Abstract
The proprotein convertases PC5, PACE4 and furin contain a C-terminal cysteine-rich domain (CRD) of unknown function. We demonstrate that the CRD confers to PC5A and PACE4 properties to bind tissue inhibitors of metalloproteinases (TIMPs) and the cell surface. Confocal microscopy and biochemical analyses revealed that the CRD is essential for cell surface tethering of PC5A and PACE4 and that it colocalizes and coimmunoprecipitates with the full-length and C-terminal domain of TIMP-2. Surface-bound PC5A in TIMP-2 null fibroblasts was only observed upon coexpression with TIMP-2. In COS-1 cells, plasma membrane-associated PC5A can be displaced by heparin, suramin, or heparinases I and III and by competition with excess exogenous TIMP-2. Furthermore, PC5A and TIMP-2 are shown to be colocalized over the surface of enterocytes in the mouse duodenum and jejunum, as well as in liver sinusoids. In conclusion, the CRD of PC5A and PACE4 functions as a cell surface anchor favoring the processing of their cognate surface-anchored substrates, including endothelial lipase.
INTRODUCTION
Posttranslational processing of numerous secretory proteins generating biologically active moieties is accomplished by the proprotein convertases (PCs), which are serine proteinases related to bacterial subtilisin and yeast kexin. These proteinases perform critical functions in a variety of physiological and pathological processes. There are seven known basic amino acid (aa)-specific PC family members that cleave various secretory precursors following basic residues: furin, PC1/3, PC2, PC4, PACE4, PC5/6, and PC7 (Seidah and Chretien, 1999). Recently, two other nonbasic aa-specific convertases implicated in cholesterol metabolism have been identified, namely, SKI-1/S1P (Seidah and Prat, 2002) and NARC-1/PCSK9 (Abifadel et al., 2003; Seidah et al., 2003; Benjannet et al., 2004). All basic aa-specific convertases contain the same N-terminal organization starting with a signal peptide, followed by a prodomain, catalytic, and a β-barrel P-domain, whereas the C-terminal architecture is specific to each convertase (Seidah and Chretien, 1999). After the P-domain, three of the basic-aa-specific convertases, furin, PACE4, and PC5, contain a cysteine-rich domain (CRD) (Seidah and Chretien, 1999). PC5 is the only member of this convertase family that exists as two isoforms: soluble PC5A (Lusson et al., 1993) and membrane-bound PC5B, the latter having an extended C-terminal CRD (Nakagawa et al., 1993). PC5A is sorted to both the constitutive and regulated secretory pathways, whereas PC5B is localized only within the constitutive secretory pathway (De Bie et al., 1996), mainly in a Golgi compartment communicating with endosomes (Xiang et al., 2000). The CRD of PC5A contains four N-linked glycosylation sites and 44 cysteine residues arranged in five tandem repeats with the consensus motif (Lusson et al., 1993): Cys-Xaa2-Cys-Xaa3-Cys-Xaa5-7-Cys-Xaa2-Cys-Xaa8-15-Cys-Xaa3-Cys-Xaa9-16. This five-tandem repeats motif is conserved between frog and mammalian PC5A (Gangnon et al., 2003). PACE4 also possesses five-tandem repeats within its CRD, whereas furin exhibits only two shortened repeats (Nakagawa et al., 1993). So far, the function of the CRD of each PC is unknown, although it was suggested that secreted PC5A and PACE4, but not soluble furin, could bind heparin within the extracellular matrix (ECM), likely via a cationic stretch of amino acids within their CRD (Tsuji et al., 2003). Finally, in the regulated corticotroph AtT20 cells, it was previously observed that the membrane-bound PC5B (∼210 kDa) can be shed into the medium (∼170 kDa) and that both PC5A and PC5B are processed at their C termini before and/or during secretion to produce a shorter, secreted form, PC5-ΔC (∼65 kDa) lacking the CRD (De Bie et al., 1996).
Although the in vivo functions of the soluble PC5A are not well delineated, it was shown to be implicated in the processing of a number of membrane-bound cell surface precursors such as adhesion molecules, including integrin α-chains (Lissitzky et al., 2000; Bergeron et al., 2003; Stawowy et al., 2004), the neural adhesion protein L1 (Kalus et al., 2003), transforming growth factor (TGF)-β like proteins (Nachtigal and Ingraham, 1996; Ulloa et al., 2001; Stawowy et al., 2003), a receptor protein tyrosine phosphatase RPTPμ (Campan et al., 1996), and membrane-bound metalloproteinases such as ADAM-17 (Srour et al., 2003) and possibly the membrane type-1 matrix metalloproteinase MT1-MMP (Yana and Weiss, 2000). The latter plays an essential role in extracellular matrix remodeling and signaling (Tam et al., 2004) after the activation of its zymogen proMT1-MMP by a furin-like convertase (Yana and Weiss, 2000).
The degradation of ECM proteins is often performed by matrix metalloproteinases (MMPs). This family of proteases is composed of 26 members (Sternlicht and Werb, 2001; Overall and Lopez-Otin, 2002). MMPs are not only involved in ECM remodeling but also can regulate the function of an increasing number of important signaling proteases through limited proteolysis (McQuibban et al., 2000; Tam et al., 2004). The activity of MMPs can be regulated by specific endogenous inhibitors known as tissue inhibitors of metalloproteinases (TIMPs), four of which are known (TIMP-1, -2, -3, and -4) (Baker et al., 2002; Jiang et al., 2002). The activation of proMMP-2 is mediated via MT1-MMP at the cell surface and it involves the formation of a ternary complex that requires TIMP-2. At the plasma membrane, the N-terminal inhibitory domain of TIMP-2 binds to the catalytic site of MT1-MMP, thus inactivating this metalloprotease. The hemopexin C-domain of the soluble proMMP-2 interacts with this binary complex by specifically binding to the C-terminal domain of TIMP-2 (Overall et al., 1999; Kai et al., 2002), thus forming a ternary complex at the plasma membrane. The prodomain of proMMP-2 is then cleaved and the enzyme activated into MMP-2 by a second MT1-MMP molecule (Butler et al., 1998; Zucker et al., 1998). Although at low concentrations of TIMP-2 its proMMP-2 activation effects outweigh its inhibitory actions, at high concentrations TIMP-2 is an inhibitor of MMP-2, thereby preventing tumor cell invasion and metastasis. However, it was recently shown that TIMP-2 is also able to inhibit endothelial cell proliferation (antiangiogenic role) through a mechanism that is independent of MMP-2 but that implicates binding of its N-terminal domain to integrin α3β1 (Seo et al., 2003).
The results of the present study center on the elucidation of the functional importance of the CRD of PC5A and PACE4 and include 1) the demonstration that excision of the CRD of PC5A is catalyzed by a metalloprotease(s); 2) the involvement of the CRD in the cell surface localization of PC5A and PACE4; 3) the role of full-length and the C-terminal domain of TIMP-2 in recruiting PC5A to the cell surface through binding of the complex to heparan sulfate proteoglycans (HSPG), displaceable by heparin; 4) the importance of the CRD of PC5A in the processing of the proteoglycan-bound endothelial lipase; 5) the colocalization of TIMP-2 and PC5 within mouse intestinal crypts and villi as well as in liver sinusoids; and finally, 6) the association of PC5A with other TIMP family member(s). To accomplish these objectives, we used recombinant wild-type PC5A, PACE4, and TIMP-2 and various deletions thereof, and/or mutants as well as cellular, immunocytochemical/histochemical, and coimmunoprecipitation analyses. We found that the CRD of PC5A binds to the C-terminal domain of TIMP-2, resulting in a complex bound to cell surface HSPGs, likely enhancing the cleavage of proteoglycan-bound substrates such as endothelial lipase.
MATERIALS AND METHODS
Vectors Constructs
Mouse PC5A was cloned into EcorI/AgeI digested pIRES2-EGFP vector (BD Biosciences Clonetech, Palo Alto, CA) with a C-terminal V5-tag. Mutagenesis was done by PCR using the pIRES2-PC5A-V5 cDNA template to generate the K613A, R616A, R618A, Y619P, S620P, R621A, and E623A mutants. The pIRES2-PC5A-ΔC-V5 (aa 1–612) construct was generated by PCR using the primers 5′ GGGCGGTAGGCGTGTACGGTGG/3′ ACCGGTGGGAAACTCGTTGGTTGGGGAG. Similarly, we also obtained a recombinant pIRES2-PACE4-ΔC-V5 (aa 1–679) lacking the CRD. The pIRES2-PC5ACRD-V5 was also generated by PCR using the following primers: 5′ GGGCGGTAGGCGTGTACGGTGG/3′ TGGCTGTACCGTCCGGCATACCGGGAG and 5′ TGCCGGACGGTACAGCCATACTCCCCAAC/3′ CTTCGGCCAGTAACGTTAGGGG, respectively. The primers used to generate the final cDNA in the second PCR reaction were 5′ GGGCGGTAGGCGTGTACGGTGG/3′ CTTCGGCCAGTAACGTTAGGGG. The pIRES2-PACE4-CRD-V5 was generated as explained above for pIRES2-PC5A-CRD-V5 except that different primers were used. The first PCR reactions were done with primers 5′ GGGCGGTAGGCGTGTACGGTGG/3′ GGTGTGGTACGGGTGCTCGGAGCAGGC and 5′ CTGCCTGCCGCCTGCTCCGAGCACCCG/3′ CTTCGGCCAGTAACGTTAGGGG using as cDNA template pIRES2-PACE4-V5. The following PCR reaction to produce the final cDNA used the primers 5′ GGGCGGTAGGCGTGTACGGTGG/3′ CTTCGGCCAGTAACGTTAGGGG. Human TIMP-1–4 were cloned into the digested Ecor1/Sma1 phCMV3 vector. The TIMP-[1–4]-Lamp1 proteins (TIMP[1-4]-LP) were obtained by fusion of C terminus of each human TIMP to the transmembrane-cytosolic tail of human Lamp1 (TM-CT-Lamp1; Figure 3A, 9), as described previously (Conesa et al., 2003). The myc-tagged cDNA (pcDNA3.1-Myc-His vector; Invitrogen, Carlsbad, CA) of human endothelial lipase-myc was a generous gift of Drs. Dan Rader and Weijun Jin (University of Pennsylvania, Philadelphia, PA) (Jaye et al., 1999). The recombinants of the N-terminal (NT-) and C-terminal (CT-) segments of TIMP-2 were made in pcDNA3 (Invitrogen). The TIMP-2-NT recombinant codes for the fragment 1–153 of human TIMP-2 fused at the C terminus with the V5-epitope. The TIMP-2-CT recombinant consists of the signal peptide of β-secretase (BACE1) (Benjannet et al., 2001) fused to a hemagglutinin (HA) tag followed by residues 154–220 of human TIMP-2. The pIRES2-furin and NARC-1 recombinants were described previously (Nour et al., 2003; Seidah et al., 2003).
Figure 3.
Effects of TIMP-2 and TIMP-2-LP on the levels of secreted convertases. (A) Schematic representation of TIMP-2 and TIMP-2-LP and results of their similar expression in HEK293 cells, as measured by their cellular [35S]Met/Cys immunoprecipitation levels. Western blots (Ab:V5) of the media and cell extracts of COS-1 cells expressing various forms of (B) PC5A and (C) CRD-PC5, CRD-PACE4, and NARC-1/PCSK9 in the presence or absence of TIMP-2 or TIMP-2-LP.
Figure 9.
PC5A/TIMP-2 complex binds HSPGs at the cell surface and the CRD enhances processing of proEL by PC5A. (A) Displacement of cell surface PC5A by exogenous heparin or heparinase-I as revealed by confocal microscopy with Ab:V5, in COS-1 cells transfected with pIRES-EGFP recombinant of FL-PC5A. The green EGFP fluorescence indicates nuclei of transfected cells. Bar, 10 μm. (B) HEK293 cells stably expressing human proEL-myc (see cartoon, left) were transiently transfected with indicated PC-constructs in pIRES-EGFP, where Vector represents an empty control pIRES-EGFP vector. The cells were pulse labeled with [35S]Cys/Met for 2 h and then chased for 4 h. Cell lysates were immunoprecipitated with the anti-myc antibody and analyzed by SDS-PAGE on 8% Tricine gels. The migration positions of proEL and its C-terminal cleavage product (CT-EL) are emphasized together with those of the molecular mass standards.
Cell Lines and Transfections
COS-1, HT1080, and human embryonic kidney (HEK)293 cell lines were grown in DMEM medium with 10% fetal bovine serum (FBS), whereas Chinese hamster ovary (CHO)-K1 and FD11 cells were grown in F12K medium with 10% FBS. Cells were transfected with Lipofectamine 2000 (Invitrogen) using a 2:1 ratio to cDNA, except for HEK293 cells that were transfected with Effectene (QIAGEN, Valencia, CA) at a 10:1 ratio of Effectene:cDNA. TIMP-2 null cells were maintained as reported previously (Morrison et al., 2001). These cells were transfected using the calcium phosphate precipitation technique as described in the commercially available protocol (BD Biosciences, San Jose, CA).
Protease Inhibitors and Microsequencing
Twenty-four hours posttransfection, HEK293 cells were treated for 6 h with different chelators and protease inhibitors in serum-free media. EDTA and EGTA (Sigma-Aldrich, St. Louis, MO) were used at a final concentration of 2 mM, whereas GM6001 (Chemicon International, Temecula, CA) was used at 25 μM. Captopril was used at a final concentration of 0.1 mM, whereas TAPI and phosphoramidon (Roche Diagnostics, Indianapolis, IN) were used at 10 μM final. Microsequencing of the [3H]Tyr-labeled R621A-PC5A was performed as described previously (Benjannet et al., 2001).
Western Blotting, Antibodies, Biosynthesis, and Immunoprecipitations
For Western blotting, the cells were washed with serum-free media 24 h posttransfection and incubated with serum-free media for the remaining 24 h. After 48 h, the media were collected, and cells were lysed with radioimmune precipitation assay buffer containing a cocktail of mixed protease inhibitors as described previously (Benjannet et al., 2001). Proteins from the media and 30 μg of proteins from cell extracts were resolved on 8% SDS-PAGE gel, electrotransferred onto nitrocellulose membrane, incubated with specific primary and secondary antibodies, and revealed by chemiluminescence as described previously (Nour et al., 2003).
Biosynthesis were performed 24 h posttransfection, and the cells were washed and pulse labeled for 4 h with 250 μCi/ml [35S]Met/Cys in RPMI 1640 medium containing 0.01% dialyzed serum. After the pulse, the media were recovered, and the cells lysed as mentioned previously, and the immunoprecipitated proteins were resolved on a 10% SDS-PAGE gel and then autoradiographed.
All antibodies were tested for specificity and optimal dilutions before use. Detection by Western blotting of PC5A and PC5B was done using a primary polyclonal rabbit anti-PC5 antibody directed against the N terminus of active PC5 (Ab:PC5-NT; 1:2000) (Nour et al., 2003). The secondary antibody used was an anti-rabbit IgG coupled to horseradish peroxidase (HRP) (Sigma-Aldrich; 1:10,000). The various truncated forms of PC5A and the different PC5A mutants were detected by Western blot using a monoclonal mouse anti-V5-HRP antibody (Ab:V5; 1:5000; Invitrogen). The actin levels were detected with the use of a rabbit anti-actin antibody (1:1000; Sigma-Aldrich).
The immunoprecipitations were done on both conditioned media and cell lysates. Cell lysis was performed in radioimmune precipitation assay buffer containing a cocktail of mixed protease inhibitors as described previously (Benjannet et al., 2001). The IgGs used for immunoprecipitations were rabbit polyclonals against TIMP-1 (Ab:TIMP-1; in house antibody), TIMP-2 (Ab: TIMP-2, recognizing the N terminus; Abcam, Cambridge, United Kingdom), TIMP-3 (Ab:TIMP-3; Chemicon International), and TIMP-4 (Ab:TIMP-4; Abcam International). The monoclonal antibody (mAb) mAb:HA was a gift from G. Boileau (University of Montreal, Montreal, Quebec, Canada), and mAb: Myc was reported previously (Rovere et al., 1999). Incubations with antibodies were done overnight at 4°C, and those of the protein A/G PLUS-agarose (Santa Cruz Biotechnology, Sanata Cruz, CA) were performed to 2–3 h at 4°C. For immunoprecipitations followed by Western blotting of TIMP-2 in COS-1 cells, we used Ab:TIMP-2, followed by goat anti-rabbit immunoglobulinbeads. The immune complex was detected on Western blots by an HRP-labeled anti-rabbit IgG (eBioscience, San Diego, CA) that preferentially binds native immunoglobulins, thereby preventing detection on Western blots of denatured heavy and light chain bands.
Immunocytochemistry and Immunohistochemistry
For cell surface immunocytochemical labeling, we used nonpermeabilizing conditions. Thus, COS-1 and TIMP-2 null cells were coated on microscope coverglasses no. 0 and transfected the following day. Forty-eight hours posttransfection, the cells were washed three times with 1× phosphate-buffered saline (PBS) and fixed with 3% paraformaldehyde for 1 h at 4°C. After fixation, cells were washed three times with PBS and incubated for 1 h with 5% normal goat serum (blocking buffer) at 4°C. The cells were then incubated overnight at 4°C with monoclonal mouse Ab:V5 (1:200) with or without rabbit polyclonal Ab:TIMP-2 or Ab:TIMP-2-CT (against the C terminus of TIMP-2; Chemicon International) 1:200 in blocking solution. After the incubation, the primary antibodies were removed and cells were washed four times with PBS and incubated for 60 min with Cy5-conjugated goat anti-rabbit IgG (1:1000; GE Healthcare, Baie d'Urfé, Québec, Canada) and with Alexa Fluor 555-conjugated goat anti-mouse IgG (10 μg/ml; Molecular Probes, Eugene, OR). Once secondary antibodies removed, the cells were washed four times with PBS and mounted in glycerol + 1,4-diazabicyclo[2.2.2]octane (DABCO; Sigma-Aldrich). Immunofluorescence analyses were performed with a Zeiss LSM-510 confocal microscope. For competition experiments, COS-1 cells were transfected with FL-PC5A-V5, and 48 h later the cells were incubated in serum-free DMEM media at 37°C for 1 h with various concentrations (4–20 μM) of recombinant human full-length TIMP-2 produced in Pichia pastoris (Mort, unpublished data). For displacement, we also used the same cells incubated for either 1 h with either heparin (1 mg/ml) or suramin (0.5 mg/ml) or for 24 h with 1 U/ml heparinase-I or 0.5 U/ml heparinase-III (all obtained from Sigma-Aldrich). Cell surface localization of PC5A was visualized by confocal microscopy with Ab:V5, as described above.
For immunohistochemistry, tissues (duodenum, jejunum, and liver) dissected from adult C56BL6 mice were embedded in OCT medium (Marcinkiewicz et al., 1993) and frozen at –20°C. Cryosections (10 μm) were fixed in methanol:acetone (1:1) for 5 min at –20°C, washed in PBS, blocked with 1% bovine serum albumin (15 min), and incubated with a monoclonal mouse mAb:TIMP-2 (1:100; Abcam) and a rabbit polyclonal antibody directed against mouse PC5 catalytic subunit (1:100; Ab:mPC5; Alexis Biochemicals, San Diego, CA) overnight at 4°C. After three washes in PBS, the mouse and rabbit primary antibodies were detected with Alexa Fluor 647-conjugated goat anti-mouse IgG and Alexa Fluor 555-conjugated anti-rabbit IgG (10 μg/ml; Molecular Probes), respectively. Sections were mounted in glycerol/DABCO and analyzed by confocal microscopy.
RESULTS
Excision of the CRD via C-Terminal Processing of PC5A and PC5B
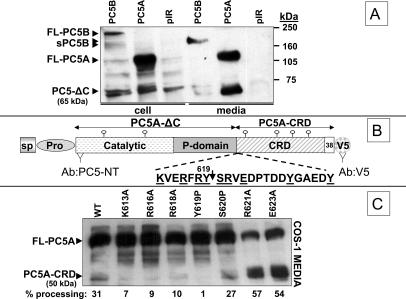
COS-1 cells were transiently transfected with either an empty pIRES2-EGFP vector, or the same construct expressing a recombinant soluble PC5A or membrane-bound PC5B (Figure 1A). The conditioned media and cell extracts were analyzed by Western blotting using an N-terminal PC5-specific antibody (Ab:PC5-NT) (De Bie et al., 1996). In cell lysates, PC5B is found in two forms, the full-length membrane-bound PC5B (FL-PC5B; ∼210 kDa) and a shorter form (sPC5B; ∼170 kDa), also found in the medium, together with an ∼65-kDa product, both likely representing shed soluble forms (Figure 1A). On the other hand, mostly mature full-length PC5A (FL-PC5A; ∼110 kDa) was detected in the cell extract, a form also found in the medium together with a significant amount of a C-terminally processed ∼65-kDa product (Figure 1A). The latter represents the N-terminal part of PC5A lacking the CRD, herein called PC5A-ΔC (Figure 1, A and B). The ∼110- and ∼65-kDa PC5A-derived proteins were observed previously in the regulated AtT20 cells (De Bie et al., 1996) and are presently detected in transfected COS-1 (Figure 1A), HEK293 (Figure 2A), and CHO (Figure 2B) cells, suggesting that such C-terminal truncation of PC5A is widespread and is not restricted to endocrine cells. Pulse-chase analyses revealed that the protease that cleaves the CRD acts late along the secretory pathway, as cleavage becomes apparent only after at least 1 h of chase, a time where PC5A has already reached the trans-Golgi network (TGN) and/or the cell surface (De Bie et al., 1996). We think that such processing occurs either after exit from the TGN, at the cell surface or in recycling endosomes.
Figure 1.
C-terminal cleavage of PC5 and the effect of various mutations on this processing. (A) Western blot (Ab:N-terminal of active PC5) of the SDS-PAGE-resolved conditioned media and lysates of COS-1 cells expressing either the empty vector pIRES2-EGFP (pIR), or recombinant PC5A and PC5B. (B) Schematic representation of PC5A and the identified C-terminal cleavage site. The recognition sites of the two antibodies used (Ab:PC5-NT and Ab:V5) are depicted. (C) Western blot of conditioned media of COS-1 cells expressing the different mutants of PC5A using an anti-V5 antibody. The percentage of processing of each mutant into the ∼50-kDa CRD was obtained by Image Quant analysis.
Figure 2.
Effect of protease inhibitors and overexpression of proteases on the C-terminal cleavage of PC5A. (A) V5-tagged WT PC5A and the R621A mutant were expressed with or without α1-PDX in HEK293 cells. Western blots were done on conditioned media using the Ab:V5. (B) Ab:V5-Western blot analysis of the conditioned media of CHO-K1 cells, FD11, and FD11-furin (stably overexpressing furin) expressing WT PC5A or its R621A mutant. (C) Ab:V5-Western blot analysis of the media of HEK293 cells transfected with WT PC5A and incubated with different protease inhibitors.
A recent study revealed that the inactive, secreted proPC5A-R84A mutant was still processed into the ∼65-kDa form, implying that the C-terminal truncation of PC5A is not autocatalytic (Nour et al., 2003). To identify the cleavage site, we analyzed in COS-1 cells the processing of various point mutants of PC5A around the suspected processing site (within aa 613–623; Figure 1, B and C). Because all constructs contained a C-terminal V5-epitope, we were able to visualize the ∼50-kDa C-terminal CRD fragment released (PC5A-CRD; Figure 1C). Whereas the Y619P mutant is not cleaved at all, processing of the K613A, R616A, and R618A mutants is reduced by >70%. Furthermore, the S620P mutation did not have a significant effect on processing, whereas both the R621A and the E623A mutants are approximately twofold better cleaved that the wild-type (WT) enzyme (Figure 1C). This experiment suggested that Tyr619 is important for cleavage. We do not think that Tyr619 is critical for folding because the Y619P mutant is secreted in normal amounts, suggesting it passed the quality control tests in the endoplasmic reticulum (Figure 1C). Arguably, Tyr619 may well be important for presentation of the scissile bond, as is the P1 position in many enzyme substrates. To confirm this, we immunoprecipitated (with the Ab:V5) the ∼50-kDa C-terminal CRD of PC5A released from the full-length PC5A (Figure 1B). The N-terminal Edman sequencing of the [3H]Tyr-labeled ∼50-kDa PC5A-CRD obtained from the well-processed R621A mutant revealed Tyr residues at positions 10 and 15 (in bold and underlined in Figure 1B, and S1). However, we cannot rule out the possibility that cleavage occurred at a preceding residue, e.g., Arg618 and that an aminopeptidase may have removed the exposed N-terminal amino acids, e.g., Tyr619. A similar type of N-terminal aminopeptidase trimming was recently reported for the β-secretase (BACE1) processing of a membrane-bound sialyltransferase (ST6Gal I), resulting in a three-amino acid loss from the N terminus of the cleaved product (Kitazume et al., 2004).
A Metalloprotease(s) Processes the CRD of PC5
To characterize the type of candidate protease(s) responsible for the processing of PC5A at its C terminus, we tested various classes of protease inhibitors, including the general PC-inhibitor α1-PDX (Figure 2A) (Benjannet et al., 1997; Jean et al., 1998) and metalloprotease inhibitors (Figure 2C). In HEK293 cells, the C-terminal processing of both WT PC5A and its R621A mutant were inhibited by cotransfection of α1-PDX (Figure 2A). However, further work demonstrated that PCs are not directly implicated in this C-terminal processing event. Accordingly, we analyzed the fate of PC5A and its R621A mutant in either CHO cells or in furin-deficient CHO-FD11 cells (Gordon et al., 1995), in the presence or absence of exogenous furin. Thus, whereas PC5A and its R621A mutant are processed in CHO cells, they are not significantly cleaved in CHO-FD11 and CHO-FD11 + furin cells (Figure 2B), suggesting that furin is not directly involved in this processing. Similarly, expression of each of the other PCs (PC5A, PACE4, or PC7) in FD11 cells did not affect this C-terminal cleavage (our unpublished data). Even though α1-PDX inhibits this processing in HEK293 cells, suggesting that one or more basic aa-specific PCs are needed for CRD excision (Figure 2A), the inability of furin to restore such cleavage in FD11 cells suggests that other proteases may also be affected in this chemically mutagenized cell line derived from CHO cells (Gordon et al., 1995). Finally, it is unlikely that by binding to PC5A, α1-PDX could mask its C-terminal cleavage site, because no cleavage occurs in FD11 cells, even in absence of α1-PDX (Figure 2B). These results argue against the direct implication of PCs in the C-terminal processing of PC5A, and the inhibition by α1-PDX rather suggests a role of PCs in the activation of cognate proteinase(s). Because PCs are known to cleave and activate a number of cell surface metalloproteinases at single or pairs of basic residues such as ADAMs and MT-MMPs, we hypothesized that activated MMPs or ADAMs may be involved in the C-terminal cleavage of PC5A. Hence, we tested the possible inhibitory effect of various metalloprotease inhibitors in HEK293 cells (Figure 2C). Accordingly, the metal chelators EDTA, EGTA, and the MMP inhibitor GM6001 (Elagoz et al., 2002) are effective blockers of the CRD release, whereas the ADAM17 inhibitor TAPI, the angiotensin-converting enzyme inhibitor captopril and the thermolysin and neutral endopeptidase-24.11 inhibitor phosphoramidon are not (Figure 2C). Thus, the available data suggest that a GM6001-inhibitable metalloprotease(s) is responsible for the excision of the CRD of PC5A by cleavage at Tyr619. However, metalloproteinases are usually not very specific, and it is very difficult to attribute a distinct motif to them. Some of them, though, do exhibit preferences for certain residues at specific P and especially P′ substrate positions. Examples include the MMPs that share many common features in their consensus cleavage motifs but that exhibit the presence of subtle distinctions in optimized peptide substrates (Turk et al., 2001). However, the proposed PC5A cleavage site does not clearly fit with any known metalloprotease cleavage motif, including those of the MMPs such as MMP2 (Turk et al., 2001).
TIMP-2 Interacts with PC5A via the CRD
Because furin and MT1-MMP colocalize at the cell surface of renal cells (Mayer et al., 2003), and both furin and PC5A process MT1-MMP (Yana and Weiss, 2000), it was plausible that PC5A may bind either MT1-MMP or its tethered inhibitor TIMP-2, possibly at the cell surface. To test this hypothesis, we used a cellular missorting technique previously applied to β3-integrin (Conesa et al., 2003), whereby the C termini of soluble TIMP-2 or the ectodomain of MT1-MMP were fused to a lysosomal/endosomal targeting sequence consisting of the Lamp1 transmembrane-cytosolic tail, herein named TIMP-2-LP (Figure 3A) and MT1-MMP-LP. According to this dominant negative technique, partners of TIMP-2 or MT1-MMP would be expected to cycle to the cell surface and be dragged to the endosomal/lysosomal compartments for degradation (Conesa et al., 2003). The data revealed that both MT1-MMP and MT1-MMP-LP do not significantly affect the level of C-terminal cleavage of PC5A or the secretion of its CRD (our unpublished data), suggesting that PC5 may not bind robustly MT1-MMP. Unexpectedly, although both TIMP-2 and TIMP-2-LP are well expressed (Figure 3A), only TIMP-2-LP (Figure 3B) largely eliminated secreted full length PC5A, but not PC5A-ΔC (lacking the CRD) (Figure 3B, left). Notably, there was a consistent drop in the levels of intracellular PC5A and the CRDs of PC5A and PACE4 in the presence of either TIMP-2 or TIMP-2-LP, compared with controls (Figure 3, B and C). We attributed this reproducible result to the coexpression experiment, even though in the control the same amount of vector DNA replaced the TIMP-2 constructs. Interestingly, even though both TIMP-2 and TIMP-2-LP similarly decrease the level of all intracellular forms of PC5A (Figure 3B, right), the ratios of media FL-PC5A and PC5A-CRD to that of the total PC5A (cell + medium) are always lower when these forms are coexpressed with TIMP-2-LP compared with TIMP-2. This was the first indication that TIMP-2 may possibly interact with the CRD of PC5A.
Further confirmation of this model was obtained by coexpression of TIMP-2-LP with a construct comprising the PC5A signal peptide directly fused to the CRD (herein called PC5A-CRD; Figure 5A). Thus, although PC5A-CRD is well folded and secreted, coexpression of TIMP-2-LP led to a significant reduction of the secreted protein (Figure 3, B and C, left). We also compared the effect of TIMP-2 and TIMP-2-LP on the secretion level of the CRDs of PC5A and its closest homologue PACE4. As a control, we also tested the recently described novel convertase NARC-1/PCSK9 because it exhibits a different CRD at its C terminus and possesses a unique cleavage specificity (Seidah et al., 2003; Benjannet et al., 2004). The data showed that for the full-length PC5A and the CRDs of both PC5A and PACE4, their secretion levels were reduced upon coexpression of TIMP-2-LP but not TIMP-2, whereas the secretion of full-length NARC-1/PCSK9 was not affected (Figure 3C). Thus, it seems that the CRDs of both PC5A and PACE4 can interact with TIMP-2.
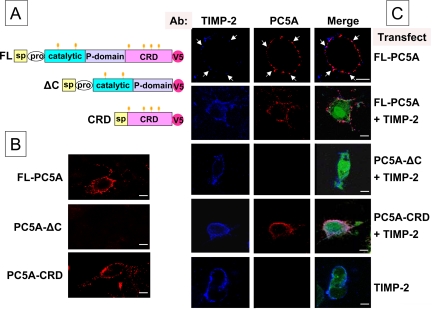
Figure 5.
Cell surface colocalization of TIMP-2 and FL-PC5A. (A) Schematic representation of the 3 constructs used for the analysis by confocal microscopy. (B) Cell surface labeling of COS-1 cells expressing FL-PC5A, PC5A-ΔC, or PC5A-CRD with Ab:V5. (C) Cell surface immunofluorescence of COS-1 cells probed with Ab:V5 and Ab:TIMP-2. TIMP-2 is in a phCMV vector and PC5A and its derivatives (all tagged with a V5 epitope) in the pIRES2-EGFP vector. Cells were transfected with either FL-PC5A and the empty phCMV vector, or with TIMP-2 plus either FL-PC5A, PC5A-ΔC or PC5A-CRD, or the pIRES2-EGFP vector alone. The green-EGFP fluorescence is a control of PC5-transfection, whereas the blue and red labeling are indicating TIMP-2 and the various forms of PC5A, respectively. Colocalization areas are seen in pink. Arrows point to colocalization areas of endogenous TIMP-2 and transfected FL-PC5A. Bars, 10 μm.
An independent verification of the potential interaction between PC5A and TIMP-2 was obtained from coimmunoprecipitation experiments of PC5A with either endogenous or coexpressed TIMP-2 and TIMP-2-LP in HT1080 cells. We first observed that in cell extracts and conditioned media overexpressed FL-PC5A coimmunoprecipitates with endogenous TIMP-2 (Control/FL-PC5A in Figure 4A). Equivalent data were also obtained when both partners are coexpressed, i.e., PC5A with either TIMP-2 or TIMP-2-LP (Figure 4A). Further evidence for the interaction of TIMP-2 and PC5A was obtained in another cell line, namely, COS-1 cells transfected with only FL-PC5A, because they express endogenously TIMP-2 (Pavlaki et al., 2002), but no PC5 or MT1-MMP. Thus, immunoprecipitation with Ab:TIMP-2 and Western blotting with Ab:V5 revealed that endogenous TIMP-2 binds PC5A in both cells and media (Control/FL-PC5A in Figure 4B, left). A similar result was also obtained when both TIMP-2 and FL-PC5A were coexpressed (TIMP-2/FL-PC5A in Figure 4B, left). However, as seen in Figure 4, A and B, we consistently observed that overexpression of TIMP-2 resulted in a larger proportion of immunoreactive FL-PC5A in cell extracts versus media. This is consistent with the possible cellular retention of PC5A by TIMP-2. For antibody specificity control, we show that immunoprecipitation with normal rabbit serum (NRS) followed by Western blotting with Ab:V5 did not reveal any PC5A immunoreactivity in cells (Figure 4B) or media (our unpublished data). The results of another control for the TIMP-2 antibody specificity are shown in Figure 4B, right. Thus, when both immunoprecipitations and Western blotting were made with Ab:TIMP-2, we observed the presence of an ∼21-kDa TIMP-2 in both cells and media. Endogenous TIMP-2 (vectors and control) was barely detectable in the cells but much more visible in the media. Consistent with the higher level of the secreted complex FL-PC5A-TIMP-2 from cells overexpressing only FL-PC5A (Figure 4B, control, left), we also find more TIMP-2 immunoreactivity in the media of these cells (Figure 4B, control, right). Here also, immunoprecipitation with NRS did not reveal any TIMP-2 on Western blots (Figure 4B, NRS, right). Finally, in Figure 4C we present evidence that in HEK293 cells the CRD of PC5A can coimmunoprecipitate with coexpressed TIMP-2 both in cell lysates and media. We note that the molecular mass of the CRD is slightly higher in the media (Figure 4C), which probably represents an effect due to the maturation of the four glycosylation chains of the PC5A-CRD.
Figure 4.
Coimmunoprecipitations of endogenous and overexpressed TIMP-2 with either FL-PC5A or PC5A-CRD. (A) HT1080 cells were transfected with either empty pIRES-EGFP + phCMV (Vectors), FL-PC5A + empty phCMV (Control), or FL-PC5A + TIMP-2 or TIMP-2-LP. The media and the cell extracts were immunoprecipitated using Ab:TIMP-2 and resolved by SDS-PAGE and then revealed by Western blot with Ab:V5. (B) Cos-1 cells were transfected with either empty pIRES-EGFP + phCMV (Vectors), FL-PC5A + empty phCMV (Control), or FL-PC5A + TIMP-2. Cell extracts and media were immunoprecipitated with Ab:TIMP-2 or NRS, resolved by SDS-PAGE, and then revealed by Western blot with Ab:V5 or Ab:TIMP-2. (C) HEK293 cells were transfected with PC5A-CRD and TIMP-2. The cell extract and medium were analyzed as in A.
In an effort to define the region within the CRD critical for its interaction with TIMP-2, we attempted to test whether 1) the loss of the granule sorting domain of PC5A, corresponding to its last C-terminal 38 aa (De Bie et al., 1996); or 2) N-glycosylation of the CRD, could affect the binding of PC5A to TIMP-2. Our data revealed that PC5A-Δ38 (lacking the C-terminal 38 aa) was equally immunoprecipitable with TIMP-2 and TIMP-2-LP as the full-length PC5A (our unpublished data). Finally, we also demonstrated that mutagenesis of the four potential N-glycosylation sites of PC5A-CRD (Asn667, Asn754, Asn804, and Asn854) to Ala did not affect its secretion or binding to TIMP-2 (Ponamarev and Seidah, unpublished data). Therefore, we conclude that neither the C-terminal 38 aa nor the N-glycosylation of the CRD is required for the binding of PC5A to TIMP-2.
PC5A and TIMP-2 Colocalize at the Cell Surface
Cell surface immunofluorescence was performed on nonpermeabilized monkey-derived kidney epithelial COS-1 cells that reportedly express TIMP-2 (Pavlaki et al., 2002). Accordingly, expression of FL-PC5A, PC5A-ΔC, or PC5A-CRD (Figure 5A) demonstrated that only those constructs containing the CRD were detectable at the cell surface (Figure 5B), possibly interacting with endogenous TIMP-2. In line with this notion, we found that endogenous TIMP-2 largely colocalizes at the cell surface of COS-1 cells with overexpressed FL-PC5A (Figure 5C, top), consistent with their deduced partnership from biochemical evidence (Figure 4). However, in view of its overexpression, we also observed some PC5A cell surface sites devoid of detectable endogenous TIMP-2, which may be due either to our TIMP-2 antibody detection sensitivity or to the presence of other TIMPs or cell surface PC5-binding molecules. Because coexpression of TIMP-2 with FL-PC5A led to higher cellular levels of both proteins (Figure 4B), we decided to use this paradigm to define the critical domain of PC5A that binds TIMP-2. Thus, we cotransfected human TIMP-2 with V5-tagged recombinant PC5A constructs (Figure 5C). Cell surface labeling was first performed using an anti-TIMP-2 antibody and a secondary antibody coupled to fluorolink Cy5 (blue color). In all cases, overexpressed TIMP-2 was well detected at the cell surface (Figure 5C, bottom), even though these cells lack MT1-MMP. This suggested that in COS-1 cells TIMP-2 can bind other plasma membrane molecules. To test whether PC5A via its CRD colocalizes with cell surface TIMP-2, we coexpressed TIMP-2 with various PC5A constructs. Immunolocalization of PC5A expression was obtained using an anti-V5 antibody followed by the addition of a secondary antibody conjugated to Alexa Fluor 555 (red color). Confocal cell surface analysis revealed that FL-PC5A and PC5A-CRD colocalize extensively with TIMP-2. In contrast, even in the presence of excess TIMP-2, PC5A-ΔC was not detectable at the cell surface (Figure 5C).
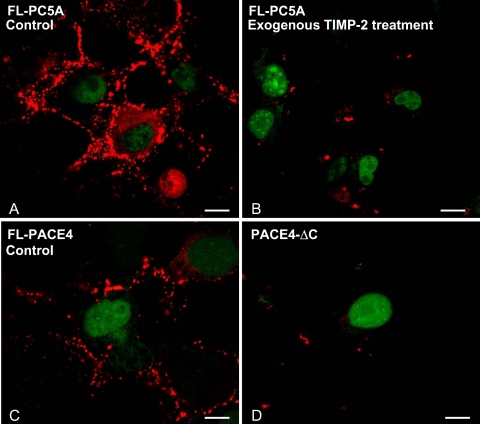
Both PC5A and PACE4 Localize to the Cell Surface and Exogenous TIMP-2 Can Displace PC5A
Immunofluorescence analysis of overexpressed FL-PC5A or FL-PACE4 in COS-1 cells revealed that both convertases, which share a similar CRD organization, localize to the cell surface (Figure 6, A and C). Furthermore, like PC5A-ΔC (Figure 5, B and C), PACE4-ΔC also does not bind to the cell surface of COS-1 cells (Figure 6D). In an effort to further prove that cell surface FL-PC5A can be displaced by its partner TIMP-2, we set up a competition experiment. Here, COS-1 cells transfected only with FL-PC5A were incubated for 1 h at 37°C with various concentrations (4–20 μM) of purified recombinant human FL-TIMP-2. Although similar data were obtained at lower concentrations, at 20 μM exogenous TIMP-2, it was evident that cell surface labeling of PC5A was greatly diminished (Figure 6B). Thus, TIMP-2 can displace cell surface FL-PC5A, likely by competing with endogenous molecules that retain it at the cell surface.
Figure 6.
Displacement of cell surface PC5A by exogenous TIMP-2 and cell surface localization of FL-PACE4 but not PACE4-ΔC. COS-1 cells were transfected with (A and B) FL-PC5A-V5, (C) FL-PACE4-V5, or (D) PACE4-ΔC-V5. Forty-eight hours, later the cells were incubated for 1 h (A) without (Control) or (B) with recombinant human TIMP-2 (20 μM) added exogenously. In all cases cell surface expression of PC5A, PACE4, and PACE4-ΔC were visualized by confocal microscopy with Ab:V5. Bars, 10 μm.
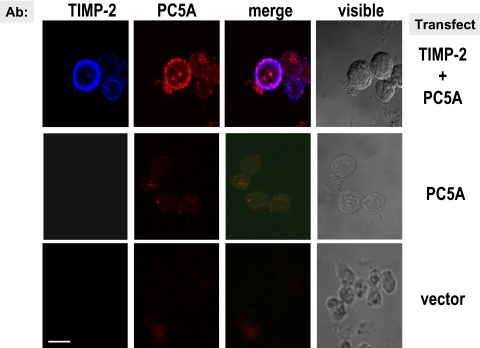
TIMP-2 Is Required for the Cell Surface Localization of PC5A
TIMP-2 null cells and TIMP-2 null cells overexpressing MT1-MMP (Morrison et al., 2001) are useful tools to probe the critical importance of the expression of TIMP-2 for the cell surface localization of PC5A. Both cell types were transiently transfected with FL-PC5A with and without TIMP-2. Transfected cells were treated with concanavalin A (ConA) to allow for the cell surface localization of TIMP-2 (Morrison et al., 2001). In agreement, in absence of ConA treatment in these cells, TIMP-2 does not localize to the cell surface (our unpublished data). Immunofluorescence analysis revealed that only in the presence of TIMP-2 could PC5A localize to the cell surface (Figure 7, top). Interestingly, PC5A was mostly found at the surface of cells that showed a high TIMP-2 labeling and revealed an uneven punctate staining, indicating that the cell surface localization of PC5A may depend on the presence of optimal amounts of TIMP-2 and/or specific lipid–protein complexes.
Figure 7.
PC5A requires TIMP-2 for its anchorage at the cell surface. TIMP-2 null fibroblast cells were transfected with the empty vectors or with FL-PC5A in the presence or absence of TIMP-2 and then treated with concanavalin A overnight (Morrison et al., 2001). Cell surface labeling was performed using Ab:V5 (red labeling) and Ab:TIMP-2 (blue labeling). Bar, 10 μm.
The C-Terminal Domain of TIMP-2 Binds to PC5A and Enhances its Cell Surface Localization
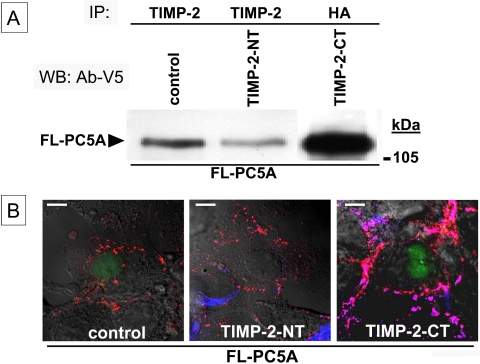
To define the domain of TIMP-2 responsible for the binding to PC5A through its CRD, and based on the TIMP-2 crystal structure that shows a bilobular conformation (Fernandez-Catalan et al., 1998), we divided the molecule into two segments. Accordingly, we generated a 153-aa N-terminal fragment (denoted TIMP-2-NT) starting from the signal peptide up to the sequence... MGCE153 that is fused to the V5-epitope. The C-terminal fragment (called TIMP-2-CT) consisted of a β-secretase signal peptide (Benjannet et al., 2001) followed by an HA tag and then the C-terminal 67-aa segment of TIMP-2, starting from the sequence C154KITR... up to the end, i.e.,... DIEDP220-stop. We then coexpressed, in COS-1 cells, FL-PC5A-V5 with either the vector alone (control), TIMP-2-NT, or TIMP-2-CT. Immunoprecipitation of the media of the control and TIMP-2-NT cells with Ab: TIMP-2 (that recognizes the N-terminal segment) or that of the TIMP-2-CT cells with mAb:HA, followed by Western blotting with mAb:V5 revealed that it is the TIMP-2-CT that is mainly responsible for the binding of PC5A with TIMP-2 (Figure 8A). Clearly, the level of coimmunoprecipitating FL-PC5A with TIMP-2-CT media is much enhanced over that of the control and TIMP-2-NT media. The small amount of coimmunoprecipitating FL-PC5A with either the control or TIMP-2-NT media is likely due to the presence of endogenous TIMP-2 in COS-1 cells (Figure 5C). Finally, we also showed that TIMP-2-CT coimmunoprecipitated with either PC5A-CRD or PACE4-CRD (our unpublished data), confirming that it is indeed the CRD of PC5A and PACE4 that binds TIMP-2-CT.
Figure 8.
PC5A binds to the C-terminal domain of TIMP-2 for its anchorage at the cell surface. (A) COS-1 cells were cotransfected at a DNA ratio of 1:3 with FL-PC5A-V5 and either empty pcDNA3 (Control), NT-TIMP-2, or CT-TIMP-2 expressing recombinants in pcDNA3. Cell media were immunoprecipitated with either Ab: TIMP-2 (Control and NT-TIMP-2) or mAb:HA (CT-TIMP-2), resolved by SDS-PAGE, and then revealed by Western blot with Ab:V5. (B) COS-1 cells were transfected as in A and analyzed by confocal microscopy for colocalization of TIMP-2 and FL-PC5A. Cell surface labeling was performed using Ab:V5 (red labeling) and Ab:TIMP-2 (left and middle; blue labeling) or Ab:TIMP-2-CT (right; blue labeling). The merged images reveal pink areas representing colocalized PC5A and TIMP-2 at the cell surface. Bar, 10 μm.
To further investigate whether it is the TIMP-2-CT that is also responsible for the plasma membrane localization of PC5A, the cell surface levels of PC5A and TIMP-2 were analyzed by confocal microscopy in these same cells as described above (Figure 8B). Again, it is clear that the expression of TIMP-2-CT (but not TMP-2-NT) greatly enhances the level of PC5A at the surface of COS-1 cells, which colocalize with TIMP-2-CT. We conclude that the CRD of PC5A and PACE4 bind the C-terminal segment of TIMP-2 and that the latter is also responsible for the attachment of this complex to the cell surface through an as yet undefined binding protein/receptor.
Heparan Sulfate Proteoglycans Are Critical for the Retention of PC5A to the Cell Surface: Physiological Relevance
Because TIMP-2-CT binds PC5A, this would exclude MT1-MMP (Overall et al., 1999; Kai et al., 2002) and α3β1 (Seo et al., 2003) as possible cell surface receptors of this complex, because they are known to bind TIMP-2-NT. Because it was reported that heparin could bind TIMP-2 (McCauley et al., 2001) as well as the CRD of secreted PC5A and PACE4, but not soluble furin (Tsuji et al., 2003), we tested whether heparin could displace out the cell surface FL-PC5A-TIMP-2 complex. The data showed that indeed incubation of COS-1 cells overexpressing FL-PC5A-V5 with heparin or heparinase-I resulted in markedly diminished levels of PC5A at the cell surface (Figure 9A). Similar results were also obtained upon incubation of these cells with either heparinase-III or the negatively charged sulfonated suramin (1,3,5-naphthylenetrisulfonic acid) (our unpublished data). We therefore conclude that the PC5A/TIMP-2 complex is mainly retained at the cell surface by HSPGs, possibly interacting with basic residues in the TIMP-2-CT and/or PC5A-CRD moieties.
This unexpected result led us to investigate the physiological significance of the surface localization of PC5A by looking at possible substrates that could also be retained at the cell surface via HSPGs. One such substrate is heparin-binding epidermal growth factor (HB-EGF) that is indeed predicted to be processed by a cell surface PC as it is cleaved at the sequence RDRKVR↓DL (No et al., 1994; Seidah and Prat, 2002). Another potential substrate was the soluble endothelial lipase (EL) first discovered by Rader's group (Jaye et al., 1999), a major endothelial enzyme regulating high-density lipoprotein metabolism. This was a potential substrate as it is very rich in endothelial cells, it colocalizes with PC5 on endothelial cells (Figure S2 available upon request), is retained at the cell surface by HSPGs, and can be displaced by heparin (Fuki et al., 2003). Furthermore, recent data suggested that it can be inactivated by the convertases through cell surface cleavage at the motif KMRNKR330↓NS (Gauster et al., 2005; Jin et al., 2005). Coexpression of human proEL with either furin, PACE4, FL-PC5A, or PC5A-ΔC in HEK293 cells revealed that although PACE4 is the best proEL to EL processing enzyme, followed by PC5A and furin, the lack of the CRD in PC5A-ΔC significantly decreases the efficacy of processing of proEL (Figure 9B). This is the first demonstration that the CRD is indeed important in the processing of a potential cell surface PC5A substrate.
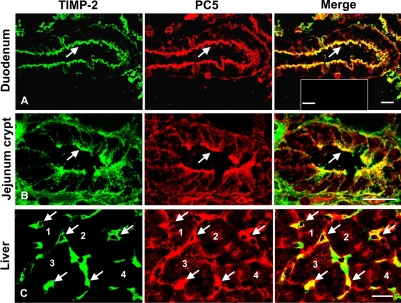
TIMP-2 and PC5 Colocalize in Various Mouse Tissues
Because both PC5A and TIMP-2 are widely distributed, we tested their endogenous in vivo expression by immunohistochemistry in selected mouse tissues where PC5A is prominently expressed, e.g., small intestine and liver (Lusson et al., 1993; Seidah et al., 1994). Although PC5A is abundant in both tissues, the PC5B isoform is mostly in intestine, but less so in endothelial cells of the liver (our unpublished data). The data show that PC5 and TIMP-2 colocalize over the apical surface of enterocytes found in the villi and crypts of the duodenum and jejunum as well as in liver sinusoids (Figure 10), in accordance with the high expression level of PC5 in gut (Lusson et al., 1993; Seidah et al., 1994) and in endothelial cells (Beaubien et al., 1995), respectively. In intestine, we note that most of the immunoreactivities of both PC5 (representing the sum of the levels of PC5A and PC5B, both of which are recognized by the antibody used) and TIMP-2 are associated with the cell surface. However, in liver sinusoids, we obtained a more diffused labeling of PC5 and TIMP-2, with some areas not exhibiting colocalization. It is possible that in those areas PC5 colocalizes with another TIMP. As controls, omission of the primary antibodies did not reveal any fluorescence signal (Figure 10, inset, top right). The data are consistent with the partnership of PC5 and TIMP-2 deduced from cell studies and extend this notion toward the realm of a whole animal.
Figure 10.
Cell surface colocalization of endogenous TIMP-2 and PC5 in mouse tissues. Frozen sections of adult mouse duodenum (A), jejunum (B), and liver (C) were coimmunostained with mAb: TIMP-2 (green) and Ab:mPC5 (red). Areas where both immunogens colocalize are seen in yellow. Arrows point to enterocyte apical cell surface of the duodenum's villi (A), apical cell surface of crypts of Lieberkühn in the jejunum (B), and liver sinusoids (C). In C, selected hepatocytes (indicated 1–4), surrounded by sinusoids (arrows) are emphasized. Bars, 20 μm. Controls lacking primary antibodies are shown in the inset of the merged picture in (A; bar, 40 μm).
PC5A Interacts with Other TIMPs
The above-mentioned results clearly demonstrated that the CRD of PC5A binds TIMP-2. We next investigated whether such binding is limited to TIMP-2 or whether other members of the TIMP family can also bind PC5A. We sought answers to this question using cotransfections of FL-PC5A-V5 with either TIMP-1, TIMP-3, or TIMP-4 or their CT-Lamp1-chimeras in COS-1 cells. Thus, from these cellular mislocalization experiments, we show that expression of TIMP-1,3,4-LP constructs results in a significant reduction in the level of cellular and secreted FL-PC5A (Figure 11A). These data suggest that all four TIMPs could interact with FL-PC5A. To further confirm this, we showed that each of the TIMP-1, -3, and -4 coimmunoprecipitates with FL-PC5A-V5 (Figure 11B). Controls using NRS instead of the specific TIMP antibodies did not reveal any coimmunoprecipitating PC5A. These data and those mentioned above demonstrate that PC5A can bind all four TIMPs.
Figure 11.
PC5A interacts with all TIMP family members. (A) Effects of overexpressed TIMP-1, -3, and -4 and their TIMP-LP derivatives on the levels of coexpressed FL-PC5A. Cell extracts and media were resolved by SDS-PAGE and then revealed by Western blot with Ab:V5. (B) Coimmunoprecipitations of endogenous and overexpressed family members TIMP-1, -3, and -4 with PC5A in COS-1 cells coexpressing FL-PC5A-V5. Cells were transiently transfected with either empty phCMV (Vector) or recombinant phCMV-TIMP-1, -3, and 4. Cell extracts were immunoprecipitated using Ab:TIMP-1, -3, and -4 or control NRS, resolved on 8% SDS-PAGE, and then revealed by Western blot with Ab:V5.
DISCUSSION
The present study demonstrated that PC5A is processed by a cell surface metalloproteinase, leading to the excision of its CRD, a process that may modulate its substrate(s) recognition and/or regulate its extracellular activity. Because many of the substrates of PC5A are membrane-bound cell surface proteins (Kalus et al., 2003), it was of interest to evaluate whether such cleavage affects the cellular localization of PC5A, and the possible cleavage of plasma membrane-associated substrates. Data in COS-1 cells do not support a significant role for the metalloproteases MT1-MMP and MT2-MMP or MMP2 in the processing of PC5A into PC5A-ΔC, consistent with the fact that overexpression of TIMP-2 did not affect this cleavage (our unpublished data), and suggesting that the cognate PC5-processing metalloproteinase is not inhibitable by TIMP-2. The next question was whether TIMP-2 could recruit PC5A to the cell surface, possibly in proximity to a protein that binds the complex TIMP-2/PC5A. Accordingly, we used a cell-based missorting approach (Conesa et al., 2003) as well as coimmunoprecipitation and immunocytochemical colocalization techniques. The data demonstrated that the C-terminal segment of TIMP-2 binds the CRD of PC5A (Figure 8) and that cell surface immobilized TIMP-2 is required for the plasma membrane colocalization of PC5A with TIMP-2 (Figure 7). It is possible that processing of the CRD regulates the level of the cell surface PC5-TIMP-2 heterodimer, resulting in the release of PC5-ΔC.
Previous studies demonstrated that the majority of protein precursors cleaved by the soluble/secreted PC5A are membrane-bound proteins, e.g., adhesion molecules including integrin α-chains (Lehmann et al., 1996; Lissitzky et al., 2000; Stawowy et al., 2004) and the neural adhesion protein L1 (Kalus et al., 2003) as well as TGFβ-like precursors (Dubois et al., 2001; Ulloa et al., 2001). Recently, it was shown that PCs can also process cell surface proteins bound to proteoglycans including proHB-EGF (No et al., 1994; Seidah and Prat, 2002), and proEL (Gauster et al., 2005; Jin et al., 2005). Our data show that the CRD enhances the efficacy of processing of proEL to EL by PC5A (Figure 9B), likely because of a proximity effect resulting from the close juxtaposition of the proteinase and its substrate through interactions with cell surface HSPGs. Indeed, both proEL (Fuki et al., 2003) and PC5A (Figure 9A) can be displaced form the cell surface by heparin, suramin, and by treatment with heparinase-I or -III. This is the first evidence for a physiological relevance of the cell surface concentration of PC5A and PACE4 (Figure 6). Other potential cell surface HSPG-bound precursors that require PC-cleavage include Glypican-3 that inhibits cell proliferation (tumor suppressor), and regulates cell survival during development (De Cat et al., 2003), and Lubricin, a secreted mucin-like proteoglycan that is a major lubricant in articulating joints (Rhee et al., 2005).
TIMP-2 was recently shown to bind α3β1 and to signal an endothelial cell antiproliferative response through this receptor (Seo et al., 2003), whereas PC5A has been reported to process a number of α-chain integrins (Lehmann et al., 1996; Lissitzky et al., 2000; Stawowy et al., 2004). It is therefore conceivable that the TIMP-2/PC5A complex may play a role in the observed inhibition of angiogenesis by TIMP-2 (Seo et al., 2003). This finding is consistent with the high expression level of PC5A in endothelial cells lining blood vessels (Beaubien et al., 1995) (Figures 10 and S2) and its reported transcriptional regulation through contact inhibition (Campan et al., 1996).
Before this work, the function of the CRD of PC5, PACE4, and furin was unknown. It was suggested that the CRD of these convertases could allow them to access plasma membrane precursors (Oliva et al., 2000), interact with the extracellular matrix (Tsuji et al., 2003), affect cell growth and/or localization (Lusson et al., 1993) as well as enhance the stability of PC5B (Wang et al., 2004). In view of specific motifs within its cytosolic tail, PC5B cycles between the TGN and the cell surface (De Bie et al., 1996; Xiang et al., 2000), and because it is PC5A that is present in all PC5-positive cells (Lusson et al., 1993), including endothelial cells (Beaubien et al., 1995), whereas PC5B is mostly in intestinal cells (Lusson et al., 1993), it was thus of interest to define the role of the PC5A's CRD. The present study demonstrated that the CRD of the soluble PC5A targets this enzyme to the plasma membrane. Moreover, we showed that in contrast to NARC-1/PCSK9, the CRD of PC5A and PACE4 can specifically interact with TIMP-2. The TIMP-2/CRD complex formation of PC5A or PACE4 was shown to implicate the C-terminal domain of TIMP-2 (Figures 8 and 9), and an as yet undefined motif within the CRD of these PCs, which is not found within the CRD of NARC-1/PCSK9 (Figure 3). The crystal structure of TIMP-2 revealed it to contain two domains, an N-terminal and a negatively charged C-terminal domain linked by a single glutamic acid residue Glu153 (Fernandez-Catalan et al., 1998; Morgunova et al., 2002). Although the inhibitory N-terminal domain binds to the active sites of MT1-MMP (Butler et al., 1998; Zucker et al., 1998), and integrin α3β1 (Seo et al., 2003), the C-terminal domain interacts with the hemopexin domain of proMMP-2 (Overall et al., 1999; Kai et al., 2002). It will be of interest to define the residues within the C-terminal domain of TIMP-2 that interact with the CRD of PC5A and PACE4, and with HSPGs, and whether these are distinct from those implicated in its binding to the hemopexin domain of proMMP-2.
We also tested the possible interaction of PC5 with other TIMPs (e.g., TIMP-1, -3, and -4), especially in view of the fact that the mRNA levels of TIMP-3 and PC5A are coordinately regulated, and both are coexpressed during placental embryonic implantation and decidualization (Rancourt and Rancourt, 1997; Wong et al., 2002) and are possibly involved in the cell surface processing of zona pellucida domain proteins (Jovine et al., 2004). In addition, PC5A can process TGFβ-like (usually antiproliferative) proteins (Dubois et al., 2001; Ulloa et al., 2001) and active TGFβ regulates the expression of TIMP-3 (Leco et al., 1994). Our present data suggest that PC5A can interact with all four TIMPs (Figure 11). In view of the high affinity of TIMP-3 for the ECM (Leco et al., 1994), it is also possible that PC5A binds TIMP-3 in the ECM.
The conservation of the Cys motif within the CRD of PC5A and PACE4 was also found in nonconvertase proteins such as in the epidermal growth factor receptor (Ward et al., 1995) and in an ECM protein known as Fras1 (McGregor et al., 2003). All these proteins are localized at the plasma membrane and are implicated in development and/or in cellular growth. A recent study on the function of the second CRD of the EGFR demonstrated its importance for the targeting of this receptor to caveolae/rafts at the plasma membrane (Yamabhai and Anderson, 2002). These studies are in accord with the role presently ascribed to the CRD of PC5A as a critical domain for the plasma membrane localization of this enzyme. The observed punctate labeling of the cell surface PC5A (Figures 5, 6, 7, 8) resembles that of the cell surface distribution of caveolae/rafts domains (Puyraimond et al., 2001), and our preliminary results seem to confirm the cell surface localization of PC5A within lipid rafts (Mayer and Seidah, unpublished data).
In conclusion, the data presented in this work reveal a new function for the CRD of the convertases PC5 and PACE4 and provide a link between mammalian serine subtilase-like proprotein convertases and cell surface proteins through TIMPs. This may also lead the way toward the future identification of other interacting molecules with the CRDs of furin and NARC-1/PCSK9.
Supplementary Material
Acknowledgments
We thank A. Chamberland, J. Hamelin, S. Benjannet, M.-C. Asselin, L. Wickham, and F. Sirois for technical assistance and M. Ponamarev for performing the PC5A-CRD N-glycosylation mutants and coimmunoprecipitation experiment with TIMP-2. Many thanks to all the other members of the Seidah laboratory for discussions and encouragement, and to M. Brigitte for secretarial assistance. This work was supported by a Canadian Institutes of Health Research (CIHR MGP-44363), a Canada Chair #201652, and by the Protein Engineering Network of Centers of Excellence (PENCE). G. M. was supported by Conseil de la Recherche en Sciences Naturelles et Génie du Canada and Institut de Recherches Cliniques de Montréal-Pizzagalli postdoctoral fellowships.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–06–0504) on August 31, 2005.
Abbreviations used: aa, amino acid; Ab, antibody; ConA, concanavalin A; CRD, cysteine-rich domain; EL, endothelial lipase; ECM, extracellular matrix FBS; HSPG, heparan sulfate proteoglycan; MMP, matrix metalloproteinase; PC, proprotein convertase; TIMP-2-LP, TIMP-2-Lamp1 protein; TIMP, tissue inhibitor of metalloproteinase; TM-CT, transmembrane-cytosolic tail; WT, wild type; NT-, N-terminal; CT-, C-terminal.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abifadel, M., et al. (2003). Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156. [DOI] [PubMed] [Google Scholar]
- Baker, A. H., Edwards, D. R., and Murphy, G. (2002). Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J. Cell Sci. 115, 3719–3727. [DOI] [PubMed] [Google Scholar]
- Beaubien, G., Schafer, M. K., Weihe, E., Dong, W., Chretien, M., Seidah, N. G., and Day, R. (1995). The distinct gene expression of the pro-hormone convertases in the rat heart suggests potential substrates. Cell Tissue Res. 279, 539–549. [DOI] [PubMed] [Google Scholar]
- Benjannet, S., et al. (2001). Post-translational processing of β-secretase (β-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-β production. J. Biol. Chem. 276, 10879–10887. [DOI] [PubMed] [Google Scholar]
- Benjannet, S., et al. (2004). NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279, 48865–48875. [DOI] [PubMed] [Google Scholar]
- Benjannet, S., Savaria, D., Laslop, A., Munzer, J. S., Chretien, M., Marcinkiewicz, M., and Seidah, N. G. (1997). α1-antitrypsin Portland inhibits processing of precursors mediated by proprotein convertases primarily within the constitutive secretory pathway. J. Biol. Chem. 272, 26210–26218. [DOI] [PubMed] [Google Scholar]
- Bergeron, E., Basak, A., Decroly, E., and Seidah, N. G. (2003). Processing of α4 integrin by the proprotein convertases: histidine at position P6 regulates cleavage. Biochem. J. 373, 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, G. S., Butler, M. J., Atkinson, S. J., Will, H., Tamura, T., van Westrum, S. S., Crabbe, T., Clements, J., d'Ortho, M. P., and Murphy, G. (1998). The TIMP2 membrane type 1 metalloproteinase “receptor” regulates the concentration and efficient activation of progelatinase A. A kinetic study. J. Biol. Chem. 273, 871–880. [DOI] [PubMed] [Google Scholar]
- Campan, M., Yoshizumi, M., Seidah, N. G., Lee, M. E., Bianchi, C., and Haber, E. (1996). Increased proteolytic processing of protein tyrosine phosphatase μ in confluent vascular endothelial cells: the role of PC5, a member of the subtilisin family. Biochemistry 35, 3797–3802. [DOI] [PubMed] [Google Scholar]
- Conesa, M., Prat, A., Mort, J. S., Marvaldi, J., Lissitzky, J. C., and Seidah, N. G. (2003). Down-regulation of αv/β3 integrin via misrouting to lysosomes by overexpression of a β3Lamp1 fusion protein. Biochem. J. 370, 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie, I., Marcinkiewicz, M., Malide, D., Lazure, C., Nakayama, K., Bendayan, M., Seidah, N. G. (1996). The isoforms of proprotein convertase PC5 are sorted to different subcellular compartments. J. Cell Biol. 135, 1261–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cat, B., Muyldermans, S. Y., Coomans, C., Degeest, G., Vanderschueren, B., Creemers, J., Biemar, F., Peers, B., and David, G. (2003). Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J. Cell Biol. 163, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, C. M., Blanchette, F., Laprise, M .H., Leduc, R., Grondin, F., and Seidah, N. G. (2001). Evidence that furin is an authentic transforming growth factor-β1-converting enzyme. Am. J. Pathol. 158, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagoz, A., Benjannet, S., Mammarbassi, A., Wickham, L., and Seidah, N. G. (2002). Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J. Biol. Chem. 277, 11265–11275. [DOI] [PubMed] [Google Scholar]
- Fernandez-Catalan, C., Bode, W., Huber, R., Turk, D., Calvete, J. J., Lichte, A., Tschesche, H., and Maskos, K. (1998). Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 17, 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuki, I. V., Blanchard, N., Jin, W., Marchadier, D. H., Millar, J. S., Glick, J. M., and Rader, D. J. (2003). Endogenously produced endothelial lipase enhances binding and cellular processing of plasma lipoproteins via heparan sulfate proteoglycan-mediated pathway. J. Biol. Chem. 278, 34331–34338. [DOI] [PubMed] [Google Scholar]
- Gangnon, F., Jegou, S., Vallarino, M., Vieau, D., and Vaudry, H. (2003). Molecular characterization of the cDNA and localization of the mRNA encoding the prohormone convertase PC5-A in the European green frog. J. Comp. Neurol. 456, 60–72. [DOI] [PubMed] [Google Scholar]
- Gauster, M., Hrzenjak, A., Schick, K., and Frank, S. (2005). Endothelial lipase is inactivated upon cleavage by the members of the proprotein convertase family. J. Lipid Res. 46, 977–987. [DOI] [PubMed] [Google Scholar]
- Gordon, V. M., Klimpel, K. R., Arora, N., Henderson, M. A., and Leppla, S. H. (1995). Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect. Immun. 63, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaye, M., Lynch, K. J., Krawiec, J., Marchadier, D., Maugeais, C., Doan, K., South, V., Amin, D., Perrone, M., and Rader, D. J. (1999). A novel endothelial-derived lipase that modulates HDL metabolism. Nat. Genet. 21, 424–428. [DOI] [PubMed] [Google Scholar]
- Jean, F., Stella, K., Thomas, L., Liu, G., Xiang, Y., Reason, A. J., and Thomas, G. (1998). α1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc. Natl. Acad. Sci. USA 95, 7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Goldberg, I. D., and Shi, Y. E. (2002). Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene 21, 2245–2252. [DOI] [PubMed] [Google Scholar]
- Jin, W., Fuki, I. V., Seidah, N. G., Benjannet, S., Glick, J., and Rader, D. J. (2005) Proprotein convertases are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. (in press). [DOI] [PubMed]
- Jovine, L., Qi, H., Williams, Z., Litscher, E. S., and Wassarman, P. M. (2004). A duplicated motif controls assembly of zona pellucida domain proteins. Proc. Natl. Acad. Sci. USA 101, 5922–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, H. S., Butler, G. S., Morrison, C. J., King, A. E., Pelman, G. R., and Overall, C. M. (2002). Utilization of a novel recombinant myoglobin fusion protein expression system to characterize the tissue inhibitor of metalloproteinase (TIMP)-4 and TIMP-2 C-terminal domain and tails by mutagenesis. The importance of acidic residues in binding the MMP-2 hemopexin C-domain. J. Biol. Chem. 277, 48696–48707. [DOI] [PubMed] [Google Scholar]
- Kalus, I., Schnegelsberg, B., Seidah, N. G., Kleene, R., and Schachner, M. (2003). The proprotein convertase PC5A and a metalloprotease are involved in the proteolytic processing of the neural adhesion molecule L1. J. Biol. Chem. 278, 10381–10388. [DOI] [PubMed] [Google Scholar]
- Kitazume, S., Suzuki, M., Saido, T. C., and Hashimoto, Y. (2004). Involvement of proteases in glycosyltransferase secretion: Alzheimer's β-secretase-dependent cleavage and a following processing by an aminopeptidase. Glycoconj. J. 21, 25–29. [DOI] [PubMed] [Google Scholar]
- Leco, K. J., Khokha, R., Pavloff, N., Hawkes, S. P., and Edwards, D. R. (1994). Tissue inhibitor of metalloproteinases-3 (TIMP-3) is an extracellular matrix-associated protein with a distinctive pattern of expression in mouse cells and tissues. J. Biol. Chem. 269, 9352–9360. [PubMed] [Google Scholar]
- Lehmann, M., Rigot, V., Seidah, N. G., Marvaldi, J., and Lissitzky, J. C. (1996). Lack of integrin α-chain endoproteolytic cleavage in furin-deficient human colon adenocarcinoma cells LoVo. Biochem. J. 317, 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissitzky, J. C., Luis, J., Munzer, J. S., Benjannet, S., Parat, F., Chretien, M., Marvaldi, J., and Seidah, N. G. (2000). Endoproteolytic processing of integrin pro-α subunits involves the redundant function of furin and proprotein convertase (PC) 5A, but not paired basic amino acid converting enzyme (PACE) 4, PC5B or PC7. Biochem. J. 346, 133–138. [PMC free article] [PubMed] [Google Scholar]
- Lusson, J., Vieau, D., Hamelin, J., Day, R., Chretien, M., and Seidah, N. G. (1993). cDNA structure of the mouse and rat subtilisin/kexin-like PC 5, a candidate proprotein convertase expressed in endocrine and nonendocrine cells. Proc. Natl. Acad. Sci. USA 90, 6691–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewicz, M., Day, R., Seidah, N. G., and Chretien, M. (1993). Ontogeny of the prohormone convertases PC1 and PC2 in the mouse hypophysis and their colocalization with corticotropin and α-melanotropin. Proc. Natl. Acad. Sci. USA 90, 4922–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, G., Boileau, G., and Bendayan, M. (2003). Furin interacts with proMT1-MMP and integrin αV at specialized domains of renal cell plasma membrane. J. Cell Sci. 116, 1763–1773. [DOI] [PubMed] [Google Scholar]
- McCauley, T. C., Zhang, H. M., Bellin, M. E., and Ax, R. L. (2001). Identification of a heparin-binding protein in bovine seminal fluid as tissue inhibitor of metalloproteinases-2. Mol. Reprod. Dev. 58, 336–341. [DOI] [PubMed] [Google Scholar]
- McGregor, L., et al. (2003). Fraser syndrome and mouse blebbed phenotype caused by mutations in FRAS1/Fras1 encoding a putative extracellular matrix protein. Nat. Genet. 34, 203–208. [DOI] [PubMed] [Google Scholar]
- McQuibban, G. A., Gong, J. H., Tam, E. M., McCulloch, C. A., Clark-Lewis, I., and Overall, C. M. (2000). Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science 289, 1202–1206. [DOI] [PubMed] [Google Scholar]
- Morgunova, E., Tuuttila, A., Bergmann, U., and Tryggvason, K. (2002). Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2. Proc. Natl. Acad. Sci. USA 99, 7414–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, C. J., Butler, G. S., Bigg, H. F., Roberts, C. R., Soloway, P. D., and Overall, C. M. (2001). Cellular activation of MMP-2 (gelatinase A) by MT2-MMP occurs via a TIMP-2-independent pathway. J. Biol. Chem. 276, 47402–47410. [DOI] [PubMed] [Google Scholar]
- Nachtigal, M. W., and Ingraham, H. A. (1996). Bioactivation of Mullerian inhibiting substance during gonadal development by a kex2/subtilisin-like endoprotease. Proc. Natl. Acad. Sci. USA 93, 7711–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Murakami, K., and Nakayama, K. (1993). Identification of an isoform with an extremely large Cys-rich region of PC6, a Kex2-like processing endoprotease. FEBS Lett. 327, 165–171. [DOI] [PubMed] [Google Scholar]
- No, M., Raab, G., Lau, K., Abraham, J. A., Klagsbrun, M. (1994). Purification and characterization of transmembrane forms of heparin-binding EGF-like growth factor. J. Biol. Chem. 269, 31315–31321. [PubMed] [Google Scholar]
- Nour, N., Basak, A., Chretien, M., and Seidah, N. G. (2003). Structure-Function Analysis of the prosegment of the proprotein convertase PC5A. J. Biol. Chem. 278, 2886–2895. [DOI] [PubMed] [Google Scholar]
- Oliva, A. A., Jr., Chan, S. J., and Steiner, D. F. (2000). Evolution of the prohormone convertases: identification of a homologue of PC6 in the protochordate amphioxus. Biochim. Biophys. Acta 1477, 338–348. [DOI] [PubMed] [Google Scholar]
- Overall, C. M., King, A. E., Sam, D. K., Ong, A. D., Lau, T. T., Wallon, U. M., DeClerck, Y. A., and Atherstone, J. (1999). Identification of the tissue inhibitor of metalloproteinases-2 (TIMP-2) binding site on the hemopexin carboxyl domain of human gelatinase A by site-directed mutagenesis. The hierarchical role in binding TIMP-2 of the unique cationic clusters of hemopexin modules III and IV. J. Biol. Chem. 274, 4421–4429. [DOI] [PubMed] [Google Scholar]
- Overall, C. M., and Lopez-Otin, C. (2002). Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat. Rev. Cancer 2, 657–672. [DOI] [PubMed] [Google Scholar]
- Pavlaki, M., Cao, J., Hymowitz, M., Chen, W. T., Bahou, W., and Zucker, S. (2002). A conserved sequence within the propeptide domain of membrane type 1 matrix metalloproteinase is critical for function as an intramolecular chaperone. J. Biol. Chem. 277, 2740–2749. [DOI] [PubMed] [Google Scholar]
- Puyraimond, A., Fridman, R., Lemesle, M., Arbeille, B., and Menashi, S. (2001). MMP-2 colocalizes with caveolae on the surface of endothelial cells. Exp. Cell Res. 262, 28–36. [DOI] [PubMed] [Google Scholar]
- Rancourt, S. L., and Rancourt, D. E. (1997). Murine subtilisin-like proteinase SPC6 is expressed during embryonic implantation, somitogenesis, and skeletal formation. Dev. Genet. 21, 75–81. [DOI] [PubMed] [Google Scholar]
- Rhee, D. K., Marcelino, J., Al Mayouf, S., Schelling, D. K., Bartels, C. F., Cui, Y., Laxer, R., Goldbach-Mansky, R., and Warman, M. L. (2005). Consequences of disease-causing mutations on lubricin protein synthesis, secretion, and post-translational processing. J. Biol. Chem. 280, 31325–31332. [DOI] [PubMed] [Google Scholar]
- Rovere, C., Luis, J., Lissitzky, J. C., Basak, A., Marvaldi, J., Chretien, M., and Seidah, N. G. (1999). The RGD motif and the C-terminal segment of proprotein convertase 1 are critical for its cellular trafficking but not for its intracellular binding to integrin α5β1. J. Biol. Chem. 274, 12461–12467. [DOI] [PubMed] [Google Scholar]
- Seidah, N. G., Benjannet, S., Wickham, L., Marcinkiewicz, J., Jasmin, S. B., Stifani, S., Basak, A., Prat, A., and Chretien, M. (2003). The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA 100, 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah, N. G., and Chretien, M. (1999). Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 848, 45–62. [DOI] [PubMed] [Google Scholar]
- Seidah, N. G., Chretien, M., and Day, R. (1994). The family of subtilisin/kexin like pro-protein and pro-hormone convertases: divergent or shared functions. Biochimie 76, 197–209. [DOI] [PubMed] [Google Scholar]
- Seidah, N. G., and Prat, A. (2002). Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 38, 79–94. [DOI] [PubMed] [Google Scholar]
- Seo, D. W., Li, H., Guedez, L., Wingfield, P. T., Diaz, T., Salloum, R., Wei, B. Y., and Stetler-Stevenson, W. G. (2003). TIMP-2 mediated inhibition of angiogenesis: an MMP-independent mechanism. Cell 114, 171–180. [DOI] [PubMed] [Google Scholar]
- Srour, N., Lebel, A., McMahon, S., Fournier, I., Fugere, M., Day, R., and Dubois, C. M. (2003). TACE/ADAM-17 maturation and activation of sheddase activity require proprotein convertase activity. FEBS Lett. 554, 275–283. [DOI] [PubMed] [Google Scholar]
- Stawowy, P., Graf, K., Goetze, S., Roser, M., Chretien, M., Seidah, N. G., Fleck, E., and Marcinkiewicz, M. (2003). Coordinated regulation and colocalization of αv integrin and its activating enzyme proprotein convertase PC5 in vivo. Histochem. Cell Biol. 119, 239–245. [DOI] [PubMed] [Google Scholar]
- Stawowy, P., Kallisch, H., Veinot, J. P., Kilimnik, A., Prichett, W., Goetze, S., Seidah, N. G., Chretien, M., Fleck, E., and Graf, K. (2004). Endoproteolytic activation of α(v) integrin by proprotein convertase PC5 is required for vascular smooth muscle cell adhesion to vitronectin and integrin-dependent signaling. Circulation 109, 770–776. [DOI] [PubMed] [Google Scholar]
- Sternlicht, M. D., and Werb, Z. (2001). How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17, 463–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, E. M., Morrison, C. J., Wu, Y. I., Stack, M. S., and Overall, C. M. (2004). Membrane protease proteomics: isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. USA 101, 6917–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, A., Sakurai, K., Kiyokage, E., Yamazaki, T., Koide, S., Toida, K., Ishimura, K., and Matsuda, Y. (2003). Secretory proprotein convertases PACE4 and PC6A are heparin-binding proteins which are localized in the extracellular matrix. Potential role of PACE4 in the activation of proproteins in the extracellular matrix. Biochim. Biophys. Acta 1645, 95–104. [DOI] [PubMed] [Google Scholar]
- Turk, B. E., Huang, L. L., Piro, E. T., and Cantley, L. C. (2001). Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 19, 661–667. [DOI] [PubMed] [Google Scholar]
- Ulloa, L., Creemers, J. W., Roy, S., Liu, S., Mason, J., and Tabibzadeh, S. (2001). Lefty proteins exhibit unique processing and activate the MAPK pathway. J. Biol. Chem. 276, 21387–21396. [DOI] [PubMed] [Google Scholar]
- Wang, L., Yang, G., and Wu, X. (2004). Identification of the role of a cysteinerich region of PC6B by determining the enzymatic characteristics of its mutants. Mol. Biotechnol. 27, 15–22. [DOI] [PubMed] [Google Scholar]
- Ward, C. W., Hoyne, P. A., and Flegg, R. H. (1995). Insulin and epidermal growth factor receptors contain the cysteine repeat motif found in the tumor necrosis factor receptor. Proteins 22, 141–153. [DOI] [PubMed] [Google Scholar]
- Wong, B. S., Liu, S., Schultz, G. A., and Rancourt, D. E. (2002). Subtilisin proprotein convertase-6 expression in the mouse uterus during implantation and artificially induced decidualization. Mol. Reprod. Dev. 61, 453–459. [DOI] [PubMed] [Google Scholar]
- Xiang, Y., Molloy, S. S., Thomas, L., and Thomas, G. (2000). The PC6B cytoplasmic domain contains two acidic clusters that direct sorting to distinct trans-Golgi network/endosomal compartments. Mol. Biol. Cell 11, 1257–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai, M., and Anderson, R. G. (2002). Second cysteine-rich region of epidermal growth factor receptor contains targeting information for caveolae/rafts. J. Biol. Chem. 277, 24843–24846. [DOI] [PubMed] [Google Scholar]
- Yana, I., and Weiss, S. J. (2000). Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol. Biol. Cell 11, 2387–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker, S., Drews, M., Conner, C., Foda, H. D., DeClerck, Y. A., Langley, K. E., Bahou, W. F., Docherty, A. J., and Cao, J. (1998). Tissue inhibitor of metalloproteinase-2 (TIMP-2) binds to the catalytic domain of the cell surface receptor, membrane type 1-matrix metalloproteinase 1 (MT1-MMP). J. Biol. Chem. 273, 1216–1222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.