Abstract
The Saccharomyces cerevisiae Taz1 protein is the orthologue of human Tafazzin, a protein that when inactive causes Barth Syndrome (BTHS), a severe inherited X-linked disease. Taz1 is a mitochondrial acyltransferase involved in the remodeling of cardiolipin. We show that Taz1 is an outer mitochondrial membrane protein exposed to the intermembrane space (IMS). Transport of Taz1 into mitochondria depends on the receptor Tom5 of the translocase of the outer membrane (TOM complex) and the small Tim proteins of the IMS, but is independent of the sorting and assembly complex (SAM). TAZ1 deletion in yeast leads to growth defects on nonfermentable carbon sources, indicative of a defect in respiration. Because cardiolipin has been proposed to stabilize supercomplexes of the respiratory chain complexes III and IV, we assess supercomplexes in taz1Δ mitochondria and show that these are destabilized in taz1Δ mitochondria. This leads to a selective release of a complex IV monomer from the III2IV2 supercomplex. In addition, assembly analyses of newly imported subunits into complex IV show that incorporation of the complex IV monomer into supercomplexes is affected in taz1Δ mitochondria. We conclude that inactivation of Taz1 affects both assembly and stability of respiratory chain complexes in the inner membrane of mitochondria.
INTRODUCTION
Barth Syndrome is an X-linked recessive disorder that is characterized by cardioskeletal myopathy, neutropenia, increased urine levels of 3-methylglutaconic acid, abnormal mitochondria, and respiratory chain dysfunction (Barth et al., 1983, 1996; Kelley et al., 1991; D'Adamo et al., 1997). The disease is often fatal in infancy and early childhood because of cardiac failure and bacterial infections (Barth et al., 2004). Bione et al. (1996) identified the gene responsible, termed Tafazzin (TAZ), and localized it to region q28 on the X chromosome. Sequence analyses demonstrate that the Taz protein is highly conserved from yeast to man and that it displays sequence similarity to acyltransferases that participate in the metabolism of phospholipids (Neuwald, 1997). Phospholipid analyses of fibroblasts from patients and yeast taz1Δ mutant cells show reduced cardiolipin levels and an altered acyl chain composition (Vreken et al., 2000; Vaz et al., 2003; Xu et al., 2003; Gu et al., 2004).
Cardiolipin is formed by two glycerol-linked phosphatidyl moieties. The four acyl side chains are usually mono- and di-unsaturated fatty acids in higher eukaryotes. Remodeling of cardiolipin through a cycle of deacylation and successive reacylation leads to the final and specific acyl composition. In heart and skeletal muscle mitochondria the tetralinoleoyl species predominates (Schlame et al., 2000; Xu et al., 2003; Hatch, 2004). Cardiolipin is primarily found in the mitochondrial membranes of eukaryotes (Hoch, 1992) and the bacterial cytoplasmic membrane. Although the molecular function of cardiolipin is still unclear, several studies have suggested that it is required for the proper function of some proteins and protein complexes in the inner mitochondrial membrane (Hoch, 1992; Jiang et al., 2000; Pfeiffer et al., 2003; Palsdottir and Hunte, 2004; Zhang et al., 2005). Two functions have been proposed for cardiolipin: 1) participation in stabilization of the physical properties of the membrane (Schlame et al., 2000; Koshkin and Greenberg, 2002; Ma et al., 2004), for example, membrane fluidity and osmotic stability and 2) participation in protein function via direct interaction with membrane proteins (Schlame et al., 2000; Palsdottir and Hunte, 2004). Indeed, cardiolipin has been found in tight association with inner membrane protein complexes such as the cytochrome bc complex (complex III). As well, it has been localized to 1the contact sites of dimeric cytochrome c oxidase, and cardiolipin binding sites have also been found in the ADP/ATP carrier (AAC; for review see Palsdottir and Hunte, 2004). Recent work also suggests a role of cardiolipin in formation of respiratory chain supercomplexes (respirasomes). Analyses of the yeast respiratory chain protein supercomplexes have demonstrated that the bc1 complex (complex III) forms a dimer that associates with either one cytochrome oxidase (complex IV) monomer (III2IV supercomplex) or two monomers (III2IV2 supercomplex) (Cruciat et al., 2000; Schägger and Pfeiffer, 2000). Likewise, the F1Fo-ATPase (complex V) can dimerize to form a complex of 1.2 MDa (Arnold et al., 1998; Schägger and Pfeiffer, 2000). In higher eukaryotes, similar oligomerization has been observed (Schägger et al., 2004; Dudkina et al., 2005). The formation of supercomplexes is thought to increase the efficiency of respiration through both substrate channeling and sequestration of reactive intermediates and is thus of advantage for efficient respiration (Cruciat et al., 2000; Schägger and Pfeiffer, 2000; Zhang et al., 2005). When yeast cells are deleted for the CRD1 gene (cardiolipin synthase) cardiolipin is not synthesized and dissociation of the respiratory chain supercomplexes and oligomers of AAC is observed (Jiang et al., 2000; Zhang et al., 2002, 2005). The reduced stability of these protein complex oligomers reflects the complete loss of cardiolipin from the mitochondrial membranes (Pfeiffer et al., 2003). In contrast to crd1Δ, yeast taz1Δ mutant mitochondria still possess cardiolipin, although in reduced amounts and with altered structure (Vaz et al., 2003; Gu et al., 2004). It is so far unknown if and how the reduced levels of cardiolipin and the altered cardiolipin acyl side chain composition found in yeast taz1Δ mutant cells or in Barth patients affect the respiratory chain supercomplexes.
Here we analyze the submitochondrial localization and biogenesis of Taz1 and address the role of Taz1 for mitochondrial function. Taz1 is an integral outer mitochondrial membrane protein mainly exposed to the intermembrane space. Transport of Taz1 into mitochondria depends on the receptor Tom5 of the translocase of the outer membrane (TOM complex). In addition, Taz1 transport across the outer mitochondrial membrane requires the Tim9-Tim10 complex of the intermembrane space but is independent of the outer membrane sorting and assembly complex (SAM complex). Yeast cells deleted for TAZ1 exhibit selective growth defects indicative of respiratory chain malfunction. Thus, we assessed how the outer membrane protein Taz1 affects the function of the respiratory chain in the inner membrane. Indeed, in taz1Δ mutant mitochondria, dissociation of the respiratory chain supercomplexes is observed. A monomer of complex IV is released from the III2IV2 supercomplex, leading to increased amounts of the III2IV supercomplex. In addition to this we demonstrate that the assembly of complex IV into supercomplexes is severely affected in taz1Δ mutant mitochondria. This finding indicates that cardiolipin is critical for the biogenesis of respiratory chain supercomplexes and that the role of cardiolipin in complex oligomerization is not limited to a stabilizing effect. We suggest that the reduction of cardiolipin levels and/or altered acyl composition contribute to the pathology of Barth Syndrome patients by affecting respiratory chain supercomplexes in a similar way as described here.
MATERIALS AND METHODS
Strains and Growth Conditions
KBY1 (Taz1HA)(Mata, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, taz1::TAZ1-HA); a region encoding a hemaglutinin (HA) tag was inserted 3′ of the open reading frame (ORF) YPR140w into the chromosome to generate a fusion of Taz1 with a C-terminal HA tag as described previously (Knop et al., 1999). DAMY02 (taz1Δ) (Mata, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, taz1::kanMX4); YPH499 (WT; Mata ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; Sikorski and Hieter, 1989). Other strains used in this study have been published previously: tim10-2 (Truscott et al., 2002); tom5Δ (Dietmeier et al., 1997); and sam50-1 (Kozjak et al., 2003).
To isolate mitochondria from yeast, the strains YPH499, Δtaz1, and Taz1HA were grown on YPG medium (1% [wt/vol]) yeast extract, 2% [wt/vol] Bacto Peptone, 3% [vol/vol] glycerol) at 30°C. Mitochondria were isolated as described previously (Ryan et al., 2001).
For plate growth comparison, cells were grown in liquid medium, harvested by centrifugation, and resuspended in water, and serial dilutions of equal amounts of cells were dropped onto minimal media. Plates (SM; 6.7 g/l yeast nitrogen base [Difco, Detroit, MI] with amino acids, 25 g/l agar) containing 2% (wt/vol) glucose or 3% (vol/vol) glycerol/1% (vol/vol) ethanol as the carbon source. Growth at 30°C was determined after 2 d, and growth at 24 and 37°C after 4 d.
Membrane Potential Measurements
Membrane potential (Δψ) measurements of mitochondria were performed at a final concentration of 60 μg/ml in membrane potential buffer (0.6 M sorbitol, 0.1% [wt/vol] bovine serum albumin, 10 mM MgCl2, 0.5 mM EDTA, 20 mM KPi, pH 7.2). The Δψ was measured by using the Δψ-sensitive dye DiSC3(5) (3,3′-dipropylthiadicarbocyanine iodide, Molecular Probes; Geissler et al., 2000; Rehling et al., 2003) and was dissipated by the addition of 1 μM valinomycin.
Separation of Outer and Inner Membrane Vesicles
Isolated mitochondria were adjusted to a protein concentration of 5 mg/ml, swollen by 10-fold dilution in swelling buffer (20 mM HEPES-KOH, pH 7.2, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and incubated for 30 min on ice. After swelling, isotonic conditions were reestablished and mitochondria were sonicated. After sonication, crude membrane vesicles were isolated by centrifugation at 20,000 × g. The pellet was resuspended in 5 mM HEPES-KOH, pH 7.2, 10 mM KCl, 1 mM PMSF, loaded onto a discontinuous sucrose gradient (1.5× Vol. 0.85 M sucrose, 2.5× Vol. 1.1 M sucrose, 5× Vol. 1.35 M sucrose, 1× Vol. 1.6 M sucrose). After centrifugation at 100,000 × g for 16 h, fractions containing separated vesicles were harvested, subjected to SDS-PAGE, and analyzed by Western blotting, followed by immunodecoration.
In Vitro Transcription/Translation
PCRs were performed using genomic yeast DNA as a template for TAZ1 and COX13. PCR primers used were complementary to the 5′ (with addition of SP6 RNA polymerase binding sequence) or 3′ untranslated regions of the TAZ1 or COX13 gene. These PCR products were transcribed in vitro in the presence of SP6 RNA polymerase (Ryan et al., 2001). The RNA generated was then used to synthesize radiolabeled precursor proteins via in vitro translation in rabbit reticulocyte lysate (Amersham, Freiburg, Germany) in the presence of [35S]methionine/cysteine (Ryan et al., 2001). Cox5a was synthesized in a coupled transcription translation system (TNT Quick Coupled Transcription/Translation Systems, Promega, Madison, WI).
Miscellaneous
Blue native-PAGE analysis was performed essentially as described previously (Dekker et al., 1997). In vitro import into isolated yeast mitochondria (performed at 30°C), protease treatment of mitochondria, and carbonate extraction have been described previously (Ryan et al., 2001; Truscott et al., 2003; Frazier et al., 2004). Standard techniques were used for SDS-PAGE and Western blotting. Immune complexes were detected by enhanced chemiluminescence (Amersham). Radiolabeled proteins were detected by digital autoradiography using the Phosphor Image Technology (Amersham). In some gels irrelevant lanes have been excised digitally. Antibodies against the bc1 complex and Rieske Fe/S-protein were kindly provided by B. Trumpower (Hanover). Antibodies directed against yeast Cyt1, Cor1, and Cox4 were generated against selected peptides in rabbits. Antibodies specific for yeast Cox2 were purchased from Molecular Probes.
RESULTS
Import of Taz1 into S. cerevisiae Mitochondria
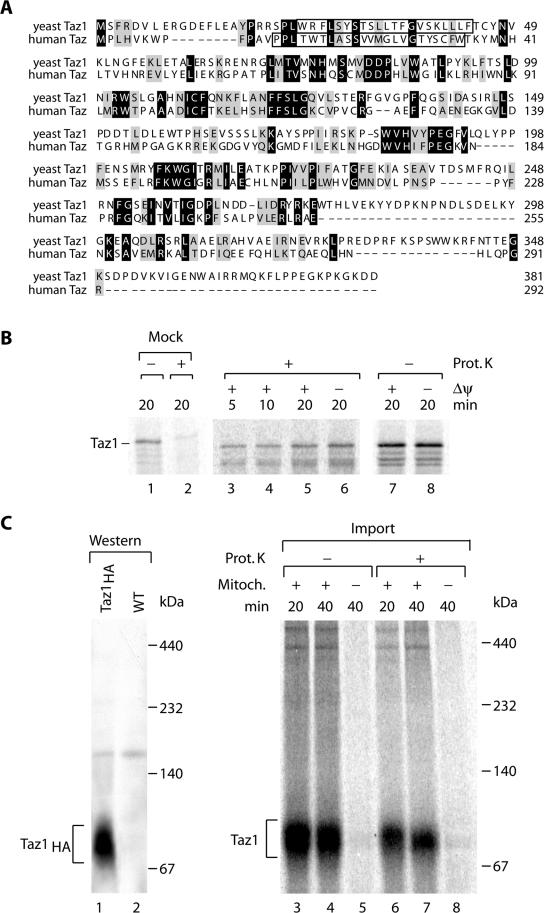
The yeast Taz1 protein (ORF YPR140w) shows significant sequence similarity to its human orthologue (Figure 1A; Neuwald, 1997; Testet et al., 2004). Taz1 has been localized to mitochondria by several methods (Sickmann et al., 2003; Ma et al., 2004; Testet et al., 2004). However, these techniques did not elucidate the mitochondrial compartment in which the protein was located. Typically, proteins that are destined for the mitochondrial matrix and some inner membrane proteins possess cleavable amino-terminal targeting signals (presequences; Neupert, 1997; Jensen and Dunn, 2002; Rehling et al., 2004). However, neither visual nor computer inspection of the primary structure of the proteins identified a predictable presequence. We synthesized Taz1 in vitro in rabbit reticulocyte lysate in the presence of [35S]methionine/ cysteine and imported it into isolated yeast mitochondria. The precursor protein associated with mitochondria (Figure 1B, lanes 3–8) and remained partially sensitive to treatment with proteinase K (Prot. K). A significant fraction remained protected against protease treatment (Figure 1B, lanes 3–6) compared with a mock reaction in the absence of mitochondria (Figure 1B, lanes 1 and 2). Accordingly, in vitro Taz1 was transported across the outer membrane of mitochondria and not processed to a faster migrating form. Dissociation of the membrane potential (Δψ) before import by the addition of the K+-ionophore valinomycin in the presence of K+ in the buffer did not affect transport of the precursor to a protease-protected location (Figure 1B, lane 6). Because protein transport across the inner mitochondrial membrane depends on Δψ (Neupert, 1997; Jensen and Dunn, 2002; Koehler, 2004; Rehling et al., 2004), we concluded that Taz1 was not transported across the inner membrane.
Figure 1.
Import of Yeast Tafazzin (Taz1) into mitochondria. (A) Sequence alignment of Tafazzins from S. cerevisiae and Homo sapiens using ClustalW. Gray, similar residues; black, identical residues. Predicted transmembrane domains are boxed. (B) Radiolabeled Taz1 was imported into isolated S. cerevisiae mitochondria for the indicated times in the presence or absence of a membrane potential (Δψ). After import mitochondria were treated with proteinase K where indicated. As control radiolabeled precursor was incubated with proteinase K in import buffer in the absence of mitochondria (lanes 1 and 2). Samples were analyzed by SDS-PAGE and digital autoradiography. (C) Import of radiolabeled Taz1 into yeast mitochondria for the indicated times in the presence or absence of mitochondria. After import, samples were treated with proteinase K where indicated, solubilized in 1% digitonin, and subjected to BN-PAGE and digital autoradiography. For comparison, solubilized mitochondria from Taz1HA were separated by BN-PAGE and analyzed by Western blotting and immunodecoration with anti-HA antibodies (lanes 1 and 2).
To further analyze if Taz1 was part of a protein complex, a yeast strain expressing a functional C-terminal HA-tagged version of Taz1 (Taz1HA) was generated (see below). After solubilization of mitochondria in digitonin and separation of protein complexes by blue native gel electrophoresis (BN-PAGE), Taz1HA migrated as a single protein complex of ∼80 kDa (Figure 1C, lane 1). When mitochondria were solubilized after import of Taz1 and subjected to BN-PAGE, a complex with similar size was observed for the radiolabeled protein (Figure 1C, right panel). As expected the Taz1-complex was protected from protease (Figure 1C, lanes 6 and 7) and complex formation depended on the presence of mitochondria in the import reaction. Based on the size, the protein complex seen on BN-PAGE might represent a dimeric form of Taz1.
Taz1 Is an Outer Mitochondrial Membrane Protein
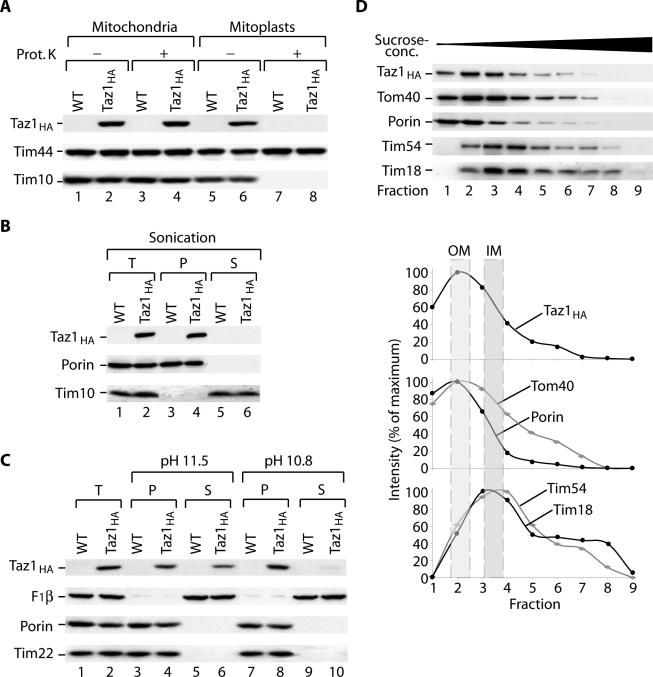
Using the Taz1HA, we further analyzed its submitochondrial localization by protease protection experiments. After treatment of intact mitochondria with proteinase K, Taz1HA, the intermembrane space protein Tim10, and the matrix protein Tim44 remained protected (Figure 2A, lanes 3 and 4). However, when mitoplasts were generated by disruption of the outer membrane before protease treatment, both Taz1HA and Tim10 became accessible to the added protease, whereas Tim44 remained protected (Figure 2A, lanes 7 and 8). Thus, Taz1 did not expose protease accessible domains to the cytosolic face of the outer membrane and the C-terminus of Taz1HA was exposed to the intermembrane space.
Figure 2.
Taz1 an outer membrane protein is exposed to the intermembrane space. (A) Wild-type and Taz1HA mitochondria were either left untreated or were swollen under hypotonic conditions before proteinase K treatment. Samples were subjected to SDS-PAGE and analysis by Western blotting and immunodecoration. (B) Mitochondria were sonicated in the presence of 500 mM NaCl and fractionated by differential centrifugation. T, total; P, pellet; S, supernatant. (C) Mitochondria were carbonate extracted at pH 11.5 or 10.8. Samples were either left untreated (T, total) or centrifuged at 100,000 × g (P, pellet; S, supernatant) and then subjected to SDS-PAGE and Western blotting. (D) Separation of outer (OM) and inner membrane (IM) vesicles. Taz1HA mitochondria were swollen and sonicated before separation of membrane vesicles on a discontinuous sucrose gradient. After centrifugation, fractions were collected from the top and analyzed by SDS-PAGE and Western blotting. Western blot signals were quantified using Scion Image 1.62.
Previous studies suggested that human Taz could be a membrane protein based on the presence of a hydrophobic domain near the N-terminus of the protein (Bione et al., 1996). Indeed, both yeast Taz1 and human Taz have a single predicted helical segment with the potential to span a membrane (Figure 1A). To determine whether Taz1 behaved as a soluble or membrane protein, we first subjected mitochondria to sonication and separated the soluble fraction from the membrane fraction by centrifugation. The outer membrane protein porin and Taz1 were only detected in the membrane fraction (Figure 2B, lane 4), whereas the soluble Tim10 was released into the supernatant (Figure 2B, lane 6). Thus, Taz1 appears to be tightly associated with membranes. To determine if Taz1 was an integral membrane protein, we performed a carbonate extraction at pH 11.5. Under these conditions, peripheral membrane proteins are typically released into the soluble fraction, and proteins that are integrated into the lipid phase remain with the membrane sheets. Porin and the multispanning inner membrane protein Tim22 remained in the carbonate pellet,,while Taz1 was partially released into the supernatant (Figure 2C, lanes 4 vs. 6). In contrast, when carbonate buffer with pH 10.8 was used, Taz1 remained entirely in the pellet fraction, whereas the soluble protein F1β, of the F1Fo-ATPase, was completely extracted (Figure 2C, lane 8 vs. 10). A similar behavior has been reported for other integral membrane proteins, such as the Rieske Fe/S-protein of the bc complex (Hartl et al., 1986; Truscott et al., 2003), which spans 1the inner membrane with a single transmembrane helix that is in close contact with other integral membrane proteins (Lange and Hunte, 2002). In addition, the inner membrane protein, Pam18 displays similar characteristics in its extractability (Truscott et al., 2003). We conclude that Taz1 is an integral membrane protein, with the predicted transmembrane helix in contact with a proteinaceous environment rather than being completely lipid-embedded. An alternative explanation however could be that Taz1 does not fully span the outer membrane but only partially inserts into one leaflet of the bilayer.
To determine the membrane localization of Taz1, we separated outer and inner mitochondrial membrane vesicles on a sucrose gradient. Taz1 migrated together with outer membrane marker proteins, such as Tom40 and porin, and not with markers for the inner membrane, such as Tim54 and Tim18 (Figure 2D). In summary, our findings indicate that Taz1 is an integral protein of the outer mitochondrial membrane and that the soluble C-terminal acyltransferase domain (Neuwald, 1997) is exposed to the intermembrane space.
Taz1 Transport into Mitochondria Requires the Tim9-Tim10 Complex of the Intermembrane Space
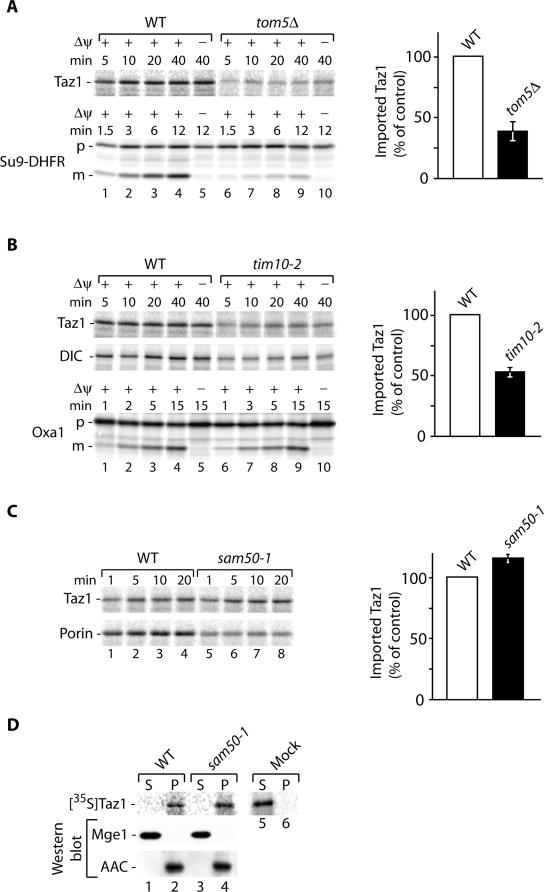
To support our finding that Taz1 is an outer membrane protein mainly exposed to the intermembrane space, we analyzed its transport pathway into mitochondria and imported radiolabeled Taz1 in vitro into mitochondria of various characterized mutant strains. First we investigated the involvement of the receptor Tom5 of the general import pore (GIP), which plays an important role in transport of proteins into all mitochondrial subcompartments (Dietmeier et al., 1997; Kurz et al., 1999). As expected, deletion of TOM5 lead to a significant reduction in Taz1 import, similar to what was seen for the preprotein Su9-DHFR (presequence of Fo-ATPase subunit 9 and DHFR; Figure 3A). Thus, Taz1 import into mitochondria appears to be a receptor-mediated process that follows the general import pathway via the GIP of the outer membrane.
Figure 3.
Biogenesis of Taz1. (A) Radiolabeled precursor proteins were imported into isolated wild-type and tom5Δ mitochondria in the presence or absence of a membrane potential (Δψ) for the indicated times. After import mitochondria were treated with proteinase K (Taz1) or left untreated (Su9-DHFR) and analyzed by SDS-PAGE and digital autoradiography. The amount of imported Taz1 in wild-type mitochondria after 10 min was set to 100% (control). SEM was calculated from at least four independent experiments. (B) Import of Taz1, dicarboxylate carrier (DIC), and Oxa1 into wild-type and tim10-2 was performed in the presence or absence of a Δψ for the indicated times, and samples were treated with proteinase K or left untreated (Oxa1) and analyzed by SDS-PAGE and digital autoradiography. Quantification was performed as in A. (C) Taz1 and porin were imported into wild-type and sam50-1 mitochondria as in A. The amount of imported Taz1 in wild-type mitochondria was set to 100% (control). SEM was calculated from at least four independent experiments. p, precursor; m, mature. (D) Radiolabeled Taz1 was imported into wild-type and sam50-1 mitochondria. Mitochondria were treated with proteinase K, sonicated in the presence of 500 mM NaCl, and fractionated by differential centrifugation at 100,000 × g into soluble and membrane fraction. As a control, reticulocyte lysate containing Taz1 was also subjected to sonication and differential centrifugation. Samples were analyzed by SDS-PAGE and digital autoradiography or Western blotting. P, pellet; S, supernatant.
Carrier proteins of the inner mitochondrial membrane and β-barrel proteins of the outer membrane are transported into the intermembrane space by a process that involves the small Tim proteins (Hoppins and Nargang, 2004; Wiedemann et al., 2004). Subsequently, the transport pathways diverge and the carrier proteins are imported in a Δψ dependent manner by the TIM22 complex into the inner membrane, whereas the β-barrel proteins insert into the outer membrane with the aid of the sorting and assembly machinery (SAM; for review see Koehler, 2004; Pfanner et al., 2004; Rehling et al., 2004). The membrane topology of Taz1 requires that the bulk of the protein is fully transported across the outer membrane, similar to carrier and β-barrel proteins. Therefore, we analyzed whether Taz1 transport required the Tim9-Tim10 complex of the IMS or the SAM complex by importing radiolabeled Taz1 into tim10-2 and sam50-1 mitochondria (Truscott et al., 2002; Kozjak et al., 2003). Similar to the transport of the dicarboxylate carrier (DIC), transport of Taz1 into mitochondria was strongly dependent on Tim10 function (Figure 3B), whereas the transport of presequence containing proteins such as Oxa1 was not affected in the mutant. Besides the Tim9-Tim10 complex a second small Tim complex, the Tim8-Tim13 complex, is found in the intermembrane space. Although carrier proteins do not depend on the Tim8-Tim13 complex for transport, an involvement of the complex in β-barrel protein biogenesis was reported (Hoppins and Nargang, 2004; Wiedemann et al., 2004). Therefore, we analyzed Taz1 import into mitochondria of a tim8Δ/tim13Δ mutant and found that its transport across the outer membrane was not affected (unpublished data). Unlike what was seen in the tim10-2 mutant mitochondria, in sam50-1 mutant mitochondria, Taz1 import was similar to wild-type, whereas transport of porin, an outer membrane β-barrel protein, to a protease protected location was significantly reduced in the mutant (Figure 3C). Similarly, Taz1 transport across the outer membrane was not affected in sam37Δ mitochondria (Wiedemann et al., 2003; unpublished data). To assess if Taz1 was associated with the membrane in sam50-1 mutant mitochondria and not in a soluble transport intermediate, we performed a fractionation analysis. After import of Taz1, mitochondria were sonicated and fractionated into a soluble and membrane fraction by differential centrifugation. Although soluble proteins such as Mge1 were efficiently released into the supernatant, Taz1 fractionated together with membranes in wild-type and mutant mitochondria (Figure 3D, lanes 2 and 4). As a control we treated radiolabeled Taz1 in reticulocyte lysate under similar conditions. However, in contrast to imported Taz1 the protein remained soluble in the absence of mitochondria (Figure 3D, lanes 5 and 6). Thus, like carrier and β-barrel proteins Taz1 appears to be translocated into the intermembrane space with the aid of the Tim9-Tim10 complex but does not require the SAM complex or the Δψ for transport. Apparently, the transport pathway for Taz1 diverges from the transport pathway of carrier proteins and β-barrel protein after the Tim9-Tim10 dependent step. These analyses further support that Taz1 localizes to the inner surface of the outer membrane.
Effect of TAZ1 Deletion on Inner Membrane Protein Complexes
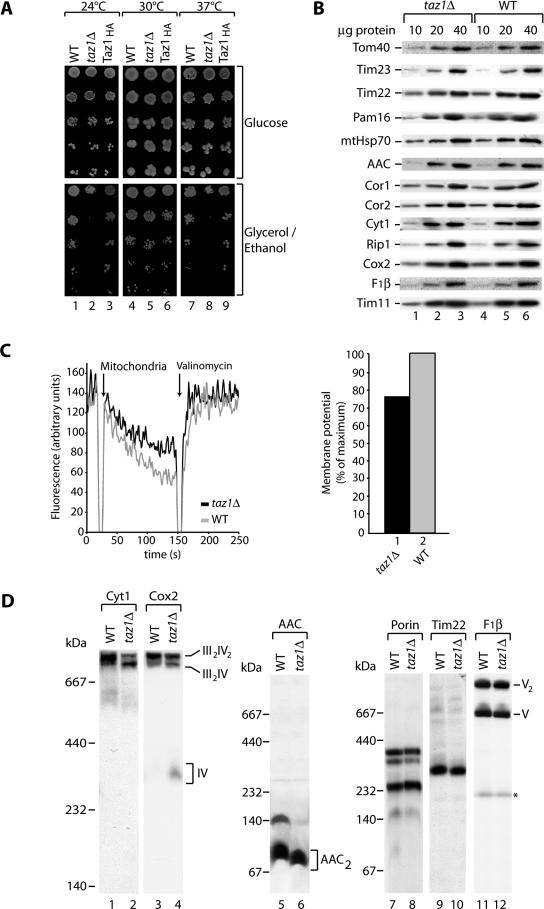
Previous studies reported a growth defect for taz1Δ cells on nonfermentable media at elevated temperature (Vaz et al., 2003; Gu et al., 2004). While analyzing the functionality of the Taz1HA at different temperatures, we compared the growth of cells expressing Taz1HA to wild type and the taz1Δ mutant. Taz1HA-expressing cells grew at rates similar to wild type under all conditions tested (Figure 4A, columns 3, 6, and 9), whereas taz1Δ cells exhibited the reported severe growth defects on the nonfermentable carbon source glycerol/ethanol at elevated temperature (37°C; Figure 4A, column 8). Interestingly, a growth defect for taz1Δ cells was also evident at 24°C (Figure 4A, column 2), indicating that taz1Δ cells are affected in respiration at both temperatures. In contrast, on the fermentable carbon source glucose, the growth of taz1Δ cells was similar to wild-type and Taz1HA cells (Figure 4A, top panels). The observed growth defect at high and low temperatures is in agreement with the idea that taz1Δ cells have a compromised membrane fluidity because of the defect in cardiolipin biosynthesis.
Figure 4.
Respiratory chain supercomplex stability is affected in taz1Δ. (A) Serial dilutions of wild-type (WT), taz1Δ, and Taz1HA-expressing cells were spotted on SM medium with glucose or glycerol/ethanol as carbon sources and incubated at the indicated temperatures. (B) Steady state protein levels of mitochondrial proteins. Isolated taz1Δ and wild-type mitochondria were subjected to SDS-PAGE and analyzed by Western blotting and immunodecoration. (C) Membrane potential (Δψ) of wild-type (gray) and taz1Δ (black) was measured at 25°C using the Δψ-dependent dye DiSC3(5). Right panel, quantification depicted as % of maximum Δψ (wild-type). (D) Wild-type and taz1Δ mitochondria were solubilized in 1% digitonin containing buffer, subjected to BN-PAGE, and analyzed by Western blotting and immunodecoration. BN-PAGE with a 5–12% acrylamide gradient was used to analyze the composition of respiratory chain complexes, whereas a 6–16.5% acrylamide gradient was used for other mitochondrial complexes. *Unspecific band.
These analyses and recent studies (Ma et al., 2004) indicate a defect in respiratory chain function in taz1Δ cells. In the light of the surprising outer membrane localization of Taz1 we wondered how defects in Taz1 would translate to a reduction of respiratory chain function at the inner membrane. To address the molecular basis for this defect, steady state protein levels from isolated mitochondria were compared between wild-type and taz1Δ mutant mitochondria (Figure 4B). Wild-type and taz1Δ mitochondria contained similar amounts of all proteins tested. These include the outer membrane protein Tom40, the inner membrane proteins Tim23, Tim22, Pam16, and AAC, and the matrix protein mtHsp70. In addition, proteins of the respiratory chain complex III (bc1 complex) such as Cor1, Cor2, Rip1 (Rieske Fe/S protein) and Cyt1 (cytochrome c1), Cox2 of the respiratory chain complex IV (cytochrome oxidase), and Tim11 and F1β of the F1Fo-ATPase were analyzed. To determine if taz1Δ mitochondria were functionally compromised, despite the lack of any obvious differences in the steady state amounts of mitochondrial proteins, we assessed the membrane potential in wild-type and taz1Δ mitochondria by fluorescence quenching (Sims et al., 1974; Geissler et al., 2000; Rehling et al., 2003). In taz1Δ mitochondria the Δψ was reduced by ∼25% compared with wild type (Figure 4C), suggesting that the activity of the respiratory chain was reduced in the mutant.
The reduction in Δψ in taz1Δ mitochondria suggested to us that respiratory complex function was impaired in these mitochondria. To determine if this was the case, we analyzed respiratory chain protein complexes in both wild-type and taz1Δ mitochondria. It has been previously shown that complexes III and IV associate into two supercomplexes of ∼750 and 1000 kDa that can be separated by BN-PAGE (Boumans et al., 1998; Cruciat et al., 2000; Schägger and Pfeiffer, 2000). The two supercomplexes are composed of a complex III dimer associated with either one (III2IV) for the 750-kDa complex or two complex IV monomers for the 1000-kDa complex (III2IV2). BN-PAGE analysis of the taz1Δ mitochondria showed decreases in the amount of the III2IV2 complex and an increase in the amount of the III2IV complex compared with the wild-type mitochondria. The III2IV complex also migrated slightly faster in the taz1Δ mutant (Figure 4D, lanes 2 and 4). In taz1Δ mitochondria, Cox2 was also detected in the ∼300-kDa complex IV monomer, which was not present in the wild-type control (Figure 4D, lane 4). The easiest explanation for these results is that the III2IV2 supercomplex is destabilized in the taz1Δ mitochondria, releasing monomeric complex IV. This interpretation would explain both the increase in the III2IV supercomplex and the presence of monomeric complex IV. In contrast to the supercomplexes, oligomerization of the outer membrane porin (Figure 4D, lanes 7 and 8), the inner membrane twin pore translocase (TIM22 complex; Figure 4D, lanes 9 and 10), and the monomeric and dimeric forms of the F1Fo-ATPase (Arnold et al., 1998; Figure 4D, lanes 11 and 12) were not affected in taz1Δ. In addition to the altered oligomerization of the respiratory supercomplexes, we also detected altered oligomerization of the inner membrane ADP/ATP carrier (AAC). AAC was still present in its dimeric form in taz1Δ, but higher oligomers were no longer detected (Figure 4D, lanes 5 and 6). This defect in AAC oligomerization is in agreement with earlier findings with regard to the phenotype of the crd1Δ mutant, which does not contain any detectable cardiolipin (Jiang et al., 2000).
Assembly of Newly Imported Subunits into Supercomplexes Is Affected in taz1Δ Mitochondria
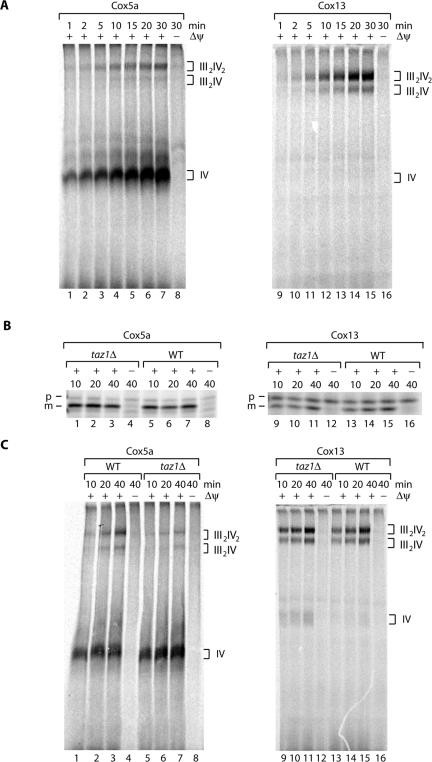
Assembly of respiratory chain complexes has been intensely analyzed by different approaches for a number of years. Most of these analyses were based on pulse-labeling experiments followed by immunoprecipitation to define subcomplexes in the assembly pathway. However, the precise assembly pathways and the role of assembly factors is largely unresolved (for review see Taanman and Williams, 2001; Herrmann and Funes, 2005). Moreover, to the best of our knowledge the question as to how newly imported nuclear encoded proteins assemble into preexisting supercomplexes has so far not been analyzed. Thus, we established an in organello assembly assay to address experimentally if, in addition to the destabilization of supercomplexes in taz1Δ mitochondria, assembly of newly imported proteins into complexes of the respiratory chain was affected in the taz1Δ mitochondria. Radiolabeled precursor proteins of complex IV subunits, Cox5a and Cox13 (CoxVIa), were synthesized in a rabbit reticulocyte lysate and imported into isolated wild-type mitochondria in the presence or absence of a Δψ for different times. Unimported precursor proteins were removed by proteinase K treatment, and then mitochondrial protein complexes were solubilized in digitonin containing buffer and subjected to BN-PAGE followed by digital autoradiography. In the presence of a Δψ newly imported Cox5a was efficiently incorporated into the monomer of complex IV and to a lesser extent into the III2IV2 and III2IV supercomplexes (Figure 5A, lanes 1–7 vs. lane 8). At steady state complex IV monomer was not detected by Western analysis of wild-type mitochondria (Figure 4D); thus, we speculate that the monomeric form of complex IV seen in the assembly assays represent a small pool of partially assembled complexes. In contrast to Cox5a, Cox13 did not assemble into the monomeric form of complex IV but instead was assembled directly into the respiratory supercomplexes in a Δψ dependent manner (Figure 5A, lanes 9–15 vs. lane 16). This is in agreement with earlier studies that demonstrated that complex IV was fully assembled in the absence of Cox13 (Taanman and Capaldi, 1993). Thus it appears that Cox5a and Cox13 follow different assembly pathways into complex IV and its higher oligomers (Figure 6). Cox5a must first assemble into complex IV monomer and the entire complex together with the radiolabeled precursor is subsequently assembled into the respiratory supercomplexes. In contrast to Cox5a, Cox13 assembles at a later step in the formation of complex IV, when it is already associated with complex III in respiratory supercomplexes.
Figure 5.
Defects in the assembly of cytochrome oxidase in taz1Δ. (A) Radiolabeled Cox5a (cytochrome oxidase subunit 5a) and Cox13 (CoxVIa) were imported into isolated wild-type mitochondria for the indicated times in the presence or absence of a membrane potential (Δψ). After import, mitochondria were treated with proteinase K and solubilized in 1% digitontin-containing buffer, and protein complexes were separated on a 5–10% BN-PAGE before digital autoradiography. (B) Cox5a and Cox13 were imported into wild-type and taz1Δ mitochondria for the indicated times and subjected to proteinase K treatment, and samples were analyzed by SDS-PAGE and digital autoradiography. p, precursor; m, mature. (C) Assembly of Cox5a and Cox13 into cytochrome oxidase complexes. Import was performed as described in A. After import, mitochondria were treated with proteinase K, reisolated, solubilized in 1% digitontin-containing buffer, and subjected to BN-PAGE and digital autoradiography.
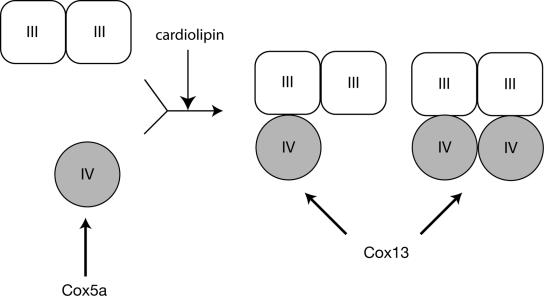
Figure 6.
Hypothetical model of Cox5a and Cox13 assembly into supercomplexes. Import analysis of Cox5a and Cox13 into isolated mitochondria showed that Cox5a efficiently assembled into complex IV monomer, whereas Cox13 directly assembled into complex IV in the supercomplexes. Defects in cardiolipin due to deletion of TAZ1 affect the incorporation of monomeric complex IV into supercomplexes.
To assess respiratory chain complex assembly in taz1Δ mitochondria, we first compared precursor import of radiolabeled Cox5a and Cox13 in wild-type and taz1Δ mitochondria on SDS-PAGE. Both proteins were imported and processed to their mature forms in a Δψ-dependent manner with similar efficiency in both strains (Figure 5B). We then analyzed assembly of Cox5a and Cox13 into complex IV in wild-type and taz1Δ mitochondria by BN-PAGE. As expected, Cox5a assembled efficiently into the complex IV monomer and with lower efficiency into the supercomplexes in wild-type mitochondria (Figure 5C, lanes 1–3). Despite the similar import efficiency for Cox5a into wild-type and taz1Δ mitochondria on SDS-PAGE, assembly of the protein into the supercomplexes was clearly reduced in taz1Δ mutant mitochondria and Cox5a remained primarily associated with the complex IV monomer (Figure 5C, lanes 5–7). In contrast, newly imported Cox13 assembled with similar efficiency into the III2IV and III2IV2 supercomplexes in both wild-type and taz1Δ mitochondria (Figure 5C, lanes 9–11 and 13–15). A small fraction of Cox13 was found in the complex IV monomer in taz1Δ mitochondria. As this protein was seen only to assemble into the supercomplexes in wild-type mitochondria, this fraction of complex IV monomer most likely represents the portion of complex IV that was released from the destabilized supercomplexes (see Figure 4D). Thus it appears that assembly of complex IV into supercomplexes is affected in the taz1Δ mitochondria.
DISCUSSION
Tafazzins mediate the late steps in the cardiolipin biosynthesis pathway, namely the acyl modifications that lead to its characteristic acyl pattern (Vreken et al., 2000; Vaz et al., 2003; Xu et al., 2003; Gu et al., 2004). Although human mitochondria predominantly contain cardiolipin with unsaturated C18 fatty acids, cardiolipin of yeast contains equal amounts of oleoyl (18:1) and palmitoleoyl (C16:1; Jakovcic et al., 1971). The typical acyl pattern is generated by hydrolysis of the phosphatidyl glycerol-derived acyl chains and subsequent reacylation. In agreement with a role of Tafazzins in the reacylation of cardiolipin, accumulation of monolysocardiolipin has been observed in yeast taz1Δ mitochondria (Vaz et al., 2003; Gu et al., 2004). Similarly, aberrant forms of cardiolipin are apparent in Barth syndrome patients (Vreken et al., 2000; Vaz et al., 2003; Xu et al., 2003; Gu et al., 2004). In higher eukaryotes, all enzymes that participate in the synthesis of cardiolipin downstream of phosphatidic acid have been localized to the inner mitochondrial membrane (for review see Schlame et al., 2000). Therefore, we expected that Taz1 would localize to the inner mitochondrial membrane, akin to other proteins involved in cardiolipin biosynthesis. However, Taz1 could be imported into yeast mitochondria independent of the inner membrane potential. Because protein translocation across the inner membrane strictly requires the Δψ, this finding indicated that the protein was not transported across the inner membrane (Neupert, 1997; Jensen and Dunn, 2002; Koehler, 2004; Rehling et al., 2004). This was corroborated by cell fractionation analyses, which showed that Taz1 was an integral membrane protein of the outer mitochondrial membrane that exposed its soluble C-terminal domain into the intermembrane space. Further support of these findings came from analysis of Taz1 biogenesis. Transport of Taz1 across the outer membrane required Tom5 of the GIP complex. Interestingly, import of Taz1 into mitochondria required the Tim9-Tim10 complex of the intermembrane space. Transport of carrier proteins and outer membrane β-barrel proteins has previously been shown to depend on small Tim proteins. These proteins are transported across the outer membrane and then integrated into the inner or outer membrane by the TIM22 complex or the SAM complex respectively (Hoppins and Nargang, 2004; Pfanner et al., 2004; Koehler 2004; Rehling et al., 2004; Wiedemann et al., 2004). In contrast to Taz1 and the β-barrel proteins, outer membrane proteins that are not fully translocated and span the outer membrane with a single α-helical membrane anchor, such as the receptor Tom20, assemble into the outer membrane independent of the small Tim proteins (Wiedemann et al., 2004). Our findings suggest that Taz1 follows the same transport pathway as carrier proteins and β-barrel proteins across the outer membrane. However, although further transport of β-barrel proteins requires the SAM complex (Wiedemann et al., 2003; Kozjak et al., 2003; Paschen et al., 2003) Taz1 transport was independent of the SAM complex. Thus, import, fractionation, and analysis of the biogenesis support an outer membrane localization for Taz1. The localization of Taz1 in the outer membrane has significant implications for cardiolipin biosynthesis because it implies that steps of cardiolipin remodeling may occur at the outer leaflet of the inner mitochondrial membrane or at the outer membrane.
Patients suffering from Barth syndrome show symptoms that are often found associated with mitochondrial respiratory chain diseases (Barth et al., 1983, 1996; DiMauro and Schon, 2003) such as altered mitochondrial morphology (Barth et al., 1983, 1996) and respiratory chain dysfunction (Barth et al., 1983, 1996). This was confirmed in our study because we observed growth defects of taz1Δ cells on a nonfermentable carbon source and a reduction of mitochondrial inner membrane potential. These results agree well with previous studies, which reported that the function of the respiratory chain was compromised in taz1Δ cells (Vaz et al., 2003; Gu et al., 2004; Ma et al., 2004). To determine how inactivation of the mitochondrial outer membrane protein Taz1 affected the respiratory chain, we visualized the respiratory chain protein complexes solubilized in the mild detergent digitonin by BN-PAGE (Cruciat et al., 2000; Schägger and Pfeiffer, 2000; Schägger et al., 2004). This technique has been used to show that respiratory complexes associate in higher order complexes, or supercomplexes. Recent analysis demonstrates that supercomplex formation between complex III and IV plays an important role for respiratory chain function in intact organelles (Zhang et al., 2005). Similarly, yeast mutants of factors required for dimerization of the F1Fo-ATPase show growth defects on nonfermentable carbon sources (Arnold et al., 1998). Thus it appears that dissociation of respiratory supercomplexes can decrease the efficiency of respiration. In a S. cerevisiae cardiolipin synthase mutant (crd1Δ), which lacks cardiolipin, oligomerization of AAC was affected (Jiang et al., 2000) and the respiratory chain supercomplexes dissociated in organello and on BN-PAGE (Zhang et al., 2002, 2005), suggesting a role for cardiolipin in the oligomerization of inner membrane protein complexes. We find that AAC oligomerization is affected in taz1Δ mitochondria, similar to that in crd1Δ; however, supercomplexes only partially dissociate on BN-PAGE. The supercomplex consisting of a dimer of complex III and two monomers of complex IV (III2IV2) is destabilized in the taz1Δ, releasing a complex IV monomer, whereas the supercomplex (III2IV), consisting of a dimer of complex III associated with monomeric complex IV seems stable. taz1Δ mitochondria have reduced amounts of cardiolipin, and the remaining cardiolipin has an altered acyl composition (Vaz et al., 2003; Gu et al., 2004), which seems sufficient to stabilize the III2IV supercomplex but not the III2IV2 supercomplex.
We next wanted to determine if taz1Δ mitochondria showed impairment in complex IV biogenesis. To do this, we developed a BN-PAGE based assay to analyze assembly of newly imported subunits of complex IV into preexisting protein complexes. Similar import and assembly assays have successfully been used to assess outer membrane protein complex biogenesis (Krimmer et al., 2001; Model et al., 2001; Kozjak et al., 2003; Paschen et al., 2003; Wiedemann et al., 2003). Newly imported Cox5a assembled into the complex IV monomer and was only seen in small amounts in the supercomplexes, whereas Cox13 assembled into respiratory supercomplexes (Figure 6). This is in agreement with the idea that Cox5a assembles into earlier forms of complex IV than Cox13 (for review see Taanman and Williams, 2001; Herrmann and Funes, 2005). In contrast to wild-type mitochondria, Cox5a accumulated in the monomeric complex IV in taz1Δ mitochondria and did not assemble into the supercomplexes, whereas Cox13 assembly into supercomplexes was not affected in the mutant. This indicates that assembly of complex IV monomers into respiratory supercomplexes is affected in taz1Δ mitochondria and provides strong evidence for a role of cardiolipin not only in the stability of supercomplexes but also supercomplex assembly (Figure 6). It is tempting to speculate that inactivation of Taz1 in Barth patients leads to similar defects in the formation and stability of supercomplexes, which in turn would contribute to the observed respiratory defects seen in these patients. Further work will be required to address this hypothesis.
Acknowledgments
We are grateful to N. Pfanner, M. van der Laan, M. McKenzie, and M. T. Ryan for many helpful discussions and comments on this manuscript and to B. Trumpower for antibodies. We thank I. Perschil for expert technical assistance. This work was supported by the Research Award of the “Wissenschaftliche Gesellschaft in Freiburg” (to P.R.) and by a fellowship from the Natural Sciences and Engineering Research Council of Canada (to R.D.T.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–03–0256) on August 31, 2005.
Abbreviations used: AAC, ADP/ATP carrier; BN-PAGE, blue native-PAGE; BTHS, Barth syndrome; Δψ, membrane potential.
References
- Arnold, I., Pfeiffer, K., Neupert, W., Stuart, R. A., and Schägger, H. (1998). Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 17, 7170–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, P. G., Scholte, H. R., Berden, J. A., Van der Klei-Van Moorsel, J. M., Luyt-Houwen, I. E., Van't Veer-Korthof, E. T., Van der Harten, J. J., and Sobotka-Plojhar, M. A. (1983). An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 62, 327–355. [DOI] [PubMed] [Google Scholar]
- Barth, P. G., Valianpour, F., Bowen, V. M., Lam, J., Duran, M., Vaz, F. M., and Wanders, R. J. (2004). X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): an update. Am. J. Med. Genet. 126, 349–354. [DOI] [PubMed] [Google Scholar]
- Barth, P. G., Van den Bogert, C., Bolhuis, P. A., Scholte, H. R., van Gennip, A. H., Schutgens, R. B., and Ketel, A. G. (1996). X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): respiratory-chain abnormalities in cultured fibroblasts. J. Inherit. Metab. Dis. 19, 157–160. [DOI] [PubMed] [Google Scholar]
- Barth, P. G., Wanders, R. J., and Vreken, P. (1999). X-linked cardioskeletal myopathy and neutropenia (Barth syndrome)—MIM 302060. J. Pediatr. 135, 273–276. [DOI] [PubMed] [Google Scholar]
- Bione, S., D'Adamo, P., Maestrini, E., Gedeon, A. K., Bolhuis, P. A., and Toniolo, D. (1996). A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 12, 385–389. [DOI] [PubMed] [Google Scholar]
- Boumans, H., Grivell, L. A., and Berden, J. A. (1998). The respiratory chain in yeast behaves as a single functional unit. J. Biol. Chem. 273, 4872–4877. [DOI] [PubMed] [Google Scholar]
- Cruciat, C. M., Brunner, S., Baumann, F., Neupert, W., and Stuart, R. A. (2000). The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275, 18093–18098. [DOI] [PubMed] [Google Scholar]
- D'Adamo, P. et al. (1997). The X-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am. J. Hum. Genet. 61, 862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, P.J.T., Martin, F., Maarse, A. C., Bömer, U., Müller, H., Guiard, B., Meijer, M., Rassow, J., and Pfanner, N. (1997). The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16, 5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmeier, K., Hönlinger, A., Bömer, U., Dekker, P.J.T., Eckershkorn, C., Lottspeich, F., Lübrich, M., and Pfanner, N. (1997). Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature 388, 195–200. [DOI] [PubMed] [Google Scholar]
- DiMauro, S., and Schon, E. A. (2003). Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656–2668. [DOI] [PubMed] [Google Scholar]
- Dudkina, N. V., Eubel, H., Keegstra, W., Boekema, E. J., and Braun, H. P. (2005). Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. USA 102, 3225–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier, A. E. et al. (2004). Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11, 226–233. [DOI] [PubMed] [Google Scholar]
- Geissler, A., Krimmer, T., Bömer, U., Guiard, B., Rassow, J., and Pfanner, N. (2000). Membrane potential-driven protein import into mitochondria: the sorting sequence of cytochrome b2 modulates the Dy-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell 11, 3977–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Valianpour, F., Chen, S., Vaz, F. M., Hakkaart, G. A., Wanders, R. J., and Greenberg, M. L. (2004). Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol. Microbiol. 51, 149–158. [DOI] [PubMed] [Google Scholar]
- Hartl, F. U., Schmidt, B., Wachter, E., Weiss, H., and Neupert, W. (1986). Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell 47, 939–951. [DOI] [PubMed] [Google Scholar]
- Hatch, G. M. (2004). Cell biology of cardiac mitochondrial phospholipids. Biochem. Cell Biol. 82, 99–112. [DOI] [PubMed] [Google Scholar]
- Herrmann, J. M., and Funes, S. (2005). Biogenesis of cytochrome oxidase—sophisticated assembly lines in the mitochondrial inner membrane. Gene 354, 43–52. [DOI] [PubMed] [Google Scholar]
- Hoch, F. L. (1992). Cardiolipins and biomembrane function. Biochim. Biophys. Acta 1113, 71–133. [DOI] [PubMed] [Google Scholar]
- Hoppins, S. C., and Nargang, F. E. (2004). The Tim8-Tim13 complex in Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279, 12396–12405. [DOI] [PubMed] [Google Scholar]
- Jakovcic, S., Getz, G. S., Rabinowitz, M., Jakob, H., and Swift, H. (1971). Cardiolipin content of wild type and mutant yeasts in relation to mitochondrial function and development. J. Cell Biol. 48, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R. E., and Dunn, C. D. (2002). Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim. Biophys. Acta 1592, 25–34. [DOI] [PubMed] [Google Scholar]
- Jiang, F., Ryan, M. T., Schlame, M., Zhao, M., Gu, Z., Klingenberg, M., Pfanner, N., and Greenberg, M. L. (2000). Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394. [DOI] [PubMed] [Google Scholar]
- Kelley, R. I., Cheatham, J. P., Clark, B. J., Nigro, M. A., Powell, B. R., Sherwood, G. W., Sladky, J. T., and Swisher, W. P. (1991). X-linked dilated cardiomyopathy with neutropenia, growth retardation, and 3-methylglutaconic aciduria. J. Pediatr. 119, 738–747. [DOI] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Koehler, C. M. (2004). New developments in mitochondrial assembly. Annu. Rev. Cell. Dev. Biol. 20, 309–335. [DOI] [PubMed] [Google Scholar]
- Koshkin, V., and Greenberg, M. L. (2002). Cardiolipin prevents rate-dependent uncoupling and provides osmotic stability in yeast mitochondria. Biochem. J. 364, 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak, V., Wiedemann, N., Milenkovic, D., Lohaus, C., Meyer, H. E., Guiard, B., Meisinger, C., and Pfanner, N. (2003). An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278, 48520–48523. [DOI] [PubMed] [Google Scholar]
- Krimmer, T. et al. (2001). Biogenesis of porin of the outer mitochondrial membrane involved an import pathway via receptors and the general import pore of the Tom complex. J. Cell Biol. 152, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, M., Martin, H., Rassow, J., Pfanner, N., and Ryan, M. T. (1999). Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 10, 2461–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, C., and Hunte, C. (2002). Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc. Natl. Acad. Sci. USA 99, 2800–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Vaz, F. M., Gu, Z., Wanders, R. J., and Greenberg, M. L. (2004). The human TAZ gene complements mitochondrial dysfunction in the yeast taz1Δ mutant. J. Biol. Chem. 279, 44394–44399. [DOI] [PubMed] [Google Scholar]
- Model, K., Meisinger, C., Prinz, T., Wiedemann, N., Truscott, K. N., Pfanner, N., and Ryan, M. T. (2001). Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nature Struct. Biol. 8, 361–370. [DOI] [PubMed] [Google Scholar]
- Neupert, W. (1997). Protein import into mitochondria. Annu. Rev. Biochem. 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Neuwald, A. F. (1997). Barth syndrome may be due to an acyltransferase deficiency. Curr. Biol. 7, R465–R456. [DOI] [PubMed] [Google Scholar]
- Palsdottir, H., and Hunte, C. (2004). Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18. [DOI] [PubMed] [Google Scholar]
- Paschen, S. A., Waizenegger, T., Stan, T., Preuss, M., Cyrklaff, M., Hell, K., Rapaport, D., and Neupert, W. (2003). Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426, 862–866. [DOI] [PubMed] [Google Scholar]
- Pfanner, N., Wiedemann, N., Meisinger, C., and Lithgow, T. (2004). Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 11, 1044–1048. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, K., Gohil, V., Stuart, R. A., Hunte, C., Brandt, U., Greenberg, M. L., and Schägger, H. (2003). Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880. [DOI] [PubMed] [Google Scholar]
- Rehling, P., Brandner, K., and Pfanner, N. (2004). Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell. Biol. 5, 519–530. [DOI] [PubMed] [Google Scholar]
- Rehling, P., Model, K., Brandner, K., Kovermann, P., Sickmann, A., Meyer, H. E., W., K., Wagner, R., Truscott, K. N., and Pfanner, N. (2003). Protein insertion into the mitochondria inner membrane by a twin-pore translocase. Science 299, 1747–1751. [DOI] [PubMed] [Google Scholar]
- Ryan, M. T., Voos, W., and Pfanner, N. (2001). Assaying protein import into mitochondria. Methods Cell Biol. 65, 189–215. [DOI] [PubMed] [Google Scholar]
- Schlame, M., Rua, D., and Greenberg, M. L. (2000). The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39, 257–288. [DOI] [PubMed] [Google Scholar]
- Schägger, H., de Coo, R., Bauer, M. F., Hofmann, S., Godinot, C., and Brandt, U. (2004). Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279, 36349–36353. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and Pfeiffer, K. (2000). Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann, A. et al. (2003). The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100, 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, P. J., Waggoner, A. S., Wang, C. H., and Hoffman, J. F. (1974). Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry 13, 3315–3330. [DOI] [PubMed] [Google Scholar]
- Taanman, J. W., and Williams, S. L. (2001). Assembly of cytochrome c oxidase: what can we learn from patients with cytochrome c oxidase deficiency? Biochem. Soc. Trans. 29, 446–451. [DOI] [PubMed] [Google Scholar]
- Taanman, J. W., and Capaldi, R. A. (1993). Subunit VIa of yeast cytochrome c oxidase is not necessary for assembly of the enzyme complex but modulates the enzyme activity. J. Biol. Chem. 268, 18754–18761. [PubMed] [Google Scholar]
- Testet, E., Laroche-Traineau, J., Noubhani, A., Coulon, D., Bunoust, O., Camougrand, N., Manon, S., Lessire, R., and Bessoule, J. J. (2004). Ypr140wp, “the yeast tafazzin,” displays a mitochondrial lyso-PC acyltransferase activity related to triglyceride and mitochondrial lipid synthesis. Biochem. J. 387, 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott, K. N. et al. (2003). A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott, K. N., Wiedemann, N., Rehling, P., Müller, H., Meisinger, C., Pfanner, N., and Guiard, B. (2002). Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol. Cell. Biol. 22, 7780–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz, F. M., Houtkooper, R. H., Valianpour, F., Barth, P. G., and Wanders, R. J. (2003). Only one splice variant of the human TAZ gene encodes a functional protein with a role in cardiolipin metabolism. J. Biol. Chem. 278, 43089–43094. [DOI] [PubMed] [Google Scholar]
- Vreken, P., Valianpour, F., Nijtmans, L. G., Grivell, L. A., Plecko, B., Wanders, R. J., and Barth, P. G. (2000). Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem. Biophys. Res. Commun. 279, 378–382. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., Kozjak, V., Chacinska, A., Schönfisch, B., Rospert, S., Ryan, M. T., Pfanner, N., and Meisinger, C. (2003). Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424, 565–571. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., Truscott, K. N., Pfannschmidt, S., Guiard, B., Meisinger, C., and Pfanner, N. (2004). Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 279, 18188–18194. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Kelley, R. I., Blanck, T. J., and Schlame, M. (2003). Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 278, 51380–51385. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Mileykovskaya, E., and Dowhan, W. (2002). Gluing the respiratory chain together. J. Biol. Chem. 277, 43553–43556. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Mileykovskaya, E., and Dowhan, W. (2005). Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280, 29403–29408. [DOI] [PMC free article] [PubMed] [Google Scholar]