Abstract
EBs and CLIPs are evolutionarily conserved proteins, which associate with the tips of growing microtubules, and regulate microtubule dynamics and their interactions with intracellular structures. In this study we investigated the functional relationship of CLIP-170 and CLIP-115 with the three EB family members, EB1, EB2(RP1), and EB3 in mammalian cells. We showed that both CLIPs bind to EB proteins directly. The C-terminal tyrosine residue of EB proteins is important for this interaction. When EB1 and EB3 or all three EBs were significantly depleted using RNA interference, CLIPs accumulated at the MT tips at a reduced level, because CLIP dissociation from the tips was accelerated. Normal CLIP localization was restored by expression of EB1 but not of EB2. An EB1 mutant lacking the C-terminal tail could also fully rescue CLIP dissociation kinetics, but could only partially restore CLIP accumulation at the tips, suggesting that the interaction of CLIPs with the EB tails contributes to CLIP localization. When EB1 was distributed evenly along the microtubules because of overexpression, it slowed down CLIP dissociation but did not abolish its preferential plus-end localization, indicating that CLIPs possess an intrinsic affinity for growing microtubule ends, which is enhanced by an interaction with the EBs.
INTRODUCTION
Microtubule (MT) plus end tracking proteins (+TIPs) are a group of MT binding factors, which associate predominantly with the ends of growing MTs and play a prominent role in regulating MT dynamics and in attachment of MTs to different cellular structures (Schuyler and Pellman, 2001; Carvalho et al., 2003; Galjart and Perez, 2003; Howard and Hyman, 2003; Akhmanova and Hoogenraad, 2005). The dynamic accumulation of +TIPs at the ends of polymerizing MTs is an evolutionary conserved phenomenon, the mechanistic basis of which is poorly understood. MT plus end accumulation of proteins may depend on their motor-driven plus end-directed transport, specific association with the freshly polymerized MT tip coupled to quick dissociation from the older lattice (often called “treadmilling”) or preferential binding to other +TIPs (“hitchhiking”; Carvalho et al., 2003; Akhmanova and Hoogenraad, 2005). Deciphering of the hierarchy of protein-protein interactions at the MT tip is needed to understand the process of MT plus end tracking and the cellular functions that depend on it.
One of the most conserved families of +TIPs includes EB1 and its homologues present in mammals, plants, and fungi (for review, see Tirnauer and Bierer, 2000). In mammalian cells this family is represented by three members: EB1, EB2 (RP1), and EB3 (EBF3; Juwana et al., 1999; Su and Qi, 2001). These proteins contain a calponin homology domain responsible for interactions with MTs (Hayashi and Ikura, 2003) and a coiled coil region, which determines their dimerization (Honnappa et al., 2005; Slep et al., 2005). In animal cells EB proteins may constitute the “core” of the plus end complex because they interact directly with most other known +TIPs including dynactin large subunit p150Glued, APC, CLASPs, spectraplakins, RhoGEF2, and a catastrophe-inducing kinesin KLP10A (Askham et al., 2002; Bu and Su, 2003; Ligon et al., 2003; Rogers et al., 2004; Honnappa et al., 2005; Mennella et al., 2005; Mimori-Kiyosue et al., 2005; Slep et al., 2005). Removal of these proteins from the MTs does not prevent specific accumulation of EBs at the distal ends of the MTs, making it unlikely that EBs “hitchhike” on any of the above-mentioned +TIPs (Berrueta et al., 1998; Komarova et al., 2002; Kodama et al., 2003; Rogers et al., 2004; Mimori-Kiyosue et al., 2005). Studies in Xenopus extracts have shown that EB1 at the MT tip is immobile with respect to the MT lattice, suggesting that it binds to the freshly polymerized MT end and detaches from the older part of the MT (Tirnauer et al., 2002).
Another +TIP conserved in animals and fungi is CLIP-170, a protein containing two MT-binding CAP-Gly domains at its N-terminus, a long coiled coil region responsible for dimerization, and two zinc finger-like domains at its C-terminus (Pierre et al., 1992; Perez et al., 1999). Similar to EB1, mammalian CLIP-170 shows treadmilling behavior (Perez et al., 1999), and recent in vitro studies have demonstrated that its localization to the growing MT ends may be due to copolymerization with tubulin oligomers (Arnal et al., 2004). However, both in budding and fission yeast, MT plus end localization of CLIP-170 homologues Bik1p and Tip1p depends on kinesin-driven transport (Busch et al., 2004; Carvalho et al., 2004). In addition to being transported to the tip, Tip1p binds directly to the EB1 homologue, Mal3p, and requires Mal3p for accumulation at the MT tip (Busch and Brunner, 2004). In contrast, the budding yeast Bik1p does not require Bim1p, the EB1 homologue, for MT end accumulation (Carvalho et al., 2004). These differences suggest that the mechanisms controlling plus end localization can vary between systems even although highly conserved proteins are involved.
The functional relationship between CLIP-170 and EB proteins in mammalian cells has not yet been fully elucidated. EB proteins do not require CLIP-170 or the other two CAP-Gly-containing +TIPs, CLIP-115 and p150Glued, for MT plus end localization (Komarova et al., 2002). On the other hand, the role of EB proteins in plus end tracking by CLIP family members has not yet been addressed. The possibility of interaction between CLIP-170 and EB proteins has been suggested by the binding of their fission yeast homologues, by the interaction between EB proteins and p150Glued, and by the identification of the Drosophila homologue of CLIP-170, D-CLIP-190, in a pulldown assay with EB1 (Askham et al., 2002; Bu and Su, 2003; Ligon et al., 2003; Busch and Brunner, 2004; Rogers et al., 2004). Further indications for EB1-CLIP-170 interaction were provided by the enhanced binding of EB1 to MTs caused by overexpression of the CLIP-170 N-terminus in cultured cells (Goodson et al., 2003).
Here we show that CLIP-170 and a closely related protein CLIP-115 bind directly to EB1 and EB3 while displaying a lower affinity for EB2. This interaction depends on the C-terminal tails of the EB proteins, which are strikingly similar to those of α-tubulin. Further, we demonstrate that simultaneous depletion of EB1 and EB3 in cultured cells causes a reduced accumulation of CLIPs at the MT distal ends due to their diminished binding and quicker dissociation from the MT lattice. Overexpression of EB1 resulting in its even distribution along the MT lattice reduces the rate of CLIP dissociation from the growing ends but does not prevent CLIPs from tip-tracking. This observation rules out “hitchhiking” on EB1 as a simple mechanism for CLIP localization to the growing MT ends.
MATERIALS AND METHODS
Antibodies
Rat monoclonal antibodies against EB proteins were generated using GST fusions of EB1-C, EB2, and EB3 by Absea (Beijing, China). We used mouse mAbs against penta-histidine tag (Qiagen, Chatsworth, CA), EB1, EB3, and p150Glued (BD Biosciences, Franklin Lakes, NY), green fluorescent protein (GFP; Roche, Indianapolis, IN), HA tag (BabCO, Richmond, CA), a rat mAb against α-tubulin (YL1/2 against EEY epitope, AbCam, Cambridge, United Kingdom), rabbit antibodies against GFP (AbCam), CLIP-170 (2360; Coquelle et al., 2002), both CLIPs (2221; Hoogenraad et al., 2000), and EB3 (02-1005-07; Stepanova et al., 2003). The following secondary antibodies were used: alkaline phosphatase-conjugated anti-rabbit and anti-mouse antibodies (Sigma, St. Louis, MO), FITC-conjugated goat anti-rabbit antibody (Nordic Laboratories, Westbury, NY), Alexa 594- and Alexa 350-conjugated anti-rat and anti-mouse antibodies (Molecular Probes, Eugene, OR), TRITC- and FITC-conjugated donkey anti-mouse and anti-rabbit and Cy5-conjugated anti-rat antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA).
Expression Constructs, Protein Purification, In Vitro Binding Assays, Coimmunoprecipitation, and Western Blotting
GST- and HIS-tagged N-terminal fragments of CLIP-170 and CLIP-115 were described previously (Lansbergen et al., 2004). GST and HIS-tagged fusions of EB1, EB2, and EB3 were generated using mouse EB1 and EB2 cDNAs and a human EB3 cDNA, which were described elsewhere (Stepanova et al., 2003). Both EB2 and EB3 cDNAs correspond to the “long” isoforms of these proteins. For GST fusions, the portions of the EB cDNAs encoding amino acids indicated in Figure 2A were obtained by PCR; the PCR primers included BamHI and EcoRI sites directly up- and downstream of the coding sequences. These sites where used to subclone the PCR products in BamHI/EcoRI-digested pGEX-3X vector; in the resulting fusion proteins the EB-encoding part was separated from the GST open reading frame by a linker composed of the Factor X cleavage site present in pGEX-3X. To generate HIS-tagged fusions, EB1 coding sequence or its portions were amplified with primers containing NdeI and EcoRI sites and subcloned into pET28a. Purification of the GST- and HIS-tagged proteins from E. coli and Western blotting were performed as described by Lansbergen et al. (2004).
Figure 2.
The C-terminal tails of EB1 and EB3 contribute to the association of these proteins with CLIP-170. (A) Schematic representation of the EB protein structure and the deletion mutants used in this study. CH, calponin homology domain; CC, coiled coil; Ac, acidic tail. Absence of the C-terminal tyrosine is indicated by asterisks. Binding to CLIP-170 N-terminus in GST-pulldown experiments is indicated: +, strong binding; +/-, weak binding; -, no binding detected; ND, not determined. In the left column the data for binding of HIS-tagged CLIP-170-N to GST-tagged EB1 mutants are indicated; in the right column the data for binding HIS-tagged EB1 mutants to GST-tagged CLIP-170-N are summarized. (B) Alignment of the C-terminal tails of the human EB1, EB2, and EB3 (Su and Qi, 2001) and α-tubulin (accession number P68363). In contrast to the other three proteins EB2 has uncharged amino acids at positions 5 and 6 counting from the C-terminus (indicated by a gray box). Similarity of the last three amino acids is emphasized by a black box. (C-E) GST pulldown assays with the indicated GST fusions. Coomassie-stained gel is shown for the GST fusions and Western blots with anti-HIS antibodies for the HIS-tagged CLIP-170 N-terminus and EB mutants. Ten percent of the input and 25% of the material bound to the beads were loaded on gel. (F) COS-1 cells were cotransfected with HA-CLIP-170 N-terminus and the indicated GFP-EB1 (or EB1-GFP) fusions. IPs were performed with anti-HA or anti-GFP antibodies and the resulting precipitates were analyzed with antibodies against HA, GFP, or the EEY epitope. The cross-reacting IgG heavy chain when present is indicated by an asterisk. Arrowheads in the lower panel indicate GFP-positive bands (GFP-EB1 and GFP-EB1-C, which coprecipitate with HA-CLIP-170 N-terminus). (G) IPs with anti-GFP antibodies from COS-1 cells transfected with the indicated GFP-EB1 (or EB1-GFP) expression constructs were analyzed by Western blotting with antibodies against GFP, CLIP-170 (2360), or p150Glued.
For GST pulldown assays individual GST fusion proteins bound to glutathione Sepharose 4B beads (Amersham Biosciences, Piscataway, NJ) were combined with dissolved purified HIS-tagged proteins in a buffer containing 20 mM Tris HCl (pH 7.5), 100-300 mM NaCl, 1 mM β-mercaptoethanol, and 0.3-1% Triton X-100. After incubation for 1 h on a rotating wheel at 4°C, beads were separated from the supernatant by centrifugation and washed four times in the same buffer, and the proteins retained on the beads (including the GST fusion and the HIS-tagged protein bound to it) were analyzed on Coomassie-stained gels and by Western blotting.
The EB-GFP expression constructs used in this study were described by Stepanova et al. (2003), HA- and GFP-CLIP-170-N and -C- by Komarova et al. (2002). GFP-EB1 and EB3 deletion mutants were generated by a PCR-based strategy and subcloned into BglII and EcoRI sites of pEGFP-C2 (Clontech, Palo Alto, CA). YFP-CLIP-170 was generated by subcloning an amplified fragment of a rat brain CLIP-170 cDNA (positions 192-4597 of the sequence with the accession number AJ237670) into XhoI and SalI sites of pEYFP-C1 (Clontech). For coimmunoprecipitation (co-IP) experiments COS-1 cells were transfected by DEAE-dextran method and lysed 2 d after transfection and the lysates were used for co-IP with anti-GFP or anti-HA antibodies (diluted 1:30) as described by Hoogenraad et al. (2000).
RNAi and Rescue Constructs
RNAi vectors for Chinese hamster ovary (CHO)-K1 cells were based on the pSuper vector (Brummelkamp et al., 2002). The used mouse target sequences for EB proteins are indicated in Figure 3B; the luciferase target sequence was CGTACGCGGAATACTTCGA. For simultaneous knockdown of EB1 and EB3, the two RNAi cassettes were combined in tandem in the same vector. To supply the RNAi cassettes with a selectable and a fluorescent marker, they were inserted into the AseI site of pEGFP-C1 or pECFP-C1. EB1 rescue constructs were prepared by a PCR-based strategy to introduce five silent substitutions in the target site of EB1 siRNA (resulting in a sequence TCTAACCAAGATCGAGCAA).
Figure 3.
Simultaneous knockdown of EB1 and EB3 proteins results in the reduction of CLIP accumulation at the MT tips. (A) Western blot analysis of the cell extracts prepared from 1) cells transfected with pECFP-C1 or cells transfected with hairpin constructs encoding RNAi to 2) Luciferase; 3) EB1; 4) EB2; 5) EB3; 6) EB1+EB3; 7) EB1+EB2+EB3. EB proteins were detected by rat mAbs KT51 (anti-EB1), KT52 (anti-EB2), and KT53 (anti-EB3; see also Supplementary Figure S1). Note that anti-EB3 antibody also reacts with an unknown species of ∼28 kDa that is insensitive to EB3 RNAi. The EB3-specific band is indicated by an arrowhead. CLIP-115 and CLIP-170 were detected with the 2221 antibody. Actin serves as a loading control. (B) Schematic representation of the relative positions and sequences of the mouse EB1, EB2, and EB3 RNAi target regions and their alignment to human sequences. Corresponding amino acid sequences are indicated below. (C) Densitometry analysis of EB proteins levels from the blots shown in A. Bar plots represent relative protein levels after expression of different hairpin constructs indicated by numbers, which are the same as in A. Protein levels in the extracts of pECFP-expressing cells (1) were assigned for 100%. (D-K) Distribution of CLIPs at the MT tips after depletion of EB family members. CHO-K1 cells were stained for EB1, CLIPs, and tubulin (D), EB1+EB3, CLIPs, and tubulin (G); EB1+EB3, CLIPs, and EB2 (I) 84 h after microinjection with hairpin construct encoding RNAi to EB1 (D); EB1 and EB3 (G); EB1, EB2, and EB3 (I). Cells with depleted proteins are indicated by dotted lines. Bars, 10 μm. Enlarged insets from the control (1) and treated cells (2) for CLIPs (grayscale) and for overlays (in color) are shown on the right; CLIPs, red; tubulin (D and G) or EB2 (I), green; EB1 (D) or EB1+EB3 (G and I); blue. Bars in the insets, 1 μm. Linescan analysis illustrates the distribution of CLIPs, EBs, and tubulin at the MT tips in control cells (E and J) after depletion of EB1 (F), after simultaneous depletion of EB1 and EB3 (H), or all three EB members (K). Note that the length of CLIP-positive structures is twice as short compared with control cells. (L-N) Quantification of the amount of CLIPs and EB proteins bound the whole tip or to its outer part after depletion of EB1 (L), EB1 and EB3 (M), or all three EB proteins (N). The integrated intensity of CLIPs and EB proteins at the MT tips in depleted cells was expressed as a percentage of the intensity in control cells from the same image, which was taken for 100% and plotted as box graphs. The boundaries of the box and whiskers indicate the 25th and the 75th percentile, and the 90th and 10th percentiles respectively. The median and mean are shown by a straight and a dotted line. The numbers on the graphs indicate the average values and standard deviations. The values for CLIPs in L are statistically different from M and N (Student's test; p < 0.0001) with 95% confidence of nonoverlapping intervals. The values for CLIPs shown in M are statistically not different from those in N. Data are derived from measurements of 146-568 individual MT tips in 10-20 cells per treatment group.
Cell Culture, Generation of the Stable Cell Line, and Introduction of RNAi Vectors
For Western blot analysis 80% confluent CHO-K1 cells were transfected using FuGene 6 (Roche) with different pECFP-RNAi plasmids. Cells were replated 16 h after transfection into glucose and sodium pyruvate-free DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and 2 mg/ml G418 (Roche) and cultured for the next 72 h.
To produce a cell line stably expressing YFP-CLIP-170, cells were transfected and cultured as described above. For colony screening, cells were plated on coverslips and clones expressing low level of YFP-CLIP-170 were selected under a microscope using polystyrene cloning cylinders (Sigma).
For microinjection, cells were plated sparsely on coverslips with photoetched locator grids (Bellco Glass, Vineland, NJ) and 12-24 h later they were microinjected with different RNAi plasmids into nuclei. The needle concentration was 100 μg/ml for RNAi plasmids and 70 μg/ml for the rescue experiments with EB1, EB2, or EB1ΔAc. For detection of microinjected cells in live imaging experiments, cells were comicroinjected with a red fluorescent protein-expressing construct to produce a soluble marker. Cells were fixed or used for live observation 84-90 h after microinjection.
Immunostaining, Linescan Analysis, and Quantification of the Protein Amount at the MT Tips
Cell fixation and staining were performed as described by Komarova et al. (2002). Briefly, cells were fixed in cold methanol (-20°C), postfixed with 3% formaldehyde, and permeabilized with 0.15% Triton X-100. Fixed samples were analyzed by fluorescence deconvolution microscopy using a Deltavision microscope system equipped with an Olympus IX70 inverted microscope (Lake Success, NY) and a PlanApo 60 × 1.4 NA objective. We used 2× binning that gave a resolution of 0.22 μm per 1 pixel. Images were prepared for presentation using Adobe Photoshop (San Jose, CA). Linescan analysis and measurements of fluorescence intensity and densitometry analysis of Western blots were performed using MetaMorph software (Universal Imaging, West Chester, PA). For linescan analysis original images were converted to 300 dpi using Adobe Photoshop software and the analysis of intensity profiles along 1 pixel depth line at the MT tips was performed in MetaMorph. For estimation of the amount of the proteins bound to the outmost MT tips or to entire tips integrated fluorescence intensities within a box of four pixels on a side (outer tip) or a rectangle covering the entire positively stained tip (total bound) were measured for each channel after subtracting external background. The integrated intensities in RNAi depleted cells were expressed as a percentage of the integrated intensities in control cells, which were taken for 100%. About 200 MTs were analyzed in 10-20 control or depleted cells for each experimental condition. Data handling was performed using SigmaPlot software (Jandel Scientific, Corte Madera, CA).
Live Cell Imaging and Quantification of CLIP Dynamics
Cells stably expressing YFP-CLIP-170 were observed at 36°C on a Nikon Diaphot 300 inverted microscope (Melville, NY) equipped with a Plan 100×, 1.25 NA objective using YFP filter set. Images of 16-bit depth were collected with a CH350 slow scan, cooled CCD camera (Photometrics, Tucson, AZ) driven by Metamorph imaging software (Universal Imaging). The images were projected onto the CCD chip at a magnification of 250×, which corresponded to a resolution of 0.09 μm per 1 pixel. Time-lapse series of 30 images were acquired with a 0.5-s interval using stream acquisition mode. To achieve high temporal resolution, we had to limit the exposure time to 0.1-0.15 s. CLIP kinetics was analyzed on 16-bit depth images after subtraction of external background. We used a kymograph function to create a cross-sectional view of average intensity values within the line of 8 px along the YFP-CLIP-170 tracks over time. Using linescan analysis on the kymograph images, we determined the intensity decay of the fluorescence signal of a single pixel over time. We plotted the intensity decay over time for each YFP-CLIP-170 positive tip using SigmaPlot software and by applying exponential curve fitting we determined the dissociation constant (koff). The half time of CLIP dissociation was calculated as ln2/koff for each CLIP-positive tip and averaged for the population. Photobleaching of the fluorescent signal due to sample illumination was not taken into account because it was negligible; the bleaching constant of YFP-CLIP-170 signal at the outer tips of growing MTs determined by exponential curve fitting to intensity decay was 0.01 ± 0.004 s-1. This value was insignificant compared with the constant of CLIP dissociation (Table 1); correction for photobleaching was therefore unnecessary.
Table 1.
Rate of YFP-CLIP-170 dissociation from the growing MT plus ends
| koff (s–1) | ½ time of YFP-CLIP-170 dissociation (s) | No. of MT plus ends | |
|---|---|---|---|
| Control cells | 0.54 ± 0.12 | 1.4 ± 0.3 | 57 |
| RNAi experiments | |||
| RNAi to Luciferase | 0.50 ± 0.10 | 1.5 ± 0.3 | 47 |
| EB1 RNAi | 0.52 ± 0.13 | 1.4 ± 0.4 | 95 |
| EB1 + EB3 RNAi | 1.13 ± 0.80 | 0.7 ± 0.2 | 111 |
| Rescue experiments after EB1 + EB3 depletion | |||
| EB1 | 0.42 ± 0.27 | 1.9 ± 0.5 | 59 |
| EB1ΔAC | 0.39 ± 0.16 | 2.0 ± 0.6 | 53 |
| Expression of EB1 or EB1 mutants in control cells | |||
| Untagged EB1 | 0.22 ± 0.08 | 3.7 ± 1.6 | 55 |
| EB1ΔCC | 0.44 ± 0.10 | 1.6 ± 0.3 | 39 |
The dissociation constants and half times of YFP-CLIP-170 dissociation were obtained for individual MT tips and the averaged values ± SDs were computed for the analyzed population.
RESULTS
CLIP-170 and CLIP-115 Bind Directly to EB Proteins
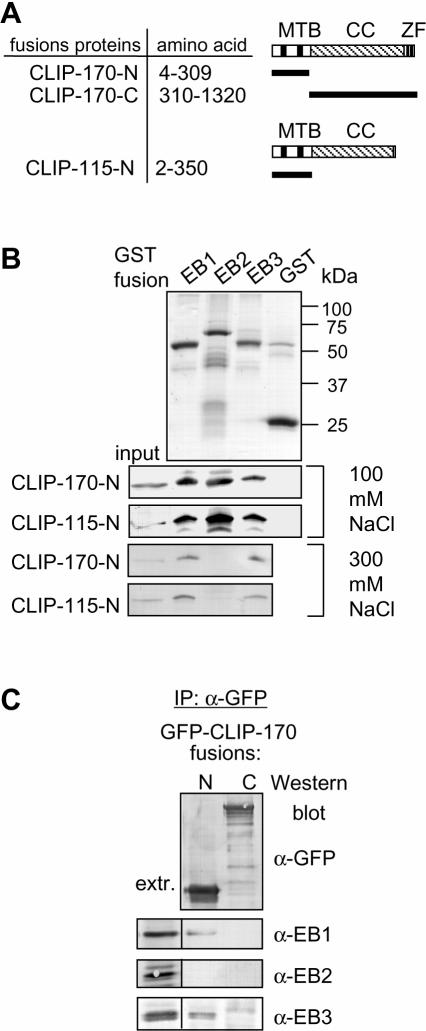
The CAP-Gly motif-containing domains of Tip1p (Schizosaccharomyces pombe homologue of CLIP-170) and p150Glued bind directly to the C-terminal parts of Mal3p and the mammalian EB1 protein, respectively (Askham et al., 2002; Bu and Su, 2003; Ligon et al., 2003; Busch and Brunner, 2004). We therefore set out to test if the N-termini of CLIP-170 and CLIP-115 can also interact directly with EB family members. We used purified 6xhistidine (HIS)-tagged N-terminal fragments of CLIP-170 and CLIP-115 (Figure 1A) in an in vitro pulldown assay with the purified glutathione S-transferase (GST) fusions of EB1, EB2, and EB3. In low-salt conditions (100 mM NaCl) both CLIP N-termini were efficiently retained by all three EB fusions but not by GST alone (Figure 1B). However, GST-EB2 could no longer bind to the CLIP N-terminal domains when the salt concentration was increased to 300 mM (Figure 1B).
Figure 1.
CLIP and EB proteins interact directly. (A) Schematic representation of the CLIP protein structure and the deletion mutants used in this study. MTB, microtubule-binding domain, CC, coiled coil, ZF, zinc finger-like domains. (B) GST pulldown assays with the indicated GST fusions. Coomassie-stained gel is shown for the GST fusions and Western blots with anti-HIS antibodies for the HIS-tagged CLIP N-termini. Ten percent of the input and 50% of the material bound to the beads were loaded on gel. (C) IP with anti-GFP antibodies from COS-1 cells transfected with the indicated GFP-CLIP-170 deletion mutants. Rat mAbs were used to detect EB1 and EB2 and rabbit polyclonal antibodies to detect EB3.
To confirm this interaction we next performed co-IPs from transfected cells. We overexpressed the N- or the C-terminal part of CLIP-170 tagged with GFP in COS-1 cells and investigated if endogenous EB proteins could be coprecipitated with these CLIP-170 fragments. To detect all three endogenous EB family proteins, we used novel monoclonal antibodies (mAbs) specific for EB1, EB2, or EB3 (Supplementary Figure S1). We found that both EB1 and EB3 but not EB2 coprecipitated with the N-terminus of CLIP-170, whereas none of the EB proteins coprecipitated with the C-terminus of CLIP-170 (Figure 1C). It should be noted that previously in a similar experiment we failed to observe a co-IP of EB1 with the CLIP-170 N-terminus (Komarova et al., 2002). This contradiction is explained by the fact that in the current study we used a new anti-EB1 mAb (mAb KT51), which is much more sensitive on Western blot than the anti-EB1 mAb from BD Biosciences used in our previous study. We conclude that CLIP-170 and CLIP-115 can bind directly to EB1 and EB3 and display a lower affinity for EB2.
To map further the CLIP interacting domains in EB1 and EB3, we used a series of EB1/EB3 deletion mutants (Figure 2A). Experiments with the N-terminus of p150Glued have shown that it binds to the C-terminal part of EB1 and that the deletion of last two amino acids of EB1 is sufficient to abolish the interaction (Askham et al., 2002; Bu and Su, 2003). The C-terminal tails of EB proteins are strikingly similar to the tail of α-tubulin because they contain several acidic residues and terminate with a tyrosine (Figure 2B). In Saccharomyces cerevisiae the CLIP-170 homologue Bik1p depends on the C-terminal aromatic residue of α-tubulin for the binding to MTs (Badin-Larcon et al., 2004). The same is likely to be true for the mammalian CLIP-170 because it is mislocalized in mouse neurons, in which the pool of C-terminally tyrosinated α-tubulin is strongly reduced because of a deficiency in tubulin tyrosine ligase (Erck et al., 2005). Taking into account all these observations, we hypothesized that the CLIP N-termini bind to the C-terminal regions of EB1 and EB3 in a manner dependent on the C-terminal tyrosine. To test this idea, we preformed GST pulldown assays with GST fusions of different EB1/EB3 deletion mutants and a HIS-tagged CLIP-170 N-terminus (Figure 2, C and D). We observed that the C-terminal half of EB1 was sufficient for the interaction with the CLIP-170 N-terminus. Removal of the tails of EB1/EB3 or their C-terminal tyrosine alone prevented HIS-tagged CLIP-170 N-terminus from binding to GST-EB fusions, confirming our hypothesis (Figure 2, C and D).
We also performed a reverse assay where we tested the in vitro binding of HIS-tagged EB1 or its mutants to GST-tagged CLIP-170 N-terminus. Full-length HIS-tagged EB1 displayed a strong affinity for GST-CLIP-170 N-terminus but not to GST-EB1, which was used as a negative control (Figure 2E). EB1 is known to form stable dimers (Honnappa et al., 2005; Slep et al., 2005), but we did not observe association between differently tagged EB1 proteins purified from bacteria. HIS-tagged EB1 mutants lacking either the C-terminal tyrosine or the whole acidic tail did show weak binding to the GST-coupled CLIP-170 N-terminus but not to GST-EB1 (Figure 2E). This observation is in line with previously published data indicating that in addition to the EB1 tail, the EB1 coiled coil region may also be important for binding to p150Glued N-terminus (Wen et al., 2004). GST-fused CLIP-170 N-terminus might have a higher affinity for EB mutants than its HIS-tagged version because it is expected to be a dimer due to self-association of GST, whereas HIS-tagged CLIP N-termini are likely monomeric.
Next we investigated if the acidic tail of EB1 and in particular its C-terminal tyrosine are important for interaction between EB1 and CLIP-170 N-terminus in cell lysates. To address this question we performed co-IPs from COS-1 cells cotransfected with constructs expressing hemagglutinin (HA)-tagged CLIP-170 N-terminus and GFP-EB1 fusion or its deletion mutants. Although GFP-EB1 and GFP-EB1 C-terminus coprecipitated with HA-CLIP-170 N-terminus (Figure 2F, lanes 1 and 4) GFP-EB1 or GFP-EB1-C lacking the whole acidic tail or its C-terminal tyrosine did not copurify with CLIP-170 N-terminus (Figure 2F, lanes 2, 5, and 7). A similar result was obtained when we used longer CLIP-170 mutants, which contain a portion of the coiled coil region and can dimerize (unpublished data). Our data indicate that the absence of the C-terminal tyrosine of EB1 makes its affinity for CLIP-170 in the extract too low to allow a co-IP.
It should be noted that in mammalian cells α-tubulin undergoes repetitive cycles of detyrosination and tyrosination. Using an antibody, which specifically recognizes the EEY epitope, we observed no tyrosine addition to detyrosinated EB1 in COS-1 cells, indicating that it is not a substrate for a tyrosine ligase in these cells (Figure 2F).
In line with the idea that the very C-terminus of EB1 is important for the interaction with CLIP-170 and p150Glued, we observed that the attachment of the GFP tag to the C-terminus of EB1 abolished its interaction with these CAP-Gly proteins when they were expressed at endogenous levels (Figure 2G) or when HA-tagged CLIP-170 N-terminus was overexpressed (Figure 2F, lanes 3 and 6). The widely used EB1-GFP fusion should therefore be treated with some caution because it might have a dominant negative effect due to its inability to interact with the CLIPs and dynactin.
Our data indicate that CLIP-170 and CLIP-115 can bind to EB1 and EB3 directly. However, we were unable to co-IP these proteins when they were both present at endogenous levels (unpublished data). We propose that the interactions between these proteins are transient and/or occur normally only in the context of MT tip binding.
EB Depletion Decreases CLIP Accumulation at MT Tips Mainly through Influencing CLIP Dynamics
Direct interactions between CLIPs and EBs suggested that these proteins may bind to the MT ends cooperatively. Expression of a dominant negative CLIP-170 mutant demonstrated that EB proteins do not require CLIPs or dynactin for the plus end binding (Komarova et al., 2002), and examination of cells after depletion of both CLIPs by RNAi confirmed this conclusion (unpublished data). To test if the EB proteins are needed for the binding of CLIPs to MT ends in mammalian cells, we used plasmid-based RNA interference (RNAi; Brummelkamp et al., 2002) in CHO-K1 cells. Target regions were selected from the open reading frames of the mouse EB1, EB2, and EB3 genes (Figure 3B), and their 100% identity with hamster sequences was confirmed by sequencing the corresponding portions of the EB cDNAs obtained by RT-PCR from CHO-K1 cells. Next we validated the effectiveness of different RNAi vectors to silence EB proteins in a population of CHO-K1 cells. Insertion of RNAi cassettes into pECFP-C1 vector, which contains a neomycin-resistance gene, permitted us to select RNAi plasmid-transfected cells for resistance to G418. As a control we used a CFP-expressing RNAi vector directed against luciferase. Starting at 16 h after transfection with RNAi vectors, cells were treated with G418 for 72 h and then collected for Western blot analysis. Using microscopic examination, we determined that after such treatment >95% of the cells in the population were CFP-positive. Immunoblotting and densitometry analysis demonstrated that each single EB species was down-regulated by ∼90% both when a single EB protein or several proteins in combination were knocked down (Figure 3, A and C). Depletion of any particular EB protein had no effect on the protein levels of other family members or on the levels of actin, tubulin, CLIP-170 and CLIP-115 (Figure 3A).
Next we analyzed whether the depletion of EB proteins had any effect on the localization of endogenous CLIPs. The distribution of CLIP-170 and CLIP-115 in correlation to the depletion of EB family members was evaluated by triple immunostaining at 84-90 h after nuclear microinjection of RNAi vectors. Microinjection allowed us to obtain a reasonably homogeneous population of cells displaying a knockdown of one or several EB proteins. In untreated CHO-K1 cells CLIP-170 and CLIP-115 visualized with an antibody, which recognizes both proteins displayed “comet”-like structures with an average length of ∼2 μm at the MT distal ends. These ends were also positive for endogenous EB1 and EB3 (Figure 3, D, G, and I, rectangles marked with “1”). Linescan analysis (plots of intensity vs. distance along the MT distal end, Figure 3E) demonstrated that the distribution of CLIPs and EBs was identical and overlapping. Knockdown of EB1 alone caused no significant change either in accumulation or in distribution of the CLIPs at the MT tip (Figure 3, D, F, and L). However, simultaneous depletion of EB1 and EB3 led to a reduction of the CLIP signal at the MT tips to ∼44%, whereas the level of remaining EB1 and EB3 proteins was only ∼11% (Figure 3, G, H, and M). Similar results were obtained when all three EB proteins were knocked down simultaneously (Figure 3, I, K, and N), suggesting that EB2 had no strong effect on the CLIP distribution.
The diminished accumulation of CLIPs at the MT tips after the depletion of EB proteins could be either due to their reduced association with the outer MT plus ends or a result of a redistribution. To test these possibilities we quantified CLIP-positive signals at the outer tips of MTs (within a 0.36-μm square box at the very end of the MT). We found that CLIP association measured in this way was slightly reduced (to ∼77% of the normal level) after knock down of EB1 and EB3 (Figure 3M). However, the length of the CLIP-positive comet was reduced approximately by half as demonstrated by linescan plots (Figure 3, E, F, H, J, and K). This effect correlated with the distribution of the remaining EB1 and EB3, which were strongly concentrated as small dots at the distal tips of the MTs with an intensity of ∼19% of their normal level at the outer tip (Figure 3M). Shortened CLIP-positive comets with a reduced intensity were also observed after knocking down EB1 and EB3 in HeLa cells with the aid of the human versions of the RNAi vectors (Figure 3B and unpublished data), indicating that the observed effects were not a peculiarity of the CHO-K1 cells.
The influence of EB proteins on CLIP distribution could possibly be explained by a change in CLIP dynamics. To analyze CLIP behavior in live cells, we generated a line of CHO-K1 cells stably expressing yellow fluorescent protein (YFP)-CLIP-170 at a level comparable to that of the endogenous CLIP-170 (Figure 4A). The average length of YFP-positive comets at the MT ends in this cell line was similar to that measured for the endogenous CLIP-170 in fixed cells, suggesting that YFP-CLIP-170 behavior reflects the dynamics of the endogenous protein. Using live imaging with a high temporal resolution, we were unable to find any evidence for transport of CLIP-170 to the MT tip in CHO-K1 cells (unpublished data). Kymograph analysis of individual growing MT plus ends (Figure 4, B and C) demonstrated very fast CLIP association with the freshly polymerized MT tip and slower dissociation from the older part of MT. Our data support previous observations on CLIP dynamics, suggesting that CLIP molecules remain stationary with respect to the MT lattice.
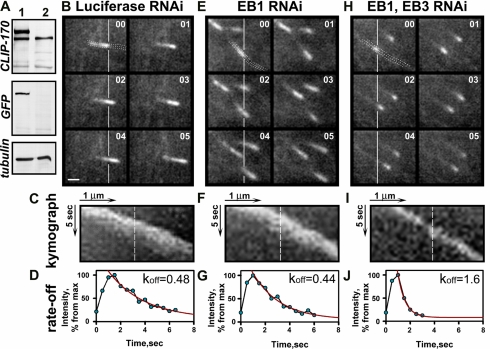
Figure 4.
Depletion of EB proteins increases the rate of CLIP-170 dissociation from the MT tips. (A) Western blots of the cell extracts prepared from CHO-K1 cells stably expressing YFP-CLIP-170 (after fluorescence-activated cell sorting) probed with CLIP-170 specific antibody 2360, anti-GFP and anti-tubulin antibodies; parental CHO-K1 cell line was used as a control. (B-J) YFP-CLIP-170 kinetics in CHO-K1 cells. The time-lapse images of YFP-CLIP-170 in the cells expressing hairpin constructs containing target sequence either for Luciferase RNAi (B); EB1 RNAi (E); or EB1+EB3 RNAi (H). Images were acquired every 0.5 s. The vertical lines correspond to very tip of CLIP-170 comets on the first image. Time is shown in seconds in the top right corner. Bar, 1 μm. A kymograph function was used to create a cross-sectional view of the YFP-CLIP-170 intensity over time (C, F, and I). The dotted lines in B, E, and H indicate the path of MT growth along which the cross-sectional views were obtained. The fluctuations of the fluorescent intensities along the dashed line represent association and dissociation of YFP-CLIP from the MT plus ends and are plotted on the graphs below (D, G, and J). The exponential curve fitting gives the dissociation constant (koff).
Analysis of the dissociation curve indicated that it could be fitted to an exponential decay (Figure 4D), which allowed us to calculate the dissociation constant and the half time of the CLIP dissociation. In control cells CLIP-170 dissociated with a half time of 1.4 ± 0.3 s (Table 1). Knockdown of EB1 alone had no effect on CLIP-170 dissociation rate (Figure 4, E-G, Table 1), while simultaneous depletion of EB1 and EB3 decreased the half time of YFP-CLIP-170 dissociation by a factor of 2 (Figure 4, H-J, Table 1). Depletion of EB proteins had no effect on the rate of MT growth as demonstrated by similar slopes of the kymographs (31.0 ± 6.6, 31.7 ± 6.0, and 32.0 ± 4.2° for control, EB1, and EB1+EB3 RNAi treatments, respectively; Figure 4, C, F, and I). Although we used only CLIP-170 in our kinetic studies, similar distribution of the endogenous CLIP-170 and CLIP-115 in fixed preparations suggested that both proteins had similar dynamics.
CLIP Accumulation at the MT Tips Is Fully Restored by EB1 and Partially Restored by EB1ΔAc
To exclude the possibility that enhanced dissociation of CLIP-170 is due to an off-target activity of our RNAi tools, we rescued the cells microinjected with EB1 and EB3 RNAi vectors with a plasmid expressing nontagged EB1. This rescue construct was resistant to the anti-EB1 small interfering (si) RNA because of five silent substitutions in the siRNA target region. EB1 expression could completely restore the normal YFP-CLIP-170 dissociation rate (Table 1).
We also performed a rescue of EB1/EB3 knockdown using EB1ΔAc mutant, which has a strongly reduced affinity to CLIP-170 (Figure 2, C-E). To detect endogenous EB1 in cells expressing EB1ΔAc we used the KT51 mAb, which does not react with EB1ΔAc (Supplementary Figure S1). The EB1ΔAc mutant was visualized with anti-EB1 mAb from BD Biosciences, which recognizes an epitope in the coiled coil domain (Askham et al., 2002). For quantification of CLIP distribution we chose cells in which EB1ΔAc was localized exclusively at the tips and not along the MT lattice. The level of EB1ΔAc expression estimated by measuring of the signal at the MT tips was 230 ± 90% of endogenous EB1 in control cells. EB1ΔAc mutant localized efficiently to the MT tips of cells depleted for EB1 and EB3 and induced an almost complete loss of the residual EB1 from the MT ends, probably due to competition (Figure 5, A-C). Total accumulation of endogenous EB1 at the MT tips of such rescued cells constituted only 3.2 ± 1.2% of those in control cells (Figure 5D). Also endogenous EB3 was almost undetectable in such cells (unpublished data).
Figure 5.
EB1ΔAc partially restores CLIP accumulation after simultaneous EB1/EB3 depletion. (A) CHO-K1 cells depleted of endogenous EB1 and EB3 proteins and rescued with EB1ΔAc were stained for EB1 with the BD Biosciences mAb (recognizes both the endogenous EB1 and the rescue construct, red), KT51 mAb (recognizes only the endogenous EB1, blue), and the CLIPs (green). Bar, 10 μm. (B) Enlarged areas of a control (1) and a rescued cell (2). Bar, 5 μm. (C) Linescan analysis demonstrates that the length of the CLIP positive structures was restored in the cells rescued with the EB1ΔAc mutant. (D) Quantification of the amount of CLIPs and EB1 bound the whole MT plus end in cells depleted of EB1 and EB3 and rescued with EB1ΔAc. The data are plotted in the same way as in Figure 3, L-N. The data are derived from 161 and 121 individual MT tips in 24 and 11 cells for control and rescue, respectively.
Surprisingly, EB1ΔAc could efficiently rescue CLIP distribution (Figure 5, A-D). It caused an increase of CLIP accumulation measured at the MT tip from 44.2 ± 12.0% in EB1/EB3 knockdown cells to 75.9 ± 14.6% of the level in control cells, suggesting that only ∼25% of CLIP accumulation may be accounted for by the CLIP-EB interaction mediated by the EB1 tail. Even more strikingly, the rate of CLIP dissociation was completely restored by the expression of EB1ΔAc (Table 1), indicating that CLIP-EB binding via the tail of EB1 has no influence on CLIP dissociation.
EB2 Cannot Rescue CLIP-170 Accumulation at the Tips of EB1/EB3-depleted Cells
The depletion of either EB1/EB3 or all three EB species caused similar effects on CLIP accumulation at the MT tips. There are two possible explanations of this result: either EB2 has a function distinct from EB1 and EB3 or endogenous EB2 level in CHO-K1 cells is too low to support normal CLIP distribution, especially given the lower affinity of this protein for the CLIPs compared with EB1 and EB3. To distinguish between these possibilities, we attempted to rescue CLIP accumulation after the simultaneous EB1 and EB3 knockdown by overexpressing EB2. To our surprise, mild overexpression (204 ± 150% compared with endogenous level) of EB2 in EB1/EB3 knockdown cells led to further loss of the CLIPs from the MT tips, reducing the level of the total MT tip-bound CLIPs to ∼27% of that in control cells (Figure 6). Linescan analysis demonstrated that the length of CLIP-stained MT tips was ∼1 μm, similar to that observed in EB1/EB3-depleted cells. Higher levels of EB2 overexpression (to an extent when it was no longer bound to the tips but rather distributed evenly along the MTs) did not improve CLIP accumulation at the tips either (unpublished data). This suggests that overexpression of EB2 in EB1/EB3-depleted cells did not lead to a change in CLIP dissociation rate compared with EB1/EB3 depletion, although very low signal of YFP-CLIP-170 at the MT tips made live imaging with high temporal resolution impossible in this case. It appears therefore, that EB2 has fundamentally different properties from EB1 and EB3 with respect to its relationship with the CLIPs.
Figure 6.
Overexpressed EB2 does not restore CLIP accumulation after depletion of EB1 and EB3. (A) CHO-K1 cells, depleted of endogenous EB1 and EB3 and expressing exogenous EB2 were stained for EB1+EB3 (blue), EB2 (green), and the CLIPs (red). Cells with depleted EB1 and EB3 proteins are indicated by dotted lines. Bar, 10 μm. (B) Enlarged areas from a control (1) and an EB1+EB3-depleted cell expressing moderate levels of exogenous EB2 (2). Bar, 1 μm. (C) Linescan analysis demonstrates the distribution of CLIPs, EB2, and EB1+EB3 at the individual MT plus ends in a control cell and in a cell rescued with EB2. (D) Quantification of the amount of CLIPs and EBs bound the whole MT plus end in cells with depleted EB1 and EB3 and overexpressing exogenous EB2. The data are plotted in the same way as in Figure 3, L-N. Data are derived from 152 and 229 individual MT tips in 17 and 13 cells for control and rescue, respectively.
Overexpression of EB1 Decreases the Rate of CLIP-170 Dissociation
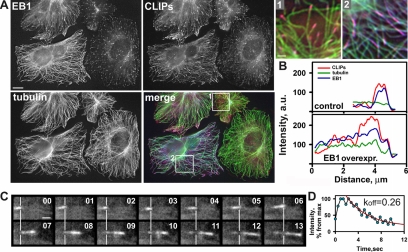
Because decreased levels of EB1 and EB3 caused quicker release of CLIP-170 from the MTs we expected an increased level of EB1/EB3 proteins to have the opposite effect. Indeed, moderate levels of EB1 overexpression, which resulted in redistribution of EB1 along the whole length of MTs without causing their stabilization or bundling, induced elongation of CLIP-positive comets at the MT ends (Figure 7, A-C). Such moderate increase in EB1 level caused no significant change in MT growth rate, whereas the half time of YFP-CLIP-170 dissociation was increased by a factor 2 compared with control cells (Figure 7D, Table 1). As a control, we used a HA-tagged EB1 mutant EB1ΔCC (1-184), which lacks the C-terminal part including the coiled coil region. EB1ΔCC decorates MTs but has no preferential affinity for their growing ends (unpublished data). Overexpression of this mutant caused no significant change in CLIP-170 dissociation rate (Table 1), indicating that the observed effect correlates with the capacity of the EB proteins for plus end association. Importantly, overexpression of EB1 did not cause complete redistribution of the CLIPs along the MTs. Although EB1 decorated MTs along their length, CLIPs still concentrated at the MT tips but dissociated with a slower rate. This indicates that CLIPs do not simply “hitchhike” on the EBs to target the growing tips but recognize these sites through another mechanism.
Figure 7.
Moderate overexpression of EB1 reduces the rate of CLIP dissociation from the MT tips. (A) CHO-K1 cells expressing exogenous untagged EB1 were stained for EB1 (blue), CLIP (red), and tubulin (green). Bar, 5 μm. Enlarged insets are from the control (1) and EB1 expressing cells (2). Bar, 1 μm. (B) Linescan analysis of individual MT plus ends from a control cell and a cell expressing exogenous EB1 (where it is evenly distributed along the MT length); CLIP-positive labeling becomes twice as long (∼4 μm) in such EB1-overexpressing cells. (C) Time-lapse images of YFP-CLIP-170 in the cell expressing exogenous EB1. Images were acquired every 0.5 s. Time is shown in seconds in the top right corner. Lines correspond to outmost CLIP-170 tips on the first row images and demonstrate that in most cases YFP-CLIP-170 is still bound to the MT lattice 7 s after it appears at the outmost tips. Bar, 1 μm. (D) A graph of intensity decay of YFP signal over time and its fitting to an exponential decay curve.
DISCUSSION
Protein-protein interactions at the growing MT plus ends create a flexible and dynamic network, which contributes to the spatial accumulation of +TIPs (for recent review, see Akhmanova and Hoogenraad, 2005). The core element of this network is likely represented by EB1 and its family members. Here we show that similar to other +TIPs, CLIPs also require EB proteins for efficient plus end localization. Direct interaction between the CLIPs and EB1/EB3 proteins provides the most straightforward explanation of this observation. EB-dependent mechanism of CLIP accumulation at the MT tips may be evolutionary conserved because Mal3p, the S. pombe EB1 homologue interacts directly with Tip1p, the CLIP-170 homologue and is strictly required for Tip1p binding to the growing MT ends (Busch and Brunner, 2004).
We have demonstrated that the acidic tail of EB1/EB3 and specifically its C-terminal tyrosine contributes to CLIP binding. CLIP-EB interaction via the tail of EB proteins plays a significant role in vivo because CLIP accumulation at the MT plus ends was higher in control cells than in cells depleted for EB1/EB3 and rescued with the EB1ΔAc mutant. The tail of EB1 or EB3, however, is not the sole site of their interaction with the CLIPs because EB1ΔAc can still bind, albeit weakly, to the CLIP-170 N-terminus in vitro. In agreement with this observation p150Glued, a CLIP family member, depends for its association with EB1 on additional more N-terminal sequences within the so-called EB1-like motif (Wen et al., 2004). The crystal structure of the EB1-like motif has been recently solved and was shown to include a dimeric parallel coiled coil and a four-helix bundle (Honnappa et al., 2005; Slep et al., 2005). This structure revealed a highly conserved surface patch with a hydrophobic cavity and a polar rim, which play an essential role in the binding to APC and spectraplakins (Honnappa et al., 2005; Slep et al., 2005). It is possible that CLIP family members may also require the EB1-like motif for their interaction. The role of the EB1 acidic tail, which is expected to be highly flexible, might primarily involve recruitment and docking of CLIP at the surface of the EB1-like motif or some other part of EB1 protein.
There are strong indications that CLIPs bind to acidic tail of tubulin (Badin-Larcon et al., 2004; Erck et al., 2005), suggesting that CLIPs may use the same sites for interaction with the EB proteins and tubulin. This does not necessarily contradict the idea that CLIPs and EBs bind to MTs cooperatively, because mammalian CLIPs contain two copies of CAP-Gly domain and are dimers, so that each CLIP molecule has four potential sites for tubulin/EB binding. Besides, in addition to the binding site accommodating tubulin/EB acidic tails with their C-terminal aromatic residue, other CLIP surfaces are likely to be involved in EB and/or tubulin binding. It is tempting to speculate that when CLIPs and EBs simultaneously bind to MTs, the highly flexible acidic tails of tubulin might substitute for the function of the EB tails. In this way, EB tails would not be essential for efficient interaction with CLIPs in a triple complex with tubulin. This might explain why EB1ΔAc can partially restore CLIP accumulation and fully restore the rate of CLIP dissociation in the absence of EB1 and EB3.
An intriguing question is why the EB2 protein, a close relative of EB1 and EB3, can only poorly interact with the CLIPs and cannot rescue their accumulation at the MT tips after depletion of EB1 and EB3. The surface residues within the EB1 motif are highly conserved between EB1, EB2, and EB3. However, the tail of EB2 differs considerably from those of EB1 and EB3. First, it contains two uncharged residues near its tip instead of acidic amino acids present in EB1, EB3, and α-tubulin (Figure 2B). Second, the tripeptide V254-I255-P256 present in the EB1 tail is substituted for GHT in human EB2 and GQT in the mouse EB2. The crystal structure of EB1 dimer showed that this tripeptide from one monomer can interact with the hydrophobic cavity of the other monomer (Honnappa et al., 2005; Slep et al., 2005). This interaction might be important for positioning of the acidic tail with respect to the rest of the molecule. Its hydrophilic character in EB2 might affect the tail conformation and, therefore, have an influence on the binding of protein partners.
Although direct interaction of CLIPs and EBs provides an attractive explanation for the observed dependence of CLIPs on EB1 and EB3 for their MT end accumulation, we cannot rule out possible indirect mechanisms. First, EB1 and EB3 may recruit additional proteins, which in turn may increase binding of the MT tip for CLIPs. Such candidate +TIPs, which can associate with both CLIPs and EBs include p150Glued and CLASPs (Askham et al., 2002; Bu and Su, 2003; Ligon et al., 2003; Lansbergen et al., 2004; Mimori-Kiyosue et al., 2005). Second, presence of EB proteins may cause displacement of certain factors, which help CLIPs to dissociate. However, our data on EB1 overexpression do not support these possibilities because in these conditions CLIP dissociates from the MTs two times slower but, unlike EB1, it does not redistribute along the MT length.
Finally, EB1 and EB3 may be influencing the MT structure itself. The polymerizing MT tip is believed to have a structure different from the rest of the MT lattice because of the presence of tubulin sheets, protruding protofilaments or a GTP cap (Carvalho et al., 2003; Howard and Hyman, 2003). It is possible that both EBs and CLIPs recognize this structure and remain at the plus ends as long as the structure is retained. If EB proteins would help to maintain this structure slowing down its conversion to the regular MT lattice, this would explain how they influence the release of CLIPs from the tips. Interesting in this respect is an observation of EB1-associated curved filamentous extensions at the MT plus ends in Xenopus extracts (Tirnauer et al., 2002).
Both direct and indirect interactions with EB proteins may contribute to the proper CLIP distribution. Nevertheless, the loss of CLIPs from the MT tips after EB knockdown is not proportional to the extent of the EB depletion. Because we were unable to achieve a complete knockdown of EB proteins, we cannot exclude that in the absence of these proteins CLIPs would fail to bind to MT tips altogether. However, because the CLIPs could still bind to the outer MT tips at a slightly reduced level when the depletion of EB proteins was quite profound, we propose that the primary mechanism of CLIP accumulation at the MT tips is EB-independent. This conclusion is further supported by the fact that CLIP-170 still tip-tracks when EB1 is distributed along the whole MT lattice because of overexpression. Several in vitro studies provided evidence for copolymerization with tubulin as the model of CLIP targeting to the MT plus ends (Diamantopoulos et al., 1999; Arnal et al., 2004). Our in vivo data are in agreement with this model. Depletion of EB family members did not cause major changes in the rate of CLIP association with the growing plus ends, which appears almost instantaneous.
EB-independent mechanism of CLIP binding to the MT plus ends is also essential in budding and fission yeast. However, unlike the mammalian system, it involves an MT plus end-directed motor, kinesin, which transports the CLIP-170 homologues to the MT ends (Busch et al., 2004; Carvalho et al., 2004). This ancient mechanism may have been replaced later in evolution as insufficient for ensuring CLIP accumulation at the fast growing MT ends in mammals. Taken together, our study emphasizes that the interactions between different +TIPs are an important contributing factor for their specific localization.
Supplementary Material
Acknowledgments
We thank Dr. J. M. Schober for the comments on the manuscript. This work was supported by the Netherlands Organisation for Scientific Research grants to A.A. and N.G., and by the National Institutes of Health Grant GM25062 to G.B.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0614) on September 7, 2005.
Abbreviations used: GFP, green fluorescent protein; GST, glutathione S-transferase; HA, hemagglutinin; HIS, 6x histidine tag; co-IP, coimmunoprecipitation; mAb, monoclonal antibody; MT, microtubule; RNAi, RNA interference; siRNA, small interfering RNA; +TIPs, plus-end tracking proteins; YFP, yellow fluorescent protein.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Akhmanova, A., and Hoogenraad, C. C. (2005). Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 17, 47-54. [DOI] [PubMed] [Google Scholar]
- Arnal, I., Heichette, C., Diamantopoulos, G. S., and Chretien, D. (2004). CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 14, 2086-2095. [DOI] [PubMed] [Google Scholar]
- Askham, J. M., Vaughan, K. T., Goodson, H. V., and Morrison, E. E. (2002). Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol. Biol. Cell 13, 3627-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badin-Larcon, A. C., Boscheron, C., Soleilhac, J. M., Piel, M., Mann, C., Denarier, E., Fourest-Lieuvin, A., Lafanechere, L., Bornens, M., and Job, D. (2004). Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc. Natl. Acad. Sci. USA 101, 5577-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrueta, L., Kraeft, S. K., Tirnauer, J. S., Schuyler, S. C., Chen, L. B., Hill, D. E., Pellman, D., and Bierer, B. E. (1998). The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc. Natl. Acad. Sci. USA 95, 10596-10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- Bu, W., and Su, L. K. (2003). Characterization of functional domains of human EB1 family proteins. J. Biol. Chem. 278, 49721-49731. [DOI] [PubMed] [Google Scholar]
- Busch, K. E., and Brunner, D. (2004). The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14, 548-559. [DOI] [PubMed] [Google Scholar]
- Busch, K. E., Hayles, J., Nurse, P., and Brunner, D. (2004). Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev. Cell 6, 831-843. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., Gupta, M. L., Jr., Hoyt, M. A., and Pellman, D. (2004). Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell 6, 815-829. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., Tirnauer, J. S., and Pellman, D. (2003). Surfing on microtubule ends. Trends Cell Biol. 13, 229-237. [DOI] [PubMed] [Google Scholar]
- Coquelle, F. M. et al. (2002). LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 22, 3089-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamantopoulos, G. S., Perez, F., Goodson, H. V., Batelier, G., Melki, R., Kreis, T. E., and Rickard, J. E. (1999). Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 144, 99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erck, C. et al. (2005). A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl. Acad. Sci. USA 102, 7853-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart, N., and Perez, F. (2003). A plus-end raft to control microtubule dynamics and function. Curr. Opin. Cell Biol. 15, 48-53. [DOI] [PubMed] [Google Scholar]
- Goodson, H. V., Skube, S. B., Stalder, R., Valetti, C., Kreis, T. E., Morrison, E. E., and Schroer, T. A. (2003). CLIP-170 interacts with dynactin complex and the APC-binding protein EB1 by different mechanisms. Cell Motil. Cytoskelet. 55, 156-173. [DOI] [PubMed] [Google Scholar]
- Hayashi, I., and Ikura, M. (2003). Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J. Biol. Chem. 278, 36430-36434. [DOI] [PubMed] [Google Scholar]
- Honnappa, S., John, C. M., Kostrewa, D., Winkler, F. K., and Steinmetz, M. O. (2005). Structural insights into the EB1-APC interaction. EMBO J. 24, 261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad, C. C., Akhmanova, A., Grosveld, F., De Zeeuw, C. I., and Galjart, N. (2000). Functional analysis of CLIP-115 and its binding to microtubules. J. Cell Sci. 113, 2285-2297. [DOI] [PubMed] [Google Scholar]
- Howard, J., and Hyman, A. A. (2003). Dynamics and mechanics of the microtubule plus end. Nature 422, 753-758. [DOI] [PubMed] [Google Scholar]
- Juwana, J. P., Henderikx, P., Mischo, A., Wadle, A., Fadle, N., Gerlach, K., Arends, J. W., Hoogenboom, H., Pfreundschuh, M., and Renner, C. (1999). EB/RP gene family encodes tubulin binding proteins. Int. J. Cancer 81, 275-284. [DOI] [PubMed] [Google Scholar]
- Kodama, A., Karakesisoglou, I., Wong, E., Vaezi, A., and Fuchs, E. (2003). ACF7: an essential integrator of microtubule dynamics. Cell 115, 343-354. [DOI] [PubMed] [Google Scholar]
- Komarova, Y. A., Akhmanova, A. S., Kojima, S., Galjart, N., and Borisy, G. G. (2002). Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159, 589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen, G. et al. (2004). Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localisation. J. Cell Biol. 166, 1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon, L. A., Shelly, S. S., Tokito, M., and Holzbaur, E. L. (2003). The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol. Biol. Cell 14, 1405-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella, V., Rogers, G. C., Rogers, S. L., Buster, D. W., Vale, R. D., and Sharp, D. J. (2005). Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7, 235-245. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y. et al. (2005). CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, F., Diamantopoulos, G. S., Stalder, R., and Kreis, T. E. (1999). CLIP-170 highlights growing microtubule ends in vivo. Cell 96, 517-527. [DOI] [PubMed] [Google Scholar]
- Pierre, P., Scheel, J., Rickard, J. E., and Kreis, T. E. (1992). CLIP-170 links endocytic vesicles to microtubules. Cell 70, 887-900. [DOI] [PubMed] [Google Scholar]
- Rogers, S. L., Wiedemann, U., Hacker, U., Turck, C., and Vale, R. D. (2004). Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 14, 1827-1833. [DOI] [PubMed] [Google Scholar]
- Schuyler, S. C., and Pellman, D. (2001). Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell 105, 421-424. [DOI] [PubMed] [Google Scholar]
- Slep, K. C., Rogers, S. L., Elliott, S. L., Ohkura, H., Kolodziej, P. A., and Vale, R. D. (2005). Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 168, 587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, T., Slemmer, J., Hoogenraad, C. C., Lansbergen, G., Dortland, B., De Zeeuw, C. I., Grosveld, F., van Cappellen, G., Akhmanova, A., and Galjart, N. (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23, 2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, L. K., and Qi, Y. (2001). Characterization of human MAPRE genes and their proteins. Genomics 71, 142-149. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J. S., and Bierer, B. E. (2000). EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 149, 761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J. S., Grego, S., Salmon, E. D., and Mitchison, T. J. (2002). EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell 13, 3614-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Y., Eng, C. H., Schmoranzer, J., Cabrera-Poch, N., Morris, E. J., Chen, M., Wallar, B. J., Alberts, A. S., and Gundersen, G. G. (2004). EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820-830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.