved in short-range dispersal, apical tethering, and fine tuning of MT-based motility (reviewed in Brown, 1999). These studies suggest the concept of “dual transport,” in which a long-range membrane transport is based on MT-dependent kinesins, in particular Kinesin-1, Kinesin-2, and Kinesin-3 members, whereas class V myosins mediate short-range traffic along F-actin (Langford, 2002). However, there are exceptions to this rule, because long-distance pigment granule traffic in retinal pigment epithelial cells is F-actin dependent (King-Smith et al., 1997).
Much of our knowledge of the molecular role of the cytoskeleton in polarized growth comes from the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe that combine a relatively simple cellular organization with powerful genetics. Indeed, the first indication for a functional interplay between kinesins and myosins was described in S. cerevisiae, where Smy1p, a distant relative of conventional kinesin (Brown, 1999), is able to compensate for the class V myosin Myo2p (Lillie and Brown, 1992). However, Smy1p might not be a motor, because its localization and function is MT independent (Lillie and Brown, 1998). In S. pombe, both F-actin and MTs participate in polar growth (Sawin and Nurse, 1998; reviewed in Hayles and Nurse, 2001). However, although class V myosins support tip growth in fission yeast (Win et al., 2001), Kinesin-1 is not involved (Brazer et al., 2000; Jeong et al., 2002), and Kinesin-3 does not even exist in both yeast species. In contrast, filamentous fungi contain Kinesin-1, Kinesin-3, and Myosin-V members (Steinberg and Schliwa, 1995; Wedlich-Söldner et al., 2002; Schoch et al., 2003) and it has been suggested that this reflects the need of long-distance transport in the highly polarized hyphal cells (Steinberg, 2000; Goldstein, 2001).
Here, we analyze the role of motors in hyphal growth of the model fungus Ustilago maydis. Using a fluorescent EB1-homologue (Straube et al., 2001), we show that the MT plus-ends are directed to the hyphal tip, and real-time PCR analysis revealed that the genes for Kinesin-1 and Kinesin-3 are up-regulated in hyphae, which is consistent with the essential role of these kinesins in long-range hyphal growth. Furthermore, we provide evidence that Kinesin-1, Kinesin-3, and Myosin-V cooperate in polarized growth and that Kinesin-3 participates in acid phosphatase secretion in hyphae.
MATERIALS AND METHODS
Strains, Plasmids, and Growth Conditions
U. maydis wild-type strains AB34 and the filamentously growing AB33 have been described previously (see Table 1 for details and references). Deletion of kin4, kin8, kin7a and kin7b, and kin14 was done by homologous replacement of the corresponding gene with either a nourseothricin or a hygromycin resistance cassette in strains FB1, FB2, and AB33 following described protocols (Brachmann et al., 2004; Kämper, 2004). Deletion of myo5 was done by homologous replacement of nucleotides –134 to +258 and that of kin3 by deletion of amino acids 12–885 with a hygromycin resistance cassette. In conditional strains AB33rKin9, AB33ΔKin1rKin3, AB33ΔKin1rKin3GT, and AB33ΔMyo5rKin1, the respective kinesin genes were expressed under the conditional crg promoter (Bottin et al., 1996) or the repressible nar-promoter (Banks et al., 1993; see Table 1 and supplemental files for details). MT organization was analyzed by expression of green fluorescent protein (GFP)-Tub1 (Steinberg et al., 2001). For localization studies, a triple GFP-tag was N-terminally fused to myo5 and kin1 and a single GFP was fused C-terminally to kin3 and expressed under control of the native myo5, kin1, or kin3 promoter, respectively. For studies on MT orientation, peb1, the plus-end binding EB1-homologue of U. maydis (Straube et al., 2003) was fused to monomeric red fluorescent protein (Campbell et al., 2002) and localized in the strain AB33GT. Yeast-like strains were grown in complete medium and on plates as described previously (Weber et al., 2003). Filamentous growth of AB33 and derivates was induced by shifting cells grown from CM-G to nitrate minimal medium (NM-G) as described previously (Weber et al., 2003). See supplemental material for further details.
Table 1.
Genotype of strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| AB33 | a2 PnarbW2 PnarbE1, bleR | Brachmann et al. (2001) |
| AB34 | a2PnarbW2 PnarbE2, bleR | Brachmann et al. (2001) |
| AB33Peb1R_GT | a2 PnarbW2 PnarbE1 peb1-mrfp, natR, bleR/potefGFPTub1 | This study |
| AB33ΔKin7a | a2 PnarbW2 PnarbE1 Δkin7a::hygR, bleR | This study |
| AB33ΔKin1 | a2 PnarbW2 PnarbE1 Δkin1::hygR, bleR | This study |
| AB33ΔKin3 | a2 PnarbW2 PnarbE1 Δkin3::hygR, bleR | This study |
| AB33ΔKin4 | a2 PnarbW2 PnarbE1 Δkin4::hygR, bleR | This study |
| AB33rKin9 | a2 PnarbW2 PnarbE1 Pcrg-kin9 natR, bleR | This study |
| AB33ΔKin8 | a2 PnarbW2 PnarbE1 Δkin8::natR, bleR | This study |
| AB33ΔKin7b | a2 PnarbW2 PnarbE1 Δkin7b::natR, bleR | This study |
| AB33ΔKin7aΔKin7b | a2 PnarbW2 PnarbE1 Δkin7a::hygR Δkin7b::natR, bleR | This study |
| FB1ΔKin14 | a1b1 Δkin14::hygR | This study |
| FB2ΔKin14 | a2b2 Δkin14::hygR | This study |
| AB33ΔKin1rKin3 | a2 PnarbW2 PnarbE1 Δkin1::hygR Pcrg-kin3 natR, bleR | This study |
| AB33ΔKin1GT | a2 PnarbW2 PnarbE1 Δkin1::hygR, bleR/potefGFPTub1 | This study |
| AB33ΔKin3GT | a2 PnarbW2 PnarbE1 Δkin3::hygR, bleR/potefGFPTub1 | This study |
| AB33ΔKin1rKin3_GT | a2 PnarbW2 PnarbE1 Δkin1::hygR Pcrg-kin3 natR, bleR/potefGFPTub1 | This study |
| AB33ΔMyo5 | a2 PnarbW2 PnarbE1 Δmyo5::hygR, bleR | This study |
| AB33ΔMyo5rKin1 | a2 PnarbW2 PnarbE Pcrg-kin1 natR, bleR | This study |
| AB33ΔMyo5rKin3 | a2 PnarbW2 PnarbE Pcrg-kin3 natR, bleR | This study |
| AB33G3Myo5_RT | a2 PnarbW2 PnarbE1, bleR/pGFP3Myo5/potefRFPTub1 | This study |
| AB33G3Kin1 | a2 PnarbW2 PnarbE1, bleR/pGFP3Kin1 | This study |
| AB33Kin3G | a2 PnarbW2 PnarbE1, bleR/pKin3GFP | This study |
| potefGFPTub1 | Potef-egfp-tub1, cbxR | Steinberg et al. (2001) |
| pGFP3Myo5 | Pmyo5-3gfp-myo5, cbxR | This study |
| potefRFPTub1 | Potef-mrfp-tub1, hygR | This study |
| pGFP3Kin1 | Pkin1-3gfp-kin1, cbxR | This study |
| pKin3GFP1 | Pkin3-kin3-gfp, cbxR | Wedlich-Söldner et al. (2002) |
a, b, mating type loci; mfa2, mating pheromone 2; Δ, deletion; P, promoter; ::, homologous replacement; -, fusion; hphR, hygromycin resistance; bleR, phleomycin resistance; natR, nourseothricin resistance; cbxR, carboxin resistance; /, ectopically integrated; otef, constitutive promoter (Spellig et al., 1996); nar, conditional nitrate reductase promoter (Banks et al., 1993); crg, conditional arabinose-induced promoter (Bottin et al., 1996); bE1, bW2, genes of the b mating type locus; gfp, enhanced green fluorescent protein; cfp, cyan-shifted fluorescent protein; yfp, yellow-shifted fluorescent protein; mrfp, monomeric red fluorescent protein (Campell et al., 2002); kin4,5,7b,8,9,14, kinesins belonging to several kinesin families; kin7a, CENP-E-like (kinesin-7), previously named `Kin1' (Lehmler et al., 1997); kin1, conventional kinesin (kinesin-1), previously named `Kin2' (Lehmler et al., 1997); kin3, Kif1A-like kinesin (kinesin-3; Wedlich-Söldner et al., 2002); myo5, class V myosin (Weber et al., 2003); tub1, α-tubulin (Steinberg et al., 2001); peb1, EB1-homologue (Straube et al., 2003).
Sequence Analysis
Kinesins in the genome of U. maydis were identified by screening the genomic sequences (www-genome.wi.edu/annotation/fungi/ustilago_maydis/) with kinesins from vertebrates and yeasts. Sequence alignment, phylogenetic analysis of the motor domains and domain analysis were performed as described previously (Weber et al., 2003). See supplemental material for further details and Web addresses.
DNA Techniques and Real-Time Polymerase Chain Reaction (PCR)
Standard DNA techniques were used. DNA and RNA isolation and real-time PCR as well as quantitative and qualitative RNA analysis was performed as described in supplemental files on the Molecular Biology of the Cell Web site at http://www.molbiolcell.org/. PCR reactions were performed in triplicates, and negative controls (in the absence of template) were included. Annealing temperatures ranged from 59 to 65°C, depending on the primers used. Target genes were normalized to the reference housekeeping gene EF-1-a (elongation factor-1-α from U. maydis). To verify the specificity of the reverse transcription-PCR reaction, the size of the amplicons was analyzed by high-resolution sizing of DNA fragments using a Bioanalyzer 2100 system and a LabChip DNA 100 assay (Agilent, Waldbronn, Germany).
Microscopy, Image Processing, and Quantitative Analysis
Cells from logarithmically growing cultures were dropped on a thin 1% agarose-layer and immediately observed using a Zeiss Axioplan II microscope. Epifluorescence was observed using filters for fluorescein isothiocyanate, DsRed, yellow-shifted fluorescent protein, and cyan-shifted fluorescent protein (see supplemental files for details). Frames were taken with a cooled charge-coupled device-camera (CoolSNAP HQ; Roper Scientific, Trenton, NJ) controlled by MetaMorph (Universal Imaging, Downingtown, PA). All image processing, including adjustment of brightness, contrast, and gamma-values, was performed with MetaMorph (Universal Imaging) and Photoshop (Adobe Systems, Mountain View, CA). All values are given as means ± SD unless otherwise stated.
Phosphatase Assay
The activities of secreted acid phosphatase were analyzed as described previously (Steinberg and McIntosh, 1998). Because the morphology of mutant strains differed drastically, cell extracts were prepared and the protein content of the used mutants was determined using with Bradford Assays (Bio-Rad, Hercules, CA) done according to manufacturer's instructions. After correlating the protein content with the OD600, the same amount of cells of each mutant strain was used in the colorimetric assays. In brief, for phosphatase assays, cells of strains AB33, AB33Δkin1, AB33Δkin3, AB33Δkin1rKin3, and AB33Δmyo5 were grown in phosphate-free medium for 1.5 h. An aliquot of cell suspension or cell-free supernatant was incubated for 3 h with p-nitrophenyl phosphate (Sigma-Aldrich, St. Louis, MO) After removal of the cells by centrifugation, the reaction was stopped by adding KPO4, pH 11.0. The release of nitrophenon was measured at OD405 and corrected for the blank.
RESULTS
Plus-Ends of Microtubules Are Oriented toward the Hyphal Tip
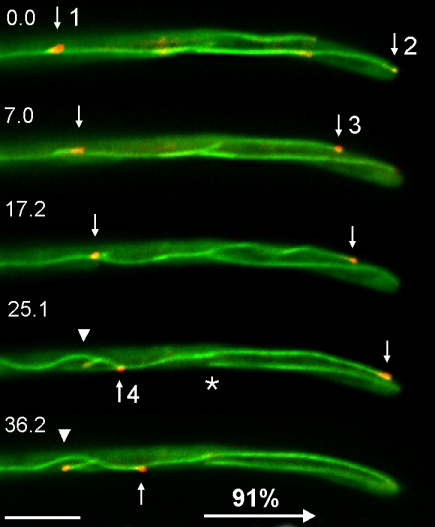
To gain a deeper understanding of the molecular machinery that supports hyphal growth, we set out to determine the orientation of MTs in hyphae of U. maydis. We fused Peb1, an EB1-homologue in U. maydis that localizes to growing MT plus-ends (Straube et al., 2003) to monomeric red-shifted fluorescent protein (RFP; Campbell et al., 2002) and expressed the fusion protein in the filamentously growing strain AB33GT that contains GFP-labeled microtubules (see Table 1 for details). In colocalization experiments, the red Peb1-RFP fusion protein localized to the ends of MTs. In 91% of all cases, these MT plus-ends elongate toward the growing apex of the hypha (Figure 1, arrows and numbers indicate growing MT ends) and only rarely did MTs polymerize toward the subapical region of the hyphae (Figure 1, arrowhead). This orientation of MTs suggests that plus-end-directed kinesins participate in polar growth of hyphae.
Figure 1.
Orientation of MT plus-ends in hyphae of U. maydis. A fusion of RFP and Peb1, an EB1-homologue that localizes to growing MT plus-ends (Straube et al., 2003) marks microtubule ends that grow toward the hyphal apex, indicating that kinesins could support polar hyphal growth. Arrows indicate four MT tips; arrowhead marks a microtubule that elongates toward the proximal end of the tip cell. Note that microtubules are not connected to the spindle pole body (asterisk marks minus-end of a cytoplasmic microtubule; the corresponding plus end is marked by “2” in first frame). Time in seconds is given. Bar, 5 μm.
The Genome of U. maydis Encodes for 10 Putative Kinesins
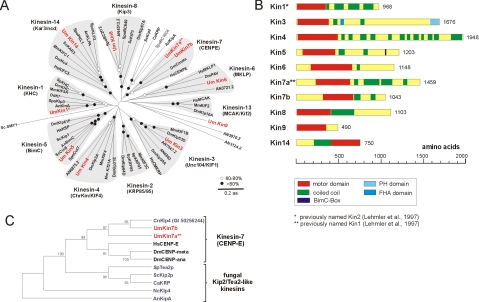
For identification of kinesin motors, we screened the published genome of U. maydis (see Materials and Methods for URL) with public sequences of kinesins of the different families (Lawrence et al., 2004). In this search, we identified 10 genes with predicted kinesin motor domains of a length of 348–498 aa (e–31 to e–175; predicted by SMART; see Materials and Methods for URL). Based on similarity in the motor domain, these putative kinsesins group in at least seven families (Figure 2A and Table 2; classification according to Lawrence et al., 2004). To avoid future confusions, the names of all kinesins in U. maydis were brought in line with the family names. Consequently, the previously published Kinesin-1 and the Kinesin-7 members (Lehmler et al., 1997) were renamed (Kin2, a Kinesin-1 member is now named Kin1; the former Kin1 is now Kin7a; Figure 2A). U. maydis has a second CENP-E-like Kinesin-7, Kin7b, that shares 45% sequence identity within the motor domain but only 17% overall identity with Kin7a. Interestingly, both U. maydis kinesins as well as a kinesin from the basidiomycete Cryptococcus neoformans that we found as a hypothetical protein in the public database (named CnKlp4, accession no. EAL18971) are more closely related to the animal CENP-E homologues than to their counterparts from ascomycete fungi (Figure 2C). Moreover, the ascomycete kinesins are involved in polar growth (Browning et al., 2000; Konzack et al., 2004), whereas neither animal CENP-E nor U. maydis kin7a and kin7b mutants show any defect in cell morphogenesis (see below), again suggesting that Kin7a and Kin7b are more closely related to animal CENP-E kinesins (Figure 2C). The domain structure of the Kinesin-1 member Kin1 (Lehmler et al., 1997) and a Kinesin-3 homologue Kin3 (Wedlich-Söldner et al., 2002) are discussed in previous publications. Kin4 and Kin6 share only weak but significant similarity with other representatives of the Kinesin-4 and Kinesin-6 families (Table 2); Kin5, Kin8, and Kin14 are highly homologous to the Kinesin-5, Kinesin-8, and Kinesin-14 families (Figure 2A and Table 2). This is further supported by the fact that Kin5 contains a family-specific C-terminal BimC-box (aa 1035–1049; 50% identity) and Kin14 has a C-terminal motor domain (Figure 2B) and a characteristic stretch of 14 residues (aa 372–385; Hirokawa et al., 1998) in front of it. In addition to these defined kinesins, we identified one orphan kinesin, Kin9, that is only weakly related to Kinesin-13 members (Figure 2A and Table 2). Furthermore, MCAK/Kif2-like kinesins have an internal motor domain important for their function in regulating MT stability (Ogawa et al., 2004), whereas Kin9 is predicted to be an N-terminal kinesin (Figure 2B and Table 2), again suggesting that this kinesin is an orphan. Together, the genome of U. maydis encodes three mitotic kinesins (Kin5, Kin6, and Kin14), two well-established membrane transporters (Kin1 and Kin3), four kinesins that, based on their counterparts in several organisms (reviewed in Schoch et al., 2003), might participate in membrane traffic or mitosis/meiosis (Kin7a, Kin7b, Kin4, and Kin8), and one orphan kinesin (Kin9).
Figure 2.
Sequence analysis of kinesins in U. maydis. (A) The genome of U. maydis encodes for 10 kinesins. Nine of these kinesin motors belong to defined subfamilies, whereas UmKin9 is ungrouped. The tree is based on an alignment of the motor domain of fungal and animal kinesins. Cn, Cryptococcus neoformans; Um, U. maydis; Hs, Homo sapiens; Mm, Mus musculus; Dm, Drosophila melanogaster; Sp, Strongylocentrotus purpuratus; Spo, S. pombe; Sc, S. cervisiae; Ca, Candida albicans; Nc, Neurospora crasssa; An/AN, A. nidulans. Note that only selected kinesins from animal and fungal species were included. Accession numbers are given in supplementary files. Closed circle, >80% bootstrap; open circle, 60–80% bootstrap. (B) Domain organization of all kinesins in U. maydis. Domain analysis was done using The SMART and PROSITE server. Note that the domain organization is based on sequence prediction and comparison with kinesin motors from other organisms. (C) Tree of members of the Kinesin-7 family. Kinesin-7 from ascomycete fungi group together, which corresponds with a role in cell polarity (Browning et al., 2000) and hyphal growth (Konzack et al., 2004). In contrast, representatives from basidiomycete fungi are more closely related to CENP-E motors from animals, where they participate in chromosome inheritance (Yen et al., 1991). For species names see Fig. 2A. Accession numbers are given in supplemental files. The tree is based on a comparison of motor domains. Bootstrap values are given in percentage.
Table 2.
Kinesins in the basidiomycete U. maydis
| Name | Subfamilya | Homologyb (head/tail) | Functionc | Motor domaind | Accession no. | Reference |
|---|---|---|---|---|---|---|
| Kin1e | Kinesin-1 (KHC) | 52/17 | Secretion/vacuole | N | U92845 | Lehmler et al. (1997) |
| Kin3 | Kinesin-3 (Unc104) | 59/20 | Endosome motility | N | AF480346 | Wedlich-Söldner et al. (2002) |
| Kin4 | Kinesin-4 (Chromokin.) | 37/10 | Unknown | N | XM_400772 | This study |
| Kin5 | Kinesin-5 (BimC) | 47/17 | Mitosis (?) | N | XM_402340 | This study |
| Kin6 | Kinesin-6 (MKLP) | 29g/13 | Mitosis (?) | N | XM_402342 | This study |
| Kin7af | Kinesin-7 (CENP-E) | 45/7 (10)g | Unknown | I | U92844 | Lehmler et al. (1997) |
| Kin7b | Kinesin-7 (CENP-E) | 35/3 (6)g | Unknown | I | XM_402139 | This study |
| Kin9 | Orphan | 25g/6 | Unknown | N | XM_404042 | This study |
| Kin8 | Kinesin-8 (Kip3) | 48/10 | Unknown | N | XM_399175 | This study |
| Kin14 | Kinesin-14 (Kar3/Ncd) | 44/13 | Mitosis (?) | C | AJ605564 | This study |
Classification according to Lawrence et al. (2004).
Percentage of identity with mouse homologue (Kin1/3/4/5/6/13), human (Kin7a/7b), and bakers yeast (Kin8).
Assumed functions indicated by (?).
I, internal; N, NH3-terminal; C, COOH-terminal; note that predicted start of the ORFs is solely based on comparison with other kinesins.
Previously named Kin2 [Lehmler et al. (1997)].
Previously named Kin1 [Lehmler et al. (1997)].
Homology after removal of insertion within the motor domain; in parentheses homology to tail of A. nidulans KipA.
Only Two Kinesin Motors Are Required for Hyphal Growth
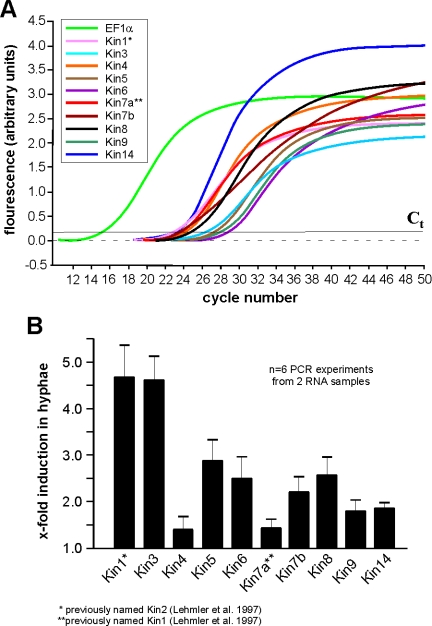
In yeast-like cells of U. maydis, MT minus-ends are focused to the growing bud (Straube et al., 2003) and in growing cultures ∼7–8% of all cells are in mitosis (Straube et al., 2005). In contrast, hyphae of U. maydis are cell cycle arrested (Garcia-Muse et al., 2003) and MT plus-ends are focused toward the growing tip, indicating that kinesin organelle transporters might play a role in tip growth. To gain insight into which kinesins might be important in both growth forms, we analyzed the expression levels of all kinesin genes in yeast-like cells and hyphae by real-time PCR. The Ct values (threshold cycle above background indicating beginning of logarithmic amplification) for all genes ranged between 20 and 30 cycles in real-time PCR on RNA from yeast-like and hyphal cells (Figure 3A). This is clearly above the nonspecific background and indicates that all genes are expressed under the conditions tested. Consistent with an assumed role in polar growth, the organelle motors Kin1 and Kin3 show a significant up-regulation in hyphal cells. None of the mitotic kinesins were down-regulated, and the putative mitotic BimC-like kinesin Kin5 and MKLP-like Kin6 were even up-regulated (Figure 3B). This is surprising, because the hyphae of U. maydis are cell cycle arrested in G2 (Garcia-Muse et al., 2003), which suggests that mitotic kinesins should not be important in these cells. However, indications exist for a mitosis-independent function of these motors in motility of MTs in axons (Ferhat et al., 1998; Yu et al., 2000), which raises the possibility that Kin5 participates in similar processes in hyphae of U. maydis.
Figure 3.
Real-time PCR analysis of expression of kinesins in U. maydis. (A) Graph showing fluorescent intensities relative to PCR cycles, by using complete RNA from the filamentously growing strain AB33, specific primers for all kinesins (Kin1–Kin14) and the housekeeping gene EF1-α. Amplification increases after 20–28 cycles indicating that all genes are expressed. Ct: threshold cycle above background indicating beginning of logarithmic amplification. (B) Comparison of the expression of all kinesins in filamentously growing AB33 and yeast-like control strain AB34 under the same growth conditions demonstrate that kin1 and kin3 are up-regulated in hyphal growth. This corresponds well with the important role of these kinesins in filamentous growth.
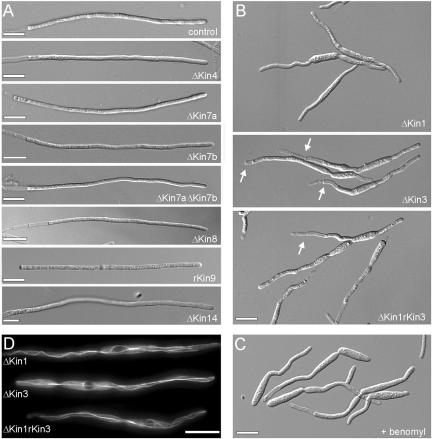
To further analyze the role of kinesins in hyphal growth, we generated null mutants of all kinesins that were implied in organelle traffic or of unknown function and characterized their morphological phenotype. All mutant strains were confirmed by Southern blotting as well as analytical PCR (our unpublished data). In yeast-like cells, only Δkin3 mutants showed a characteristic cell separation defect (Wedlich-Söldner et al., 2002), whereas all other mutants had no significant defect (our unpublished data). Even in hyphae, most kinesin null mutants also were without phenotype (Figure 4A). This was most surprising, because fungal members of the CENP-E family are involved in polar growth of S. pombe and Aspergillus nidulans (Browning et al., 2000; Konzack et al., 2004). Due to their similarity, we considered it likely that Kin7a and Kin7b have redundant functions. Therefore, we generated Δkin7aΔkin7b mutant strains (see Table 1 for details). However, even these double mutants formed regular yeast-like cells (our unpublished data) and hyphae that grew at one cell pole and formed vacuolated sections at the rear pole of the tip cell (Figure 4A), indicating that these Kinesin-7 members have no critical role in tip growth.
Figure 4.
Morphology phenotype of kinesin null mutants. (A) Most kinesin null mutants neither have defects in the yeast-like stage (our unpublished data) nor show impaired hyphal growth. Note that most mutants are in the ABB33 background (see Materials and Methods), whereas Δkin14 hyphae were obtained from crossing compatible mutant strains on charcoal plates. All bars, 10 μm. (B) Δkin1 and Δkin3 mutant hyphae often grow in a bipolar manner and are unable to form long filaments. No additional defects were seen in Δkin1rkin3 double mutants. Note that, for unknown reason, the double mutants again formed vacuolated hyphal parts. Bar, 10 μm. (C) Treatment of AB33 hyphae with 10 μM benomyl for 12 h resulted in short hyphae that showed phenotypic similarities to Δkin1 and Δkin3 mutant hyphae. Bar, 10 μm. (D) GFP-labeled MTs were normal in Δkin1 and Δkin3 mutants (see Steinberg et al., 2001 for MT organization in wild-type cells), which demonstrates that the morphological phenotype of the kinesin mutants is not due to a global defect in MT organization. Bar, 10 μm.
In contrast to all other kinesins, the deletion of Kin1 (Kinesin-1) and Kin3 (Kinesin-3) in the filamentously growing strain AB33 resulted in bipolar growth and the formation of very short hyphae, and only Δkin3 mutants formed vacuolated regions (Figure 4B, arrows). Hyphae of a conditional Δkin1rkin3 double mutant, in which the kin3 expression was repressed upon shift from arabinose to glucose containing medium (our unpublished data), showed essentially the same phenotype, raising the possibility that both motors perform functions in the same cellular pathway. To check this, we overexpressed Kin3 by growing strain AB33ΔKin1rKin3 in arabinose-containing medium. This treatment induces the crg promoter and strongly increased Kin3 levels (our unpublished data, but see Figure 6E for example of crg controlled expression of kin3). However, no rescue of the mutant phenotype was observed (our unpublished data). The morphological defects of kin1 and kin3 null mutants were reminiscent of hyphal growth in the presence of benomyl (Figure 4C), a fungicide that efficiently disrupts the MT cytoskeleton in these cells and drastically reduces hyphal growth (Fuchs et al., 2005). This indicated that the deletion of either kin1 or kin3 has severe defects that reflected growth in the absence of MTs, suggesting that both Δkin1 and Δkin3 could be defective in MT organization. We checked this possibility by labeling MTs with GFP-αtubulin (Steinberg et al., 2001) and found that MTs were not significantly altered in these mutants (Figure 4D; MTs labeled by GFP-αtubulin). This suggests that both kinesins have essential roles in MT-based traffic to the growing tip, whereas the other kinesins have either redundant functions or are not involved in hyphal growth.
Figure 6.
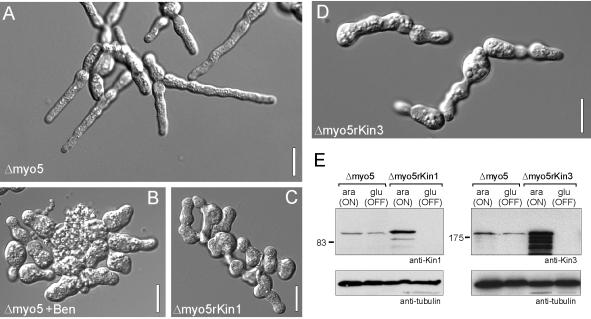
Phenotype of conditional Δmyo5rkin1 and Δmyo5rkin3 mutants. (A) Although deletion of Δmyo5 mutants impaired hyphal morphology, cells of strain AB33ΔMyo5 are still able to grow polarized. Bar, 10 μm. (B) Hyphal polarity is lost in the absence of MTs after overnight treatment with 10 μM benomyl, indicating that both F-actin and MTs are needed for polar hyphal growth. Bar, 10 μm. (C) Hyphal growth of Δmyo5rkin1 double mutants (strain AB33ΔMyo5rKin3) was induced for ∼16 h by growing cells in NM-G. This medium also repressed kin1 (see E) that is placed under the control of the crg promoter. In the absence of Kin1, myo5 null mutant lost their ability to form hyphae, indicating that both Myo5 and Kin1 activity is needed to maintain polarized hyphal growth. Bar, 10 μm. (D) In similar experiments, filamentous growth was induced, whereas kin3 was repressed in Δmyo5rkin3 mutants (strain AB33ΔMyo5rKin3; see below, E). In the absence of Kin3 polarized hyphal growth was also almost abolished, arguing that Kin3 also cooperates with Myo5 to maintain tip growth. Bar, 10 μm. (E) Western blots showing that Kin1 and Kin3 expression is repressed in CM-G (glu, OFF), whereas high expression levels were found in CM-A (ara, ON).
The Kinesins and a Class V Myosin Cooperate in Polar Growth
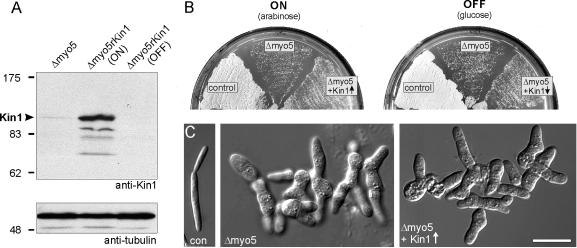
In S. cerevisiae, high expression of Smy1p, a distant relative of Kinesin-1, rescued growth defects of mutants defective in a class V myosin (Lillie and Brown, 1992). To check for a similar connection in U. maydis, we expressed kin1 under the control of the crg-promoter in a Δmyo5 mutant strain that showed severe growth defects due to the deletion of the only class V myosin in U. maydis (Weber et al., 2003). Western blots using specific anti-Kin1 antibodies raised against peptides in the motor head and tail (Straube, Hause, and Steinberg, unpublished data) revealed that, under inductive conditions, protein levels were ∼150-fold increased compared with wild-type cells (Figure 5A). However, high expression of Kin1 did not suppress the Δmyo5 growth defect (Figure 5B; con: wild-type cell; Δmyo5: FB2ΔMyo5; Δmyo5 + Kin1↑; FB2ΔMyo5rKin1) nor rescue the abnormal morphology of myo5 null mutants (Figure 5C).
Figure 5.
Effect of high levels of Kin1 on the phenotype of a class V myosin mutant. (A) An affinity-purified rabbit antibody raised against peptides in the motor domain and the tail of Kin1 recognizes a faint band of ∼110 kDa in extracts of myo5 null mutants (control). Expression of Kin1 under the control of the crg-promoter in the presence of arabinose shows strongly induced expression (Δmyo5rKin1, ON), but all expression of kin1 was repressed in CM-G (Δmyo5rKin1, OFF). (B) On CM-A plates (ON, arabinose), wild-type cells form colonies after 2 d at 28°C (control), whereas plate growth of Δmyo5 mutant cells was heavily impaired (Δmyo5). High levels of Kin1 did not rescue this phenotype (Δmyo5 + Kin1↑). (C) Yeast-like control cells form elongated “cigar-shaped” cells that grow by polar budding (con). Deletion of myo5 led to severe defects in polar growth and aggregates of cells that still have a tendency to grow in a polar manner (Weber et al., 2003). In CM-A medium, high levels of Kin1 did not rescue the Δmyo5 phenotype (Δmyo5 + Kin1↑). Bar, 10 μm.
We noticed that in the absence of Kin1 and Kin3 or even MTs hyphae are still able to undergo a certain degree of polar growth (Figure 4, B–D). This suggested that an MT-independent mechanism participates in polar growth. Indeed, indirect evidence exists that actin forms long tracks in hyphae (Fuchs et al., 2005) and Δmyo5 hyphae show defective morphology and reduced ability to grow polarized (Weber et al., 2003; Figure 6A, strain AB33ΔMyo5). Therefore, we speculated that Myo5-based transport cooperates with a MT-dependent transport mechanism in hyphal growth. Indeed, disruption of MTs abolished the residual growth of AB33ΔMyo5 hyphae (Figure 6B; Δmyo5 + Ben) and led to aggregates of rounded cells. A similar loss of cell polarity was found in conditional AB33ΔMyo5rKin1 double mutants. Growth of these cells in glucose-containing medium led to repression of kin1 and no Kin1 motor was detected in cell extracts (Figure 6E; 16 h after medium shift). The absence of Kin1 abolished hyphal growth and led to apolar cell aggregates (Figure 6C, Δmyo5rKin1). In neurons it was shown that Kinesin-1 and Myosin-V specifically interact, suggesting that the synthetic phenotype of a double mutant in both motors is due to a specific functional or structural interaction. However, when precipitating GFP-Myo5 with anti-GFP antibodies, we never copurified Kin1 (our unpublished data), which argues against a direct interaction of both motors. Alternatively, we considered it possible that the loss of hyphal growth in Δmyo5rkin1 mutants simply reflected the need for both actin- and MT-based transport in hyphal growth. Kin3 null mutant hyphae also display a defect in filamentous growth, but to our knowledge no specific interaction of Kinesin-3 and Myosin-V was reported. Thus we concluded that a double mutant in kin3 and myo5 could still grow polar in case the loss of polarity in Δmyo5rkin1 is due to a specific interaction between Kin1 and Myo5. In contrast, a loss of filamentous growth in Δmyo5rkin3 double mutants would argue for a broader cooperation between actin and microtubule motors. In extracts of Δmyo5rkin3 double mutant (AB33ΔMyo5rKin3) repression of Kin3 in CM-G medium was checked by Western Blot analysis using specific antibodies raised against recombinant motor domains (Straube, Hause, and Steinberg, unpublished data). Next, we repressed kin3 expression by switching cells to glucose medium (Figure 6E) and monitored the ability of hyphae to grow in a polarized manner. Again, cells were almost unable to form hyphae, which suggests that the cooperation of Kinesin-3, Kinesin-1, and Myosin-V is essential to maintain polarized hyphal growth.
Kinesin-1, Kinesin-3, and Myosin-V Accumulate in the Hyphal Apex
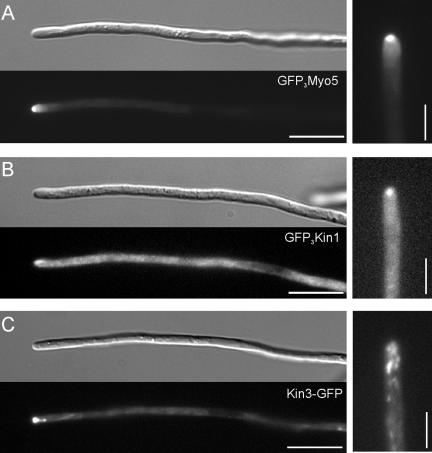
To better visualize the steady-state distribution of Kin1 and Myo5, we N-terminally tagged all motors with a triple-GFP tag, whereas for Kin3 a C-terminal single GFP was sufficient for visualization (Wedlich-Söldner et al., 2002). All GFP-tagged proteins were able to at least partially rescue the respective mutant phenotypes (our unpublished data), which demonstrated that they are biologically active. Consistent with previous reports (Weber et al., 2003), the GFP3-Myo5 fusion protein localized to a defined dot in the hyphal apex (Figure 7A). A fusion protein of triple-GFP and Kin1 also concentrates at the hyphal tip (Figure 7B), although a significant portion of GFP3-Kin1 was found equally distributed in the cytoplasm. Finally, we observed Kin3-GFP as rapidly moving dots that also accumulated near the hyphal tip. However, this cluster was much more dynamic, usually less focused and, in contrast to GFP3-Myo5 and GFP3-Kin1, not located at the very tip of the hypha (Figure 7C).
Figure 7.
Secretion and localization defects in Δkin1 and Δmyo5 mutants. (A) A fusion protein of a triple GFP-tag fused to Myo5 localizes to the hyphal apex. Note that the fusion protein localizes to a focused dot at the hyphal tip, where the Spitzenkörper is located (Lehmler et al., 1997). Bars, 10 and 3 μm. (B) A fusion protein of a triple GFP-tag fused to Kin1 also localizes to the hyphal apex, but the signal is often more dispersed and a strong cytoplasmic background was seen. Bar, 10 and 3 μm. (C) A fusion protein of a GFP-tag fused to Kin3 localized as rapidly moving dots along the length of the hyphae but also concentrates near the hyphal tip. Note that the apical Kin3-GFP does not reach the apex and is more dispersed. Bar, 10 and 3 μm.
The tip localization of GFP3-Myo5 and GFP3-Kin1 was very fragile, because the focused motors rapidly dispersed by active retrograde motility during microscopic observation (our unpublished data). This behavior as well as the apical localization is consistent with that of the Spitzenkörper, an accumulation of secretory vesicles that is typical for growing fungal hyphae (Grove and Bracker, 1970; Harris et al., 2005) and that also was described in the tip of U. maydis hyphae (Lehmler et al., 1997). In addition, it was suggested that conventional kinesin is involved in tip-ward transport of secretory vesicles and participates in polar exocytosis in Neurospora crassa (Seiler et al., 1997, 1999). Therefore, we analyzed the secretion of the marker enzyme acid phosphatase in filamentously growing strains AB33, AB33ΔKin1, AB33ΔKin3, and AB33ΔMyo5 and the double mutant AB33ΔKin1rKin3 mutant strains (Figure 8). In these experiments, Δkin1 mutants showed no significant reduction in acid phosphatase secretion (ΔKin1; P: 0.1807), whereas kin3 null mutant were reduced to ∼50% (Figure 8, ΔKin3). Consequently, a double mutant did not show additional reduction in secreted enzyme activity (Figure 8, ΔKin1rKin3; P: 0.222). Surprisingly, in Δmyo5 mutants, acid phosphatase secretion was even higher than in control cells (Figure 8, ΔMyo5), and this was also found in Δmyo5rkin1 double mutants (our unpublished data). Currently, we have no explanation for this phenomenon.
Figure 8.
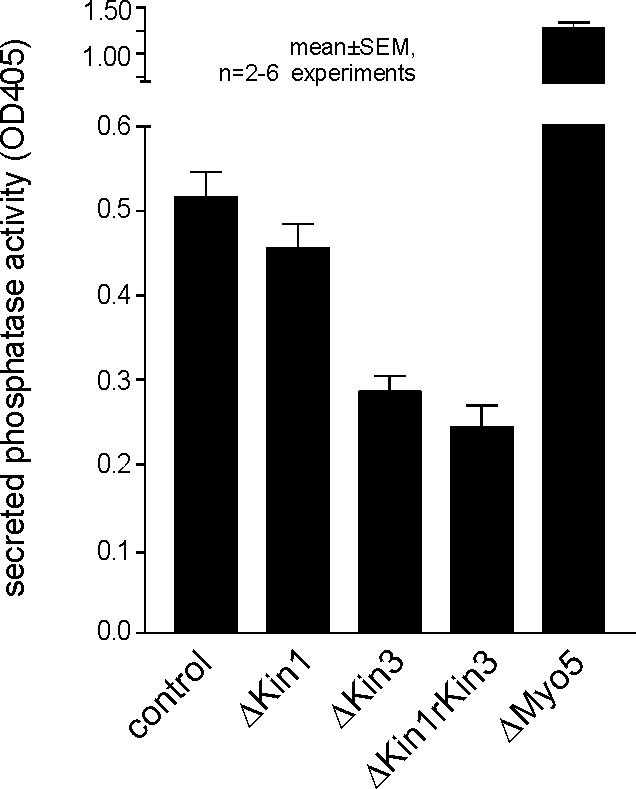
Quantitative analysis of acid phosphatase secretion in hyphae of U. maydis. For analysis of the extracellular activity of the marker enzyme acid phosphatase, cells were incubated in phosphate-free medium and the release of nitrophenon from p-nitrophenyl phosphate was measured as an indication of secreted enzyme activity. Δkin1 mutants were not significantly impaired in secretion of acid phosphatase, whereas secretion was found to be reduced in Δkin3 mutants. Consistently, Δkin1rkin3 double mutants show no additional phenotype. For unknown reasons, deletion of myo5 even increased secretion, a phenomenon that was also found in Δmyo5rkin1 mutants (our unpublished data).
DISCUSSION
Phenotypically, fungal hyphae are very similar to the long neuronal extensions. In the axon, most microtubule plus-ends elongate toward the apical terminus (Heidemann et al., 1981), and consequently plus-end-directed kinesins are most important for anterograde long-distance transport from the cell body to the synapsis (Brady, 1995; Hirokawa et al., 1998). Hyphal cells of U. maydis can reach a length of 150–160 μm (Steinberg et al., 1998), and growth occurs at the apical tip. MT plus-ends are directed toward the growing hyphal apex, and U. maydis, as well as other filamentous fungi contain more kinesins than the yeasts S. cerevisiae and S. pombe (reviewed in Schoch et al., 2003; this study). It was suggested that this could reflect the increased need for long-distance transport to the hyphal tip in filamentous fungi (Steinberg, 2000; Goldstein, 2001). Indeed, the well-established membrane transporters Kinesin-1 (Brady, 1985; Scholey et al., 1985; Vale et al., 1985) and Kinesin-3 (Otsuka et al., 1991; Aizawa et al., 1992), which deliver axonal vesicles (reviewed in Hirokawa and Takemura, 2004), are absent from S. cerevisiae, but they are described for filamentous fungi (Steinberg and Schliwa, 1995; Lehmler et al., 1997; Wu et al., 1998; Wedlich-Söldner et al., 2002). Our results add further support to this notion, because both kin1 and kin3 show increased expression in hyphae and are essential for filamentous growth. Given that Δkin1 and Δkin3 mutants have similar phenotypes, both kinesins might perform overlapping functions or depend on each other. Indeed, we have evidence that Kin1 activity is needed for Kin3-recycling back to the minus-ends of MTs for another round of tip-directed transport (Schuchardt and Steinberg, unpublished data). Thus, polarized hyphal growth seems to depend on a network of motors and kinesins have a crucial role in this.
Indications exist that fungal Kinesin-1 participates in polarized secretion in N. crassa (Seiler et al., 1999) and such a role in anterograde transport corresponds well with the disappearance of an apical vesicle cluster in Kinesin-1 null mutants in N. crassa (Seiler et al., 1997) and U. maydis (Lehmler et al., 1997). Therefore, it was tempting to speculate that Kin1 participates in tip-ward delivery of secretory vesicles in hyphae. The apical localization of GFP3-Kin1 supports this notion, because it indicates that this kinesin transports vesicles to the tip that accumulate in the apical Spitzenkörper (Figure 9). This vesicle cluster in the hyphal tip is thought to be a reservoir for secretory vesicles (Bartnicki-Garcia et al., 1995). Our localization data reveal that the GFP-fusion proteins accumulate in the growing tip of the hypha, which suggests that motors, including Kinesin-1 and Myosin-V, are mainly bound to the vesicles and thus accumulate in the Spitzenkörper. Kinesin-3, the other kinesin required for tip growth, also accumulates in the hyphal tip, but its distribution was found to be less focused, suggesting that it might not be involved in secretion (Figure 9). However, our colorimetric assays clearly indicate that Kin3 participates in acid phosphatase secretion. Such a role of Kinesin-3 in secretion would be reminiscent of Caenorhabditis elegans neurons, where Kinesin-3 is thought to deliver synaptic vesicles to the growth cone (Hall and Hedgecock, 1991). It was previously described that Kin3 is a motor for early endosome motility (Wedlich-Söldner et al., 2002). Our finding that Kin3 also participates in exocytosis indicates that this motor, and most probably also Kin1, transport a broad spectrum of organelles and other compounds to the growing tip. Thus, to understand tip growth it will be necessary to identify other cargoes of these kinesins in hyphae.
Figure 9.
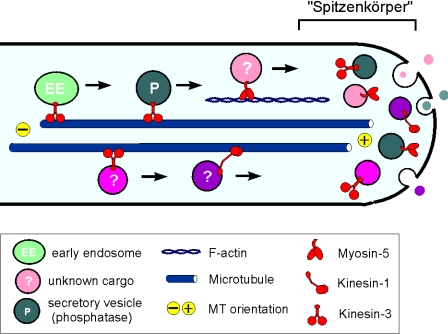
Model on the role of Kin1, Kin3, and Myo5 in polar fungal growth. Polar hyphal growth is supported by long-distance delivery of growth supplies to the expanding hyphal tip. MT plus-ends are directed to the growing tip, and Kinesin-1, Kinesin-3, and Myosin-V use cytoskeletal tracks to deliver their cargo to the growth region. The ability to grow polarized is only abolished when both Myosin-V and either Kinesin-1 or Kinesin-3 are blocked. Kinesin-3 most likely transports acid phosphatase containing vesicles, as well as early endosomes (Wedlich-Söldner et al., 2002), whereas the putative exocytic cargo of Kinesin-1 and Myosin-V is unknown. In the hyphal apex, secretory vesicles and attached motors cluster to form the fungal Spitzenkörper that is thought to mediate controlled local exocytosis and thereby determines tip growth.
The Type V Myosin Myo5 Cooperates with Kin1 and Kin3 in Polarized Hyphal Growth
Our results demonstrate that Δkin1 and Δkin3 hyphae are still able to grow polarized, and even disruption of all MT-dependent transport did not abolish filament formation, although growth rates were largely decreased and hyphae remain much shorter (Fuchs et al., 2005). This suggested that additional MT-independent mechanisms support filamentous growth. U. maydis hyphae contain long actin filaments and MTs that both are required for hyphal growth (Fuchs et al.; 2005). In addition, previous results have shown that Myo5, the only class V myosin in U. maydis (Weber et al., 2003), participates in tip growth and is therefore a good candidate for anterograde transport in hyphae. Indeed, disruption of MTs in Δmyo5 hyphae abolished cell polarity. A similar situation was found in the absence of Myo5 and Kin1 or Kin3, respectively. This additive phenotype suggests that Myosin-V and the kinesins independently deliver components to the hyphal apex that are required for directed tip growth (Figure 9, vesicles are marked by “?”). This notion is further supported by the observation that high levels of Kin1 are not able to complement for the absence of Myo5, demonstrating that Kin1 cannot replace Myo5 in anterograde transport. In mouse cells and squid neurons, Kinesin-1 and class V myosins motors directly interact (Huang et al., 1999; Stafford et al., 2000). However, in coimmunoprecipitation assays we did not find evidence for a physical interaction (our unpublished data). Thus, it is tempting to speculate that in U. maydis Kin1 and Myo5 transport different secretory vesicle, or other cargo, that are essential for hyphal growth.
CONCLUSIONS
Fungal hyphae are highly polarized cells that, similar to axons, direct MT plus-ends to the apical end of the cell. This MT organization implies fungal Kinesin-1 and Kinesin-3 in long-distance anterograde transport, and we surprisingly found that among all kinesins only these kinesins have an essential role in hyphal tip growth. Kinesin-1, as well as the class V myosin Myo5 localize to the hyphal apex, suggesting that they participate in formation of the Spitzenkörper, which is thought to be a fungal specific accumulation of secretory vesicles. Interestingly, both MT-based transport and Myo5-based transport is required for polarized hyphal growth, because double mutants lacking Kinesin-1 or Kinesin-3 and Myosin-V lose polarity. Although we found that Kinesin-3 is involved in acid phosphatase secretion, as well as endosome motility (Wedlich-Söldner et al., 2002), the actual cargoes of Kinesin-1 and Myosin-V remain to be uncovered. Further studies on the identification of these cargoes are needed to enhance our understanding of the functional interplay of the MT- and actin-based transport machinery in hyphal growth.
Supplementary Material
Acknowledgments
We are grateful to U. Fuchs and A. Straube for fruitful discussions and technical help. P. Happel is acknowledged for excellent technical support and R. Wedlich-Söldner for generating the Δkin1Δkin3 mutant strain. U. Theisen is acknowledged for critically reading the manuscript. We thank R. Kahmann for continuous support. This work was supported by a grant to G. S. (DFG, SPP 1111).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–04–0272) on August 24, 2005.
Abbreviations used: aa, amino acid; GFP, green fluorescent protein; MT, microtubule; ORF, open reading frame; RFP, red fluorescent protein.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aizawa, H., Sekine, Y., Takemura, R., Zhang, Z., Nangaku, M., and Hirokawa, N. (1992). Kinesin family in murine central nervous system. J. Cell Biol. 119, 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks, G. R., Shelton, P. A., Kanuga, N., Holden, D. W., and Spanos, A. (1993). The Ustilago maydis nar1 gene encoding nitrate reductase activity: sequence and transcriptional regulation. Gene 131, 69–78. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S., Bartnicki, D. D., Gierz, G., Lopez-Franco, R., and Bracker, C. E. (1995). Evidence that Spitzenkorper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp. Mycol. 19, 153–159. [DOI] [PubMed] [Google Scholar]
- Bottin, A., Kamper, J., and Kahmann, R. (1996). Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 253, 342–352. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., König, J., Julius, C., and Feldbrügge, M. (2004). A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272, 488. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Weinzierl, G., Kamper, J., and Kahmann, R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42, 1047–1063. [DOI] [PubMed] [Google Scholar]
- Brady, S. T. (1985). A novel brain ATPase with properties expected for fast axonal transport. Nature 317, 73–75. [DOI] [PubMed] [Google Scholar]
- Brady, S. T. (1995). A kinesin medley: biochemical and functional heterogeneity. Trends Cell Biol. 5, 159–164. [DOI] [PubMed] [Google Scholar]
- Brazer, S. C., Williams, H. P., Chappell, T. G., and Cande, W. Z. (2000). A fission yeast kinesin affects Golgi membrane recycling. Yeast 16, 149–166. [DOI] [PubMed] [Google Scholar]
- Brown, S. S. (1999). Cooperation between microtubule- and actin-based motor proteins. Annu. Rev. Cell Dev. Biol. 15, 63–80. [DOI] [PubMed] [Google Scholar]
- Browning, H., Hayles, J., Mata, J., Aveline, L., Nurse, P., and McIntosh, J. R. (2000). Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 151, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhat, L., Cook, C., Chauviere, M., Harper, M., Kress, M., Lyons, G. E., and Baas, P. W. (1998). Expression of the mitotic motor protein Eg5 in postmitotic neurons: implications for neuronal development. J. Neurosci. 18, 7822–7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, U., Manns, I., and Steinberg, G. (2005). Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell 16, 2746–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse, T., Steinberg, G., and Perez-Martin, J. (2003). Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot. Cell 2, 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann, A., and Emons, A. M. (2000). The cytoskeleton in plant and fungal cell tip growth. J. Microsc. 198, 218–245. [DOI] [PubMed] [Google Scholar]
- Goldstein, L. S. (2001). Molecular motors: from one motor many tails to one motor many tales. Trends Cell Biol. 11, 477–482. eton. J. Cell Biol. 133, 1277–1291. [DOI] [PubMed] [Google Scholar]
- Grove, S. N., and Bracker, C. E. (1970). Protoplasmic organization of hyphal tips among fungi: vesicles and Spitzenkorper. J. Bacteriol. 104, 989–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D. H., and Hedgecock, E. M. (1991). Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65, 837–847. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., Read, N. D., Roberson, R. W., Shaw, B., Seiler, S., Plamann, M., and Momany, M. (2005). Polarisome meets spitzenkorper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4, 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles, J., and Nurse, P. (2001). A journey into space. Nat. Rev. Mol. Cell Biol. 2, 647–656. [DOI] [PubMed] [Google Scholar]
- Heidemann, S. R., Landers, J. M., and Hamborg, M. A. (1981). Polarity orientation of axonal microtubules. J. Cell Biol. 91, 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N., Noda, Y., and Okada, Y. (1998). Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr. Opin. Cell Biol. 10, 60–73. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., and Takemura, R. (2004). Kinesin superfamily proteins and their various functions and dynamics. Exp. Cell Res. 301, 50–59. [DOI] [PubMed] [Google Scholar]
- Hodge, T., and Cope, M. J. (2000). A myosin family tree. J. Cell Sci. 113, 3353–3354. [DOI] [PubMed] [Google Scholar]
- Huang, J. D., Brady, S. T., Richards, B. W., Stenolen, D., Resau, J. H., Copeland, N. G., and Jenkins, N. A. (1999). Direct interaction of microtubule- and actin-based transport motors. Nature 397, 267–270. [DOI] [PubMed] [Google Scholar]
- Jeong, J. W., Rhee, D. K., Cho, S. Y., Hae, K. L., Kim, D. U., Won, M., and Kim, H. B. (2002). Cloning and characterization of the kinesin-related protein, Krp1p, in Schizosaccharomyces pombe. Mol. Cell 13, 389–398. [PubMed] [Google Scholar]
- Kämper, J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271, 103–110. [DOI] [PubMed] [Google Scholar]
- King-Smith, C., Paz, P., Lee, C. W., Lam, W., and Burnside, B. (1997). Bidirectional pigment granule migration in isolated retinal pigment epithelial cells requires actin but not microtubules. Cell Motil. Cytoskeleton 38, 229–249. [DOI] [PubMed] [Google Scholar]
- Klopfenstein, D. R., Holleran, E. A., and Vale, R. D. (2002). Kinesin motors and microtubule-based organelle transport in Dictyostelium discoideum. J. Muscle Res. Cell Motil. 23, 631–638. [DOI] [PubMed] [Google Scholar]
- Konzack, S., Rischitor, P. E., Enke, C., and Fischer, R. (2004). The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell 16, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov, S. A., Langford, G. M., and Weiss, D. G. (1992). Actin-dependent organelle movement in squid axoplasm. Nature 356, 722–725. [DOI] [PubMed] [Google Scholar]
- Langford, G. M. (2002). Myosin-V, a versatile motor for short-range vesicle transport. Traffic 3, 859–865. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmler, C., Steinberg, G., Snetselaar, K. M., Schliwa, M., Kahmann, R., and Bolker, M. (1997). Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 16, 3464–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie, S. H., and Brown, S. S. (1992). Suppression of a myosin defect by a kinesin-related gene. Nature 356, 358–361. [DOI] [PubMed] [Google Scholar]
- Lillie, S. H., and Brown, S. S. (1998). Smy1p, a kinesin-related protein that does not require microtubules. J. Cell Biol. 140, 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermall, V., Post, P. L., and Mooseker, M. S. (1998). Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279, 527–533. [DOI] [PubMed] [Google Scholar]
- Ogawa, T., Nitta, R., Okada, Y., and Hirokawa, N. (2004). A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell 116, 591–602. [DOI] [PubMed] [Google Scholar]
- Otsuka, A. J., Jeyaprakash, A., Garcia-Anoveros, J., Tang, L. Z., Fisk, G., Hartshorne, T., Franco, R., and Born, T. (1991). The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron 6, 113–122. [DOI] [PubMed] [Google Scholar]
- Pollock, N., de Hostos, E. L., Turck, C. W., and Vale, R. D. (1999). Reconstitution of membrane transport powered by a novel dimeric kinesin motor of the Unc104/KIF1A family purified from Dictyostelium. J. Cell Biol. 147, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., and Nurse, P. (1998). Regulation of cell polarity by microtubules in fission yeast. J. Cell Biol. 142, 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch, C. L., Aist, J. R., Yoder, O. C., and Gillian Turgeon, B. (2003). A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet. Biol. 39, 1–15. [DOI] [PubMed] [Google Scholar]
- Scholey, J. M., Porter, M. E., Grissom, P. M., and McIntosh, J. R. (1985). Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature 318, 483–486. [DOI] [PubMed] [Google Scholar]
- Seiler, S., Nargang, F. E., Steinberg, G., and Schliwa, M. (1997). Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16, 3025–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., Plamann, M., and Schliwa, M. (1999). Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9, 779–785. [DOI] [PubMed] [Google Scholar]
- Stafford, P., Brown, J., and Langford, G. M. (2000). Interaction of actin- and microtubule-based motors in squid axoplasm probed with antibodies to myosin V and kinesin. Biol. Bull. 199, 203–205. [DOI] [PubMed] [Google Scholar]
- Steinberg, G. (2000). The cellular role of molecular motors in fungi. Trends Microbiol. 4, 162–168. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., and McIntosh, J. R. (1998). Effects of the myosin inhibitor 2,3-butanedione monoxime on the physiology of fission yeast. Eur. J. Cell Biol. 77, 284–293. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., and Schliwa, M. (1995). The Neurospora organelle motor: a distant relative of conventional kinesin with unconventional properties. Mol. Biol. Cell 6, 1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, G., Schliwa, M., Lehmler, C., Bölker, M., Kahmann, R., and McIntosh, J. R. (1998). Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111, 2235–2246. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., Wedlich-Söldner, R., Brill, M., and Schulz, I. (2001). Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 114, 609–622. [DOI] [PubMed] [Google Scholar]
- Straube, A., Brill, M., Oakley, B. R., Horio, T., and Steinberg, G. (2003). Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol. Biol. Cell 14, 642–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube, A., Enard, W., Berner, A., Wedlich-Soldner, R., Kahmann, R., and Steinberg, G. (2001). A split motor domain in a cytoplasmic dynein. EMBO J. 20, 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube, A., Weber, I., and Steinberg, G. (2005). A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 24, 1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale, R. D., Reese, T. S., and Sheetz, M. P. (1985). Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, I., Gruber, C., and Steinberg, G. (2003). A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 15, 2826–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner, R., Straube, A., Friedrich, M. W., and Steinberg, G. (2002). A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 21, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win, T. Z., Gachet, Y., Mulvihill, D. P., May, K. M., and Hyams, J. S. (2001). Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114, 69–79. [DOI] [PubMed] [Google Scholar]
- Wu, Q., Sandrock, T. M., Turgeon, B. G., Yoder, O. C., Wirsel, S. G., and Aist, J. R. (1998). A fungal kinesin required for organelle motility, hyphal growth, and morphogenesis. Mol. Biol. Cell 9, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, T. J., Compton, D. A., Wise, D., Zinkowski, R. P., Brinkley, B. R., Earnshaw, W. C., and Cleveland, D. W. (1991). CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., Cook, C., Sauter, C., Kuriyama, R., Kaplan, P. L., and Baas, P. W. (2000). Depletion of a microtubule-associated motor protein induces the loss of dendritic identity. J. Neurosci. 20, 5782–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.