Abstract
CLIP-170 belongs to a group of proteins (+TIPs) with the enigmatic ability to dynamically track growing microtubule plus-ends. CLIP-170 regulates microtubule dynamics in vivo and has been implicated in cargo-microtubule interactions in vivo and in vitro. Though plus-end tracking likely has intimate connections to +TIP function, little is known about the mechanism(s) by which this dynamic localization is achieved. Using a combination of biochemistry and live cell imaging, we provide evidence that CLIP-170 tracks microtubule plus-ends by a preassociation, copolymerization, and regulated release mechanism. As part of this analysis, we find that CLIP-170 has a stronger affinity for tubulin dimer than for polymer, and that CLIP-170 can distinguish between GTP- and GDP-like polymer. This work extends the previous analysis of CLIP-170 behavior in vivo and complements the existing fluorescence microscope characterization of CLIP-170 interactions with microtubules in vitro. In particular, these data explain observations that CLIP-170 localizes to newly polymerized microtubules in vitro but cannot track microtubule plus-ends in vitro. These observations have implications for the functions of CLIP-170 in regulating microtubule dynamics.
INTRODUCTION
Microtubule dynamic instability plays a fundamental role in cellular processes ranging from formation of the mitotic spindle to cell polarization. Dynamic instability allows microtubules to explore space and find poorly diffusible targets, such as organelles, chromosomes, and specific regions of the cell cortex. Selective stabilization of dynamic microtubules determines the morphology of the microtubule network and likely underlies morphological change (Kirschner and Mitchison, 1986; Gundersen et al., 2004). The plus-end of the microtubule (the fast-growing end normally distal from the centrosome) is particularly important to microtubule function because its conformation governs transitions between polymerization and depolymerization and because it is often the first part of the microtubule to encounter new cargo as the microtubule probes the cytosol (Hill and Carlier, 1983; Desai and Mitchison, 1997; Vaughan, 2004). Therefore, the plus-end is a logical place to localize regulators of dynamic instability and mediators of microtubule-cargo interactions.
The first protein observed to dynamically track microtubule plus-ends was CLIP-170 (Perez et al., 1999). Other plus-end tracking proteins (+TIPs) include dynactin, EB1, and Lis1 (Morrison et al., 1998; Vaughan et al., 1999; Faulkner et al., 2000; Kaplan et al., 2001). These proteins have disparate sequences and structures, but share involvement in microtubule dynamics and/or cargo-microtubule interactions. To understand the roles of +TIPs in these processes, it is necessary to explain how they track plus-ends. For example, it has been postulated that plus-end tracking is a mechanism for delivering proteins to the cell cortex, but this is true only if the tracking behavior results from actual translocation of the protein in question. Our analysis focuses on CLIP-170, a well-conserved protein that promotes endosome-microtubule interactions in vitro (Pierre et al., 1992) and is required for processive microtubule growth in vivo (Komarova et al., 2002).
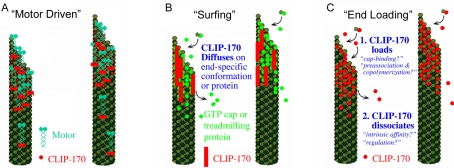
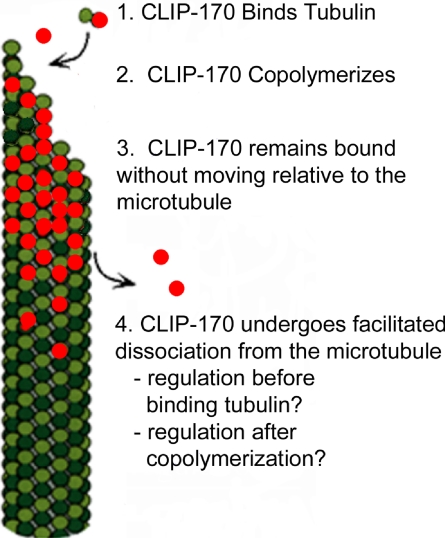
Three possible mechanisms for CLIP-170 plus-end tracking behavior are outlined in Figure 1. First, CLIP-170 could ride on a plus-end directed motor (Figure 1A). Recent evidence suggests that yeast orthologues of CLIP-170 (Saccharomyces cerevisiae Bik1p and Schizosaccharomyces pombe Tip1p) track plus-ends by this mechanism (Busch and Brunner, 2004; Busch et al., 2004; Carvalho et al., 2004). Second, CLIP-170 could diffuse, or “surf,” along a plus-end specific conformation or protein. (Figure 1B). Although some researchers have used the term “surfing” to describe all plus-end tracking behavior, we use surfing to specifically refer to movement of a +TIP on the potential energy “wave” created by the existence of an end-specific conformation or protein. S. cerevisiae Kar9p appears to track microtubule plus-ends by a surfing mechanism (Liakopoulos et al., 2003). In the case of Kar9p the surfing appears to be accomplished via Bim1p, but surfing could also be accomplished by surfing on a conformational cap at the microtubule plus-end. Both the “motor-driven” and “surfing” mechanisms involve the physical translocation of CLIP-170 during plus-end tracking behavior. The third mechanism, “end-loading,” actually describes a group of mechanisms that have in common the binding of CLIP-170 specifically at the microtubule end followed by rapid dissociation from the microtubule after loading. End-loading mechanisms (sometimes called “treadmilling” mechanisms) result in CLIP-170 being stationary with respect to the microtubule lattice.
Figure 1.
Potential mechanisms of microtubule plus-end tracking. (A) Motor-driven transport: CLIP-170 (red) is physically moved toward the microtubule tip by motors (blue). Fungal CLIP-170 homologues Bik1p and Tip1p appear to track by this mechanism; (B) “surfing”: CLIP-170 (red bars) diffuses (“surfs”) along a tip-specific conformation or another plus-end tracking protein (bright green). Note that CLIP-170 is represented by bars rather than circles because this model likely requires multiple binding sites; (C) end-loading: CLIP-170 (red) binds specifically at the microtubule plus-end and then dissociates shortly thereafter. CLIP-170 is stationary on the microtubule and would not be observed to move toward either the plus- or minus-end of the microtubule. Loading at the tip could occur by preferential binding of CLIP-170 to the cap (“cap-recognition”), or by preassociation with unpolymerized tubulin followed by copolymerization. Dissociation could occur by CLIP-170 having an intrinsically weak affinity for the “older” GDP polymer or be induced by a regulatory event that reduces the affinity of CLIP-170 for microtubules.
Previous analysis of CLIP-170 behavior has indicated that CLIP-170 associates with newly polymerized tubulin both in vivo and in vitro and has provided evidence that CLIP-170 tracks microtubule plus-ends by end-loading (Diamantopoulos et al., 1999; Perez et al., 1999). However, this work left many issues unresolved, including the specific mechanism of end-loading. In particular, the experiments indicating that CLIP-170 interacts with unpolymerized tubulin could not distinguish whether CLIP-170 binds primarily to preexisting oligomers or to dimer, and did not address whether the affinity for unpolymerized tubulin is in a physiologically significant range (Diamantopoulos et al., 1999; Arnal et al., 2004). Also, the conclusion that CLIP-170 tracks microtubule plus-ends by an end-loading mechanism has recently been called into question by evidence that fungal CLIP-170 homologues move via kinesin motor activity (Busch and Brunner, 2004; Busch et al., 2004; Carvalho et al., 2004).
Using a combination of live cell imaging at high frame rates and quantitative biochemical analysis of CLIP-170-tubulin interactions, we provide strong evidence that CLIP-170 tracks microtubule plus-ends by a preassociation, copolymerization, and regulated release mechanism. In particular, we find that CLIP-170 binds tightly and stably to both dimeric tubulin and to microtubules. Although CLIP-170 does have stronger affinity for GTP-like microtubules than for GDP-like microtubules, our kinetic analysis indicates that cap recognition cannot account for CLIP-170 plus-end tracking behavior, even if the physiologically significant cap conformation is one that has not been tested. This differential affinity for GTP-microtubules may however have implications for the mechanism CLIP-170 uses to influence microtubule dynamics.
MATERIALS AND METHODS
Plasmids, Protein Expression, and Purification
Bacterially expressed CLIP-170 H1 and H2 fragments were prepared as previously described by Scheel et al. (1999), except that they were subcloned from pET19b into pET15b. H1-pET19b was digested with NdeI and BamHI to remove the H1 fragment that was then cloned into the same pET15b restriction sites. H2 was transferred using NdeI and XhoI. The proteins were then expressed in BL21 bacteria and purified after French press lysis by the standard Novagen His-tag purification protocol (Madison, WI). The purified proteins were ∼40 kDa (H1) and ∼60 kDa (H2) as predicted by the amino acid sequence (Scheel et al., 1999). Both H1 and H2 were centrifuged for 15 min at 65,000 × g before all experiments to remove any aggregated protein. The GFP-CLIP-170 and GFP-H1 constructs were described previously (Perez et al., 1999).
Tubulin was purified from porcine brain by three cycles of glycerol-dependent polymerization in PEM buffer (100 mM Pipes, 1 mM MgCl2, 1 mM EGTA) followed by purification across a P11 column (Hyman et al., 1991). Tubulin was stored in PEM buffer in aliquots at -80°C, cycled the day experiments were performed, and spun for 15 min at 65,000 × g before all experiments. Unless otherwise noted, all protein concentrations were estimated by Bio-Rad Protein Assay (Richmond, CA).
Preparation of Microtubules
Polymerization of microtubules with Taxol (paclitaxel, Sigma, St. Louis, MO) was performed by the stepwise addition of Taxol [http://mitchison.med.harvard.edu/protocols/poly.html]. Taxol microtubules were frozen in small aliquots at -80°C. GMPCPP microtubules were prepared by three cycles of polymerization in the presence of 50 μM GMPCPP (purchased from Jena Biosciences, Jena, Germany) without any additional GTP or GDP, and they were prepared fresh from P11 tubulin for each experiment.
Coimmunoprecipitation
HeLa lysates were prepared as previously described (Rickard and Kreis, 1990). The lysate (50 μL; 7.5 mg/ml) was incubated for 90 min at 4°C with 2 μL of either a monoclonal antibody (mAb) against CLIP-170, 4D3, (Rickard and Kreis, 1991) or an mAb, 9E10, against the myc epitope (Cancer Research Technologies, London, United Kingdom) as a negative control. Reactions were then brought to 100 μL total volume with PEM. 25 μL of protein A-Agarose beads (Pierce, Rockford, IL; 20333) were added, and the reactions incubated an additional 30 min. Three 10-min washes at 4°C with PEM were then performed. Proteins were eluted from the beads with SDS sample buffer, separated by SDS-PAGE and blotted onto nitrocellulose. The resulting Western blot was probed with the T13 antibody against alpha tubulin, which was detected by chemiluminescence visualization of a secondary antibody conjugated to HRP (Pierce; 34080).
Surface Plasmon Resonance
General. Unless otherwise indicated, all experiments were conducted on a Biacore3000 machine (Piscataway, NJ) with bacterially expressed CLIP-170 fragments coupled to NTA chips via the N-terminal His-tag. All experiments were run at conditions unfavorable for tubulin polymerization: 10°C, 1 mM MgCl2, and tubulin concentrations <5 μM. For each run, one of the four flow cells on the chip was left without CLIP-170 as a negative control. The signal in this flow cell was subtracted from the experimental wells before further analysis. An additional negative control in some experiments was tubulin folding cofactor B. NTA chips were superior to standard CM5 chips for Biacore analysis of CLIP-170 behavior because: 1) coupling of H1 to the sensor chip via the His-tag produced a homogeneous binding surface and consistent binding behavior; and 2) NTA chips could be reused after being stripped with EDTA, whereas CM5-CLIP-170 chips could be used only once because of the difficulty in achieving complete dissociation of bound tubulin.
Equilibrium Binding Experiments. The CLIP-170 H1 fragment was coupled to a level of 200 response units (RUs), whereas H2 was coupled a level of 300 RUs. Experiments in which the amount of coupled CLIP-170 differed by more than 2% were discarded. After coupling, the system was primed with PEM supplemented with 10 μM GTP (Sigma) and 0.001% surfactant P20. Tubulin in PEM + 10 μM GTP was then injected at a concentration between 0.01 and 1.0 μM and at a flow rate of 10 μL/min until steady state was achieved. After a short dissociation phase the chip was stripped (“strip buffer”; Novagen, Madison, WI) and the sequence started again with variation only in the concentration of tubulin injected. The fractional saturation of the response at steady state was then plotted against the concentration of tubulin and fit to a bimolecular binding curve Y = Bmax*X/(Kd + X), where Y is the fractional saturation of the CLIP-170 fragment, X is the concentration of tubulin, and Bmax is the maximal achievable response. Curves were fit using Prism 4.00 (GraphPad software). The experiment at each tubulin concentration was repeated in triplicate, with the variation in results indicated by the error bars in Figure 4. Control experiments were performed in which the His-tag was removed from H1, and the H1 was subsequently coupled to a CM5 chip by standard amine coupling procedures. As expected, CLIP-170 does bind to tubulin under these conditions (unpublished data). However, technical difficulties that arose from the slow dissociation of the CLIP-170-tubulin complex as related to the stability of tubulin made it impractical to perform quantitative experiments in this manner. Instead it was more efficient to repeatedly strip the NTA sensor chip and recouple fresh H1.
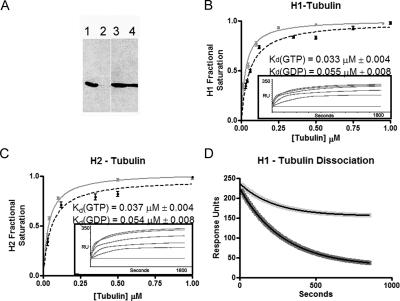
Figure 4.
CLIP-170 binds tubulin tightly and with slow kinetics. (A) Coimmunoprecipitation experiment visualized by western blot with the polyclonal antibody against tubulin, T13. Coimmunoprecipitations were performed with 4D3, a mAb against CLIP-170 (lane 1) or a mAb against myc (lane 2). Loading controls from these experiments are shown in lane 3 (4D3) and lane 4 (anti-myc); (B and C) the tubulin binding ability of both H1 and H2 were analyzed by SPR (Biacore) as described in Materials and Methods. H1 (B) and H2 (C) show nearly identical equilibrium binding abilities for soluble tubulin with respective Kd values of 0.033 ± 0.004 and 0.037 ± 0.004 μM for GTP tubulin (gray) and 0.055 ± 0.008 and 0.054 ± 0.009 μM for GDP tubulin (black). Three replicates were performed at each tubulin concentration and the variation is shown by error bars. Representative experiments for binding to GTP tubulin are shown as insets; (D) SPR experiments optimized to determine dissociation kinetics show that CLIP-170 releases both GDP tubulin dimers (dark gray) and GTP tubulin dimers (light gray) slowly. Fitting of both data sets as single phase dissociations (see Materials and Methods for details) showed that GTP tubulin and GDP tubulin dissociate with koff values of 3.5 × 10-3± 2 × 10-4 s-1 and 4.4 × 10-3± 5 × 10-4 s-1, respectively.
Kinetic Analysis. Biacore experiments for kinetic analysis were performed as above with the following modifications. Dissociation phases of the sensogram were exported and fit (using Prism 4.00) to the single-phase dissociation equation Y = Ymax*e-kx, where Y is the response (RUs) and x is the time in seconds. A common complication in SPR experiments is ligand (tubulin) rebinding, resulting in an artifactually slow dissociation rate. To control for such problems, flow rates were varied from 10 to 50 μL/min. Experiments with flow rates of <35 μL/min were judged to be subject to rebinding complications because the apparent koff increased with the flow rate. However, no difference was observed in observed koff between experiments performed at 35 and 50 μL/min. The data depicted in Figure 4 were obtained after an injection of 0.2 μM tubulin at a flow rate of 50 μL/min. Injections of tubulin at three different concentrations (0.01 μM [below Kd], 0.05 μM [∼Kd], and 0.2 μM [CLIP-170 saturated]) confirmed that the off rate constant was concentration independent.
As mentioned in the text, the dissociation of GTP-tubulin from H1 is biphasic. The off rate reported in Figure 4D is derived from the fast phase only. We have chosen to ignore the slow phase in the present analysis because the origin of this slow phase is still under investigation and because it is the fast phase that sets an upper limit on the dissociation rate.
Cosedimentation Assays
Experiments were performed with 2.0 μM CLIP-170 fragment and varying concentrations of microtubules. Reactions were brought to a final volume of 100 μL with PEM buffer, incubated at 37°C for 20 min, and then centrifuged at 65,000 × g for 30 min at 37°C. The supernatant and the pellet were separated, and equal fractions were analyzed by SDS-PAGE and Coomassie staining. A Molecular Dynamics (Sunnyvale, CA) densitometer was used to capture the image, and Image Quant 5.2 (Molecular Dynamics) used to quantify the bands. The fraction of the CLIP-170 fragment in the pellet was defined to be the fraction of CLIP-170 bound because CLIP-170 did not sediment on its own (confirmed for each set of experiments by a control experiment without microtubules). The amount of free tubulin was calculated by assuming a 1:1 binding stoichiometry (see note below) and subtracting the number of moles of bound CLIP-170 from total moles of tubulin. The fraction of CLIP-170 bound was then plotted against the concentration of free tubulin, and the data were fit to the standard bimolecular binding equation Y = Bmax*X/(Kd + X), where Y is the fraction of CLIP-170 in the pellet; X is the concentration of microtubules, and Bmax is the maximal achievable binding (Prism 4.00 was used for the fitting; GraphPad, San Diego, CA). Control experiments with the His-tag cleaved from H1 produced indistinguishable results (unpublished data). Because technical difficulties (discussed above) dictated that the SPR analysis be performed with His-tagged protein, His-tagged H1 was also used for the sedimentation experiments unless otherwise indicated.
Note about stoichiometry of H1-tubulin binding: The cosedimentation analysis presented in this article assumes a 1:1 stoichiometry for H1: tubulin binding in microtubules. This assumption may be invalid—we find a binding ratio of 1.5:1 at saturation (Supplementary Data Figure 2), and Scheel et al. (1999) found a ratio of 1.8:1. However, because CLIP-170 bundles microtubules (Arnal et al., 2004) and because bundling reduces the effective concentration of microtubules, it seems likely that deviations from 1:1 binding are in part artifacts of microtubule bundling. Therefore, we have performed our analysis under the assumption that the binding ratio is 1 H1 per polymerized tubulin dimer. If this assumption is incorrect, then the Kd values are lower (stronger) and the dissociation rates are even slower than those reported in this article. This outcome would provide even stronger support for the preferred plus-end tracking mechanism outlined in the Discussion.
Fluorescence Anisotropy
H1 was labeled with bis (2,2′-bipyridine)-4,4′-dicarboxybipyridine-ruthenium di (N-succinimidyl ester) bis (hexafluorophosphate) (Sigma; 96632). Labeling was accomplished by adding 1 mg of fluorophore to 1 ml of 1 mg/ml protein in phosphate-buffered saline buffer. The mixture was then stirred at room temperature for 3 h, and the free label was removed by purification across a 10 DG disposable chromatography column (Bio-Rad, Richmond, CA; 732-2010). The labeling ratio was estimated to be 5:1 using the respective molar extinction coefficients 16,000 M-1 cm-1 at 450 nm (Terpetschnig et al., 1995) for the fluorophore and 22,430 M-1 cm-1 at 280 nm for H1 (estimated using ExPASy Protparam). All polarization analysis was performed on a Beacon 2000 (PanVera, Madison, WI) at 25°C using the standard fluorescein filters supplied with the instrument.
Equilibrium Analysis. H1, 0.1 μM, was incubated with concentrations of microtubules or tubulin ranging from 0 to 6 μM for 10 min, and the anisotropy was then measured. Anisotropy was plotted against the concentration of microtubules, and the data were fit to the bimolecular binding equation (Y = Bmax*X/(Kd + X) + C) using Prism 4.00, where Y is the anisotropy; X is the concentration of microtubules; Bmax is the maximal achievable anisotropy; and C is the anisotropy in the absence of ligand (tubulin or microtubules).
Kinetic Analysis. Tubulin (1, 2, or 4 μM) or microtubules (5, 10, or 20 μM) was added to 0.1 μM labeled H1. The solution was briefly mixed by pipetting twice and the anisotropy was read immediately. Timing of the process revealed that it was 5 s between the addition of tubulin/microtubules and the first reading. The system was maintained at 25°C and the polarization read initially at 5 s and then every 15 s thereafter. Addition of bovine serum albumin instead of tubulin did not change the anisotropy of the H1 from the 110–120 mAnisotropy units that H1 displayed alone. Data for tubulin were fit as a single phase association (Y = Ymax*(1 - e-kX) + C) using Prism 4.00, where Y is the anisotropy; X is the time in seconds; and C is the anisotropy in the absence of ligand (tubulin). It was necessary to fit the polymer data as a two-phase association [Y = (Ymax1*(1 - e(-K1*X)) + Ymax2*(1 - exp(-K2*X))) + C], where Y is the anisotropy, X is the time in seconds, K1 is the first association constant, K2 is the second association constant, and C is the anisotropy in the absence of ligand. The existence of this second phase is intriguing (it is very slow, ∼1 × 10-5 μM-1 s-1), but at present it is unclear whether it reflects a CLIP-170-related phenomenon or is an artifact of another change in the system, such as microtubule bundling. Therefore, we have based our current analysis on the rapid association phase. Rate constants obtained by this approach are similar to those obtained from a single phase fit to the early portion of the data (unpublished data).
Live Cell Imaging and Analysis of Fluorescence Data
Microscopy was performed on a Nikon inverted microscope (Melville, NY; TE2000) fitted for immunofluorescence with HighQ filter sets from Chroma (Brattleboro, VT) in a room thermostatted at 28°C. Cells were grown on top of coverslips that were then inverted onto slides, sealed with VaLaP, mounted onto the stage, and used for no more than 45 min. Microscopy was performed using a 60× 1.4NA objective and a 1.5× optivar. Images were acquired at 0.2-s intervals with a cooled backthinned CCD camera (Photometrics, Tucson, AZ; Cascade 512B) operating in streaming mode.
Kymograph Analysis. Kymographs displaying the evolution of CLIP-170 fluorescence along a microtubule as a function of time were produced with the kymograph function of Metamorph software (Universal Imaging, West Chester, PA) set to average the intensity of two adjacent pixels. The value of the fluorescence intensity of a given position as a function of time was extracted from the kymographs by using the “linescan” function of Metamorph to log the data into a spreadsheet program (Microsoft, Redmond, WA; Excel). Dissociation constants describing the decay of CLIP-170 fluorescence were estimated from this intensity data using Prism 3.2c (GraphPad) to fit the equation for a damped single exponential function (Intensity = Span*exp(-koffapp*time)*exp(-kbleach*time) + Plateau) to the linescan data (see Figure 5). Only the data corresponding to the decay was used for the fitting. At least 10 positions in 10 microtubules in 5 separate cells (total of 50 microtubules) were examined for calculation of the average apparent dissociation constants. Bleaching constants were obtained separately for each cell by fitting the total integrated fluorescence intensity of that cell to a single exponential function. In these calculations, rebinding of CLIP-170 was ignored. Therefore, these apparent dissociation constants represent the lower limits of the actual dissociation constants.
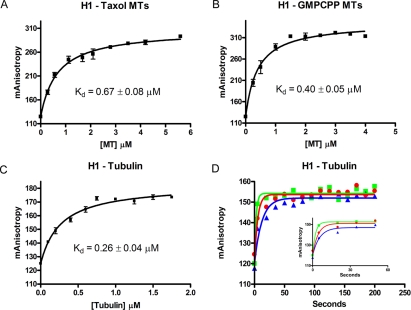
Figure 5.
Fluorescence anisotropy confirms the cosedimentation and Biacore experiments. Equilibrium binding affinities of CLIP-170 for GTP-like microtubules, GDP-like microtubules, and GTP tubulin were determined by measuring the anisotropy of labeled H1 as a function of tubulin/microtubule concentration and fitting to a rectangular hyperbola (see Materials and Methods for details). In the case of these analyses, unlike those in Figure 3, H1 binding is plotted as a function of total microtubule concentration because the technique allows us to use H1 at a concentration that is less than the Kd (0.1 μM). This results in a situation where the concentration of free ligand is approximately equal to the concentration of total ligand. (A) H1 binds GDP-like (Taxol) microtubules with a 0.67 ± 0.08 μM affinity; (B) H1 binds GTP-like (GMPCPP) microtubules with a 0.40 ± 0.05 μM affinity; (C) H1 binds GTP tubulin with a 0.26 ± 0.04 μM affinity; (D) binding kinetics were studied by adding tubulin (1 μM, blue); 2 μM, red; or 4 μM green) to labeled H1. Anisotropy was read before the addition of tubulin, 5 s after the tubulin was added, and every 15 s thereafter. The first 60 s of binding are depicted in the inset. Data were fit as a single-phase association and analysis of the kinetic data are presented in Table 1.
RESULTS
CLIP-170 Uses an End-loading Mechanism to Track Microtubule Plus-Ends
Because “motor-transport” and “surfing” both require that CLIP-170 translocates along the microtubule, we examined whether CLIP-170 moves along microtubules in vivo. The initial description of CLIP-170 plus-end tracking behavior reported that CLIP-170 does not move, but this analysis was limited to few frames obtained at large time intervals (3–5 s; Perez et al., 1999). Because plus-end tracking behavior of fungal CLIP-170 orthologues appears to be kinesin-mediated (Busch and Brunner, 2004; Busch et al., 2004; Carvalho et al., 2004), the possibility of CLIP-170 movement needed to be addressed with better time resolution and with a more comprehensive approach. We imaged GFP-CLIP-170 in vivo at 0.2-s intervals and used kymographs to analyze the behavior of heterogeneities (“fluorescent speckles”) in the comet-like fluorescence signal on single microtubule plus-ends (Figure 2; Supplementary Data Figure 1a). Because the y-axis of the kymographs corresponds to frame number (time) and the x-axis corresponds to position, movement of CLIP-170 would be expected to result in diagonal lines of fluorescence (Figure 2D). However, both bright and dim regions of CLIP-170 fluorescence fall on vertical lines, indicating that most CLIP-170 maintains its position over time (Figure 2E).1
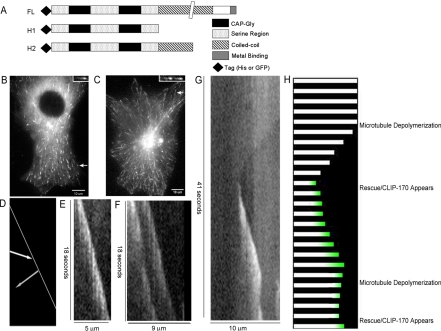
Figure 2.
Dynamic behavior of CLIP-170 in vivo. (A) CLIP-170 constructs used in this work. As previously described by Scheel et al. H1 acts as a monomer and H2 as a dimer; (B and C) Cos7 cells were transfected with GFP-CLIP-170 (B) or GFP-CLIP-170-H1 (C), imaged by streaming video (0.2 s/frame; see movies in Supplementary Data), and the behavior of CLIP-170 on individual microtubules was examined by making kymographs; (D) Cartoon depicting the pattern expected if CLIP-170 were moving along a microtubule via motors. The white line indicates the plus-end of the microtubule, the gray arrow predicts what would be observed if CLIP-170 were moving toward the plus-end and the white arrow predicts what would be observed if CLIP-170 is moving away from the microtubule plus-end; (E–G) Kymographs of comets in cells transfected with GFP-CLIP-170 (E) and GFP-CLIP-170-H1 fragment (F and G). The kymographs depict position on the x-axis (increasing to the right) and time on the y-axis (increasing toward the bottom). Notice that both bright and dim heterogeneities form vertical lines indicating that they maintain their position (they do not move to the left or right) while they decay over time. (G) An example of GFP-H1 behavior during microtubule rescue. The microtubules corresponding to the kymographs are indicated by arrows in B and C and in the corresponding movies (see Supplementary Data); (H) cartoon interpreting the kymograph of the rescue event (2G). The microtubule seen in white is shrinking for several seconds. Suddenly CLIP-170 (green) appears at the plus-end of the shrinking microtubule at approximately the same time that the microtubule is rescued. The most important aspect of the CLIP-170 appearance is that it is not seen traveling to the plus-end on the microtubule but instead appears de novo from the cytosol.
GFP-H1, a construct consisting only of the CLIP-170 “head,” behaved similarly to GFP-CLIP-170 at low levels of expression (Figure 2F, Supplementary Data Figure 1b). At intermediate levels, GFP-H1 began to label the microtubule lattice dimly while still labeling tips brightly, allowing the imaging of rescue events. Analysis of these events reveals that CLIP-170 appears de novo on the rescued microtubule: regions of the microtubule distal to the rescue event remain dim before the rescue (Figure 2G, Supplementary Data Figure 1, c and d). This observation implies that the CLIP-170 observed on newly rescued microtubules originates in the cytosol and is not translocating along the microtubule. The lack of observable CLIP-170 movement toward the microtubule plus-end, together with the behavior of CLIP-170 during rescue, eliminates “motor-transport” and “surfing” as major mechanisms for CLIP-170 plus-end tracking and leaves end-loading as the most plausible mechanism.
Dissection of the End-loading Mechanism
The most likely mechanisms for binding specifically at the microtubule plus-end are “cap-recognition” and “preassociation and copolymerization” (Figure 1). Both mechanisms, which are nonexclusive, require that CLIP-170 have the ability to “sense” changes in tubulin conformation, with stronger affinity for certain conformations. They differ in which conformations should be preferred. Specifically, cap-recognition requires that CLIP-170 have a strong affinity for caplike polymer, and preassociation and copolymerization requires that CLIP-170 have a strong affinity for tubulin dimer. Therefore, we conducted quantitative biochemical analysis of the interactions between CLIP-170, tubulin, and microtubules to distinguish between these end-loading mechanisms.
Three techniques were used to study CLIP-170 binding to tubulin and microtubules. Cosedimentation, the classic technique for studying interactions with microtubules, was used to measure binding of CLIP-170 to microtubule polymer. Surface plasmon resonance (SPR) on a Biacore3000 was used to measure interactions between CLIP-170 and tubulin dimer. Finally, interactions of CLIP-170 with both tubulin and microtubules were studied by fluorescence anisotropy. Bacterially expressed N-terminal fragments of CLIP-170 (H1 and H2; Figure 2A) were used for this analysis because they track plus-ends in vivo (Perez et al., 1999; Figure 2), are easily expressed and purified (Scheel et al., 1999) and are homogeneous in splice isoform and regulatory state, unlike native CLIP-170 (Rickard and Kreis, 1991; Griparic and Keller, 1999).
CLIP-170 Has Stronger Affinity for GTP-like Polymer than GDP-like Polymer
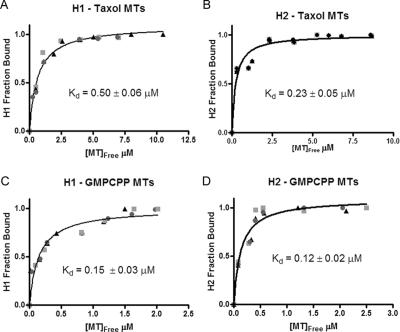
Cosedimentation assays were used to investigate interactions between CLIP-170 and polymerized tubulin, with Taxol microtubules used as mimics of GDP microtubule polymer and GMPCPP microtubules used as mimics of the GTP microtubule polymer in the plus-end cap (Hyman et al., 1992; Severin et al., 1997). Assuming a 1:1 binding ratio (see Materials and Methods), the affinities of H1 and H2 for GDP-like microtubules are 0.50 ± 0.06 and 0.23 ± 0.05 μM, respectively (Figure 3, A and B). In contrast H1 and H2 bind GTP-like microtubules with affinities of 0.15 ± 0.03 and 0.12 ± 0.02 μM, respectively (Figure 3, C and D). Shearing the microtubules did not change the affinities, implying that CLIP-170 can bind the length of the microtubule and not merely the microtubule end (unpublished data). The subtle preference of H1 for GTP-like polymer suggests that CLIP-170 can sense differences in microtubule conformation and provides initial support for the cap recognition hypothesis (it should be noted that it is difficult to use microtubule cosedimentation to measure affinities stronger than 0.2 μM, so H2 may show a greater preference for GTP-like microtubules than reported here). However, even though CLIP-170 has stronger affinity for “caplike” conformations, it could still load onto the microtubule plus-end by preassociation and copolymerization if CLIP-170 binds sufficiently tightly to unpolymerized tubulin. Therefore, interactions between CLIP-170 and tubulin were investigated.
Figure 3.
CLIP-170 can distinguish between GTP- and GDP-like microtubules. Cosedimentation assays were used to analyze CLIP-170 binding to microtubules. Microtubules stabilized with Taxol were used to mimic the GDP lattice of the microtubule. Microtubules stabilized with the slowly hydrolyzed GTP analog GMPCPP were used to mimic the GTP tubulin in the cap. The binding of 2 μM CLIP-170 fragments was measured as a function of microtubule concentration. Three separate experiments were performed and are indicated by squares, triangles, and circles. The fraction of CLIP-170 bound (in the pellet) was plotted against the concentration of unbound polymerized tubulin, and the three experiments then simultaneously fit to the bimolecular binding curve to obtain the apparent Kd (see Materials and Methods for details). Both H1 and H2 bind GDP microtubules with a strong affinity, 0.5 ± 0.1 μM (A) and 0.2 ± 0.1 μM (B), respectively. However, each construct binds the GTP-like GMPCPP microtubules more tightly, with respective Kd values of 0.15 ± 0.3 μM (C) and 0.12 ± 0.02 μM (D).
CLIP-170 Binds to Tubulin Dimer with Strong Affinity
To determine whether CLIP-170 can bind tubulin dimers, we first tested whether His-tagged H1 binds soluble tubulin in pull-down assays and found that it did (unpublished data). Coimmunoprecipitation of tubulin from HeLa cytosol using a mAb against CLIP-170, 4D3, confirmed that the interaction between CLIP-170 and tubulin can occur in a physiological context (Figure 4A). However, these approaches give no information about the strength or characteristics of the interaction. SPR was used to obtain the quantitative estimate of the CLIP-170 affinity for dimeric tubulin that is necessary to test the preassociation and copolymerization model. These experiments revealed that both H1 and H2 bind tubulin dimer very tightly, with respective affinities of 0.033 ± 0.004 μM (Figure 4B) and 0.037 ± 0.004 μM (Figure 4C). These affinities are ∼10 times stronger than those measured for GDP microtubules (Figure 3, A and B), and ∼4 times stronger than those measured for GMPCPP microtubules (Figure 3, C and D). Given that the cellular concentrations of tubulin dimer and microtubule polymer are approximately equal (∼10 μM; Hiller and Weber, 1978; Rodionov et al., 1999; Bulinski et al., 2001) and that the conformational cap is likely very small, the strong preference of CLIP-170 for tubulin dimer suggests that CLIP-170 will be heavily biased toward unpolymerized tubulin at physiological concentrations.
To provide further support for these conclusions, we utilized fluorescence anisotropy to analyze interactions between CLIP-170 and different conformations of tubulin. The apparent affinities of H1 as measured by fluorescence anisotropy were 0.67 ± 0.08 μM for GDP-like microtubules, 0.40 ± 0.05 μM for GTP-like microtubules, and 0.26 ± 0.04 μM for tubulin (Figure 5). The measurements made by fluorescence anisotropy differ from those made by cosedimentation and SPR, but the overall trends are similar: H1 binds GTP-like microtubules more tightly than GDP-like microtubules, but binds tubulin more strongly than either microtubule conformation. The only major discrepancy between methods is in the affinity of H1 for tubulin dimer. Our analysis focuses on dimer affinities as determined by SPR because tubulin and H1 form oligomers under the conditions in fluorescence anisotropy experiments (Diamantopoulos et al., 1999; Arnal et al., 2004) (Folker and Goodson, unpublished results), and so the fluorescence anisotropy experiments likely do not reflect H1 binding to tubulin dimer (see Discussion).
The Importance of Kinetic Analysis
The likely saturation of CLIP-170 by tubulin suggests that end-loading is achieved by a preassociation and copolymerization mechanism. However, equilibrium analysis alone is not sufficient to define the end-loading mechanism: association and dissociation rates could impact the distribution of CLIP-170 because the microtubule cytoskeleton is not a system at equilibrium. Kinetic analysis is needed for an additional reason: GMPCPP microtubules may not accurately mimic the physiological microtubule cap. Cap recognition could potentially drive plus-end tracking if the “real” cap conformation has a strong enough affinity for CLIP-170. However, tracking by cap recognition would require that CLIP-170 rapidly “sample” different conformations of tubulin: given that the cap conformation has a very short lifetime and that CLIP-170 is likely saturated by free tubulin (explained above), CLIP-170 would have to exchange off of tubulin dimer quickly to enable it to bind the transient cap. Using this logic, measurement of the kinetics of CLIP-170-tubulin interactions is essential to establishing the mechanism of plus-end loading.
CLIP-170 Dissociates from Tubulin Dimer Slowly
To analyze the possibility that CLIP-170 rapidly samples its environment, we performed additional SPR experiments optimized for measuring dissociation kinetics. GTP tubulin dissociates from CLIP-170 H1 in a two-phase process that has a fast phase with an apparent koff of 3.5 × 10-3 ± 2 × 10-5 s-1 (t1/2 ∼ 190 s; Figure 4D) and a slow phase that is not measurable (t1/2 > hours; tubulin never dissociates completely and is not stable over this time period). We have chosen to ignore the slow phase in the present analysis because the origin of this slow phase is still under investigation and because the fast phase sets an upper limit on the dissociation rate. To confirm that dissociation of tubulin from H1 can be analyzed using SPR methods, we measured the dissociation of GDP-tubulin from H1. Unlike GTP tubulin, GDP tubulin dissociates completely and fits well as a single phase with an apparent koff of 4.4 × 10-3± 5 × 10-4 s-1 (t1/2 ∼170 s).2
The slow dissociation rate of tubulin from H1 was further supported by measuring the association kinetics by fluorescence anisotropy. Tubulin was added to fluorescently labeled H1, and the anisotropy was measured 5 s after the tubulin was added and every 15 s thereafter. Experiments were performed at three tubulin concentrations (1, 2, and 4 μM; Figure 5D). Fitting each data set as a single phase association results in a linear increase in the observed association rate (kobserved) with respect to tubulin concentration (Table 1). This linear relationship results in constant apparent kon values of ∼0.09 ± 0.02 μM-1 s-1 (Table 1). Using this kon and the Kd as measured by the fluorescence anisotropy experiments (0.26 ± 0.04 μM; Figure 5C), dissociation was calculated to occur with a half-life of ∼30 ± 9 s (Table 1).
Table 1.
Association of CLIP-170 with tubulin
| [Tubulin] (μM) | kobserved (s–1) | Apparent kon (μM–1 s–1) | t1/2 dissociation (s–1) |
|---|---|---|---|
| 1 | 0.08 ± 0.02 | 0.08 ± 0.02 | 33 ± 9 |
| 2 | 0.19 ± 0.04 | 0.10 ± 0.02 | 27 ± 7 |
| 4 | 0.37 ± 0.08 | 0.09 ± 0.02 | 30 ± 9 |
The data presented in Figure 5D were fit as a single-phase exponential association (details in Materials and Methods) to determine the observed rate of association (kobserved). This value was then related to the concentration of tubulin to determine the apparent kon. Finally the koff was calculated from the measured kon value, and the measured Kd (Figure 5C) and is presented as the t1/2 of dissociation.
Although the koff values obtained by SPR and fluorescence anisotropy differ significantly (see Discussion), both are slow. These observations indicate that the CLIP-170-tubulin complex is stable, and that exchange of CLIP-170 off of dimeric tubulin is insignificant during the lifetime of the GTP cap. This kinetic analysis argues strongly that cap recognition does not play a major role in plus-end loading and indicates that preassociation and copolymerization is the primary mechanism used by CLIP-170 to bind microtubule plus-ends.
Dissociation of CLIP-170 from Microtubules
Plus-end loading is only half of the plus-end tracking mechanism: after loading, CLIP-170 must rapidly dissociate from “older” polymer. Two mechanisms seem plausible. First, CLIP-170 could dissociate because it has an intrinsically weak affinity for older polymer (“intrinsic release” mechanism). Second, a regulatory event could weaken the affinity of CLIP-170 for microtubules, allowing CLIP-170 to dissociate (“regulated release” mechanism; Figure 1C).
The observation that CLIP-170 binds more weakly to GDP-like polymer than to GTP-like polymer (Figure 2, A and B) provides initial support for the intrinsic release hypothesis. However, the affinity of CLIP-170 for GDP-like microtubules is similar to that of other microtubule binding proteins (e.g., Butner and Kirschner, 1991). This suggests that CLIP-170 does not dissociate from microtubules simply by having “weak affinity” and that CLIP-170 should behave like other MAPs in the absence of regulation and bind the length of the microtubule. Consistent with this reasoning, Diamantopoulos et al. have observed that in vitro CLIP-170 binds along the length of preformed microtubules and also along the length of microtubules polymerized off of asters in the presence of CLIP-170 (Diamantopoulos et al., 1999). This suggests that a regulatory mechanism is involved in the dissociation of CLIP-170 from microtubules.
However, just as consideration of kinetic parameters was necessary to establish the mechanism of CLIP-170 loading, the kinetic parameters are essential to dissecting the mechanism of CLIP-170 dissociation. Given that CLIP-170 has a 10× (2.5× if fluorescence anisotropy measurements are considered) stronger affinity for dimer than for GDP polymer and that the apparent kon for dimer is 0.09 μM-1 s-1, CLIP-170 should become sequestered by dimer quickly (∼1 s) after dissociating from the microtubule. The intrinsic release model could be valid, despite the strong affinity of CLIP-170 for microtubules, if the koff is fast enough. Therefore comparison of the rate of CLIP-170 dissociation from microtubules in vivo and in vitro is necessary to support or exclude this model.
Dissociation In Vivo and In Vitro Differ by More Than an Order of Magnitude
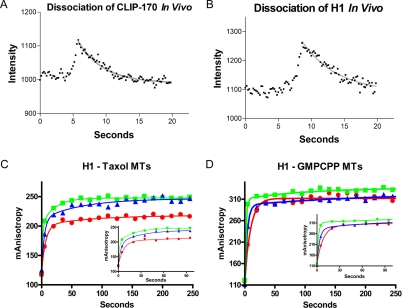
Line scan analysis of kymographs was used to measure the decay of CLIP-170 fluorescence at a given position on the microtubule over time in vivo. Single position examples of this line scan analysis for CLIP-170 and H1 are seen in Figure 6, A and B. Analysis of more than 250 positions on 25 different microtubules coming from five different cells showed that CLIP-170 dissociates from microtubules in vivo with an apparent koff of 0.3 ± 0.2 s-1. Within the errors, H1 behaves the same (koff = 0.4 ± 0.2). These data indicate that the apparent half-life of CLIP-170 on the microtubule is in vivo is ∼2 s.
Figure 6.
Binding of CLIP-170 to microtubules is characterized by slow kinetics in vitro and fast kinetics in vivo. (A and B) Linescan analysis of kymographs that were generated from the microtubules indicated in Figure 2 was used to measure the decay of CLIP-170 fluorescence (A) or H1 fluorescence (B) at a particular position on the microtubule over time. Fitting the decay as a single-phase dissociation gave koff values of 0.21 s-1 and 0.16 s-1 for both CLIP-170 and H1, respectively. Note that though these single measurements appear to give different rates, the average values obtained from the full analysis (see text) show that H1 and CLIP-170 have indistinguishable off rates in our experiments; (C and D) the rate of H1 association with microtubule polymer was measured by fluorescence anisotropy. Microtubules, 5 μM (blue), 10 μM (red), and 20 μM (green), stabilized by either Taxol (C) or GMPCPP (D) were added to 0.1 μM labeled H1. The anisotropy was measured before microtubule addition, 5 s after microtubule addition, and every 15 s thereafter; insets show the first 100 s of binding. Data sets were fit as a two phase association (see Materials and Methods for details) and analysis of this data are presented in Table 2.
Because the CLIP-170 fragments bundle microtubules, it is technically difficult to measure the rate of CLIP-170 dissociation from microtubules in vitro. Therefore, we measured kon and then calculated koff from the kon and Kd. Fluorescence anisotropy experiments similar to those used to measure the kinetics of H1 binding to tubulin (Figure 5D) were used to measure the kinetics of H1 binding to preformed microtubules. Microtubules at varying concentrations (5, 10, and 20 μM) were added to fluorescently labeled H1. The anisotropy was then measured 5 s after microtubule addition and every 15 s thereafter (Figure 6, C and D). As can be seen in Figure 6, C and D, the anisotropy rises quickly in a fast initial phase (this is expected to represent binding of CLIP-170 to microtubules), and more slowly in a second phase (the origin of this phase is unknown, but it may be related to bundling). Therefore, we fit the data to a two-phase exponential (see Materials and Methods). Because distinguishing between dissociation mechanisms requires that we set an upper limit on the dissociation constant, we used only the fast phase for our analysis of association and dissociation rates.
To extract kon from association rates in a bimolecular reaction, one needs to confirm that the conditions are “pseudo-first-order” (i.e., that the observed association rate has a linear dependence on the concentration of ligand). As can be seen in Table 2, this was true for the association of H1 with GMPCPP microtubules, resulting in an apparent kon of 0.026 μM-1 s-1. Using this number together with the affinity determined by fluorescence anisotropy gives a koff of 0.01 s-1 (half-life of dissociation = ∼66 s; Table 2). The situation for the binding of H1 to Taxol microtubules was not as straightforward: the apparent rate constant decreased as the concentration of microtubules increased (Table 2). This trend is opposite from that expected when pseudofirst order conditions are not met (i.e., when H1 molecules are competing for tubulin). We attribute this deviation to microtubule bundling. Bundling of microtubules would reduce their effective concentration and would therefore result in an artifactually slow apparent rate of association (kobserved). Taking this into consideration, we proceeded to use the fastest observed association rate (0.026 μM-1 s-1). Calculating the off rate through the Kd results in a koff of 0.017 s-1 or a half-life of 41 s. The slow rates of dissociation from the microtubule in vitro compared with the rapid rates of dissociation observed in vivo (Figure 6) suggest that some component of the cellular environment, likely regulatory, is required to facilitate CLIP-170 dissociation from the microtubule during plus-end tracking.
Table 2.
Association of CLIP-170 with microtubule polymer
| Taxol microtubules
|
GMPCPP microtubules
|
|||||
|---|---|---|---|---|---|---|
| [MT] μM | kobserved (s–1) | Apparent kon (μM–1 s–1) | t1/2 dissociation (s) | kobserved (s–1) | Apparent kon (μM–1 s–1) | t1/2 dissociation (s–1) |
| 5 | 0.13 ± 0.02 | 0.026 ± 0.004 | 41 ± 6 | 0.13 ± 0.03 | 0.026 ± 0.006 | 66 ± 17 |
| 10 | 0.22 ± 0.03 | 0.022 ± 0.003 | 53 ± 7 | 0.25 ± 0.06 | 0.025 ± 0.006 | 69 ± 19 |
| 20 | 0.31 ± 0.03 | 0.016 ± 0.005 | 63 ± 20 | 0.51 ± 0.09 | 0.026 ± 0.005 | 66 ± 15 |
The data presented in Figure 6, C and D, were fit as a two-phase exponential association (details in Materials and Methods) to determine the observed rate of association (kobserved). This value was then related to the concentration of microtubule polymer to determine the apparent kon. Finally the koff was calculated from the measured kon value and the measured Kd (Figure 5, A and B) and is presented as the t1/2 of dissociation.
DISCUSSION
We have used a combination of live cell imaging and quantitative biochemical analysis of purified proteins to address the mechanism of CLIP-170 plus-end tracking behavior. The kymograph analysis of GFP-CLIP-170 fluorescence in movies acquired at high frame rates (Figure 2) confirms that CLIP-170 tracks microtubule plus-ends by an end-loading mechanism: CLIP-170 undergoes a repeated cycle of loading at the microtubule plus-ends and dissociating from “older” areas of the polymer (Figure 1). It is important to point out that the apparent movement of CLIP-170 is an illusion, similar to that which would be observed if the GTP cap itself could be visualized. Although our experiments cannot rule out the possibility that some fraction of CLIP-170 tracks microtubule plus-ends by other mechanisms such as “hitchhiking” on motors, it is clear from the kymographs (Figure 2) that the bulk of GFP-CLIP-170 loads specifically at the microtubule end and does not move.
To dissect the specific end-loading mechanism, we quantitatively analyzed the interactions between CLIP-170 and different conformations of polymerized and unpolymerized tubulin. Our experiments lead to the following conclusions. First, CLIP-170 has strong affinity for tubulin dimer (30 nM as determined by SPR; ∼0.26 μM by fluorescence anisotropy), indicating that CLIP-170 should be considered a tubulin-binding protein. Second, CLIP-170 has strong (but weaker) affinity for GTP- and GDP-like microtubule polymer (∼0.40 and 0.76 μM, respectively, as determined by fluorescence anisotropy; 0.2 and ∼0.5 μM, respectively, by cosedimentation assays). The affinity for GDP-like microtubules is similar to that observed for bona fide microtubule-binding proteins and is consistent with previous observations that CLIP-170 binds preformed microtubules (Rickard and Kreis, 1991; Diamantopoulos et al., 1999). Although the absolute affinities obtained by fluorescence anisotropy were weaker than those determined by SPR (see discussion of this point below), the relative affinities were similar: CLIP-170 binds tightly to unpolymerized tubulin, and more weakly (but still firmly) to microtubule polymer.
These relative affinities imply that CLIP-170 loads specifically at the microtubule plus-end by preassociating with soluble tubulin and copolymerizing. They initially suggest that CLIP-170 dissociates because of its relatively lower affinity for microtubule polymer. However, although equilibrium-based affinity values are useful to understand CLIP-170 behavior, they are not sufficient: knowledge of kinetic rate constants is essential to dissecting the plus-end tracking mechanism because the microtubule cytoskeleton is not an equilibrium system. Our experiments indicate that the intrinsic rate constants governing interactions between CLIP-170, tubulin, and microtubules are all slow: in vitro, the half-life of CLIP-170 dissociation from GTP tubulin is ≥170 s (Figure 4D), and the half-life of dissociation from GDP-like polymer in vitro is ∼40 s (Figure 6C, Table 2). In contrast, the dissociation of CLIP-170 from microtubules in vivo is fast (half-life ∼2 s, Figure 6, A and B). The large difference between the dissociation rate in vitro and in vivo indicates that cellular factors, likely regulatory, are involved in the dissociation phase of CLIP-170 plus-end tracking behavior.
These kinetic constants are important for an additional reason: One could argue that GMPCPP microtubules are a poor mimic of the “real” cap conformation and that CLIP-170 tracks plus-ends by having an even stronger affinity for this hypothetical conformation. However, the koff for the CLIP-170-tubulin interaction suggests that CLIP-170 exchanges off of tubulin too slowly to interact significantly with the short-lived cap conformation, regardless of its exact nature.
The sum of these observations suggests that the following is the predominant mechanism for CLIP-170 plus-end tracking behavior in human cells: CLIP-170 associates tightly and stably with unpolymerized tubulin, it copolymerizes with tubulin, and then it dissociates rapidly from the microtubule polymer in a process facilitated by cellular factors. After dissociation, CLIP-170 binds again to free tubulin, and the cycle repeats (Figure 7).
Figure 7.
Model for the plus-end tracking mechanism of CLIP-170. (1) CLIP-170 binds dimeric tubulin; (2) CLIP-170 copolymerizes onto the growing microtubule plus-end; (3) CLIP-170 remains bound to the microtubule without moving toward either the plus-end or the minus end; (4) CLIP-170 dissociates from the microtubule with the help of other cellular factors.
We speculate that the cellular factors that promote CLIP-170 dissociation are kinases because it has already been established that phosphorylation can reduce the ability of CLIP-170 to cosediment with taxol microtubules (Rickard and Kreis, 1991). Moreover, phosphorylation has been implicated in the plus-end tracking mechanism of dynactin p150, a +TIP that contains a microtubule-binding domain related to that of CLIP-170 (Vaughan et al., 2002).
Our work argues strongly against the hypothesis that cap recognition contributes significantly to CLIP-170 plus-end tracking behavior, but it does leave open the possibility that other mechanisms, such as “hitchhiking” on motors or binding to other plus-end tracking proteins, could contribute to plus-end tracking behavior. One might contend that overexpression leads to an enhanced end-loading signal that obscures the signal of CLIP-170 tracking by these other mechanisms. While this is possible, the strength and stability of the CLIP-170-tubulin interaction suggests that preassociation and copolymerization are behaviors intrinsic to the CLIP-170 microtubule binding domain, and that these behaviors will be dominant unless the tubulin/microtubule binding domain is down-regulated.
Comparison of Different Methods for Measuring Interactions between CLIP-170 and Tubulin
Because accuracy of the affinity and kinetic measurements is essential to the validity of our conclusions, the binding of CLIP-170-H1 to each conformation of tubulin/microtubules was measured by two independent methods. Binding to microtubule polymer was measured by cosedimentation, binding to dimer was measured by SPR, and fluorescence anisotropy was used as a single method by which we could measure the binding of H1 to both tubulin and microtubule polymer. Fluorescence anisotropy produced values similar to the other methods used with one significant exception: the affinities and rate constants for binding to unpolymerized tubulin.
One explanation for this discrepancy is that the binding site of H1 is affected by the addition of the fluorescent label. However, the binding to polymer is similar by cosedimentation and fluorescence anisotropy, arguing against this possibility. The fact that different analyses were conducted at different temperatures may also contribute: SPR was performed at 10°C, cosedimentation experiments at 37°C, and fluorescence anisotropy at 25°C. Temperature dependence of these interactions is a subject for future investigation.
Another explanation, one that we favor, is that this discrepancy is due to tubulin oligomerization in the fluorescence anisotropy experiments. It is well established that CLIP-170 induces oligomerization of tubulin (Diamantopoulos et al., 1999; Arnal et al., 2004). The SPR experiments were conducted under conditions that reduce or prevent oligomerization (cool temperatures, coupling of H1 to the surface, low coupling levels). In contrast, both tubulin and CLIP-170 were free to participate in supramolecular assemblies in the fluorescence anisotropy experiments. This oligomerization could lead to artifactually weak affinities in two ways. First, oligomerization will reduce the concentration of tubulin available to H1 below that which is expected. Second, and more interesting, is the possibility that oligomeric tubulin may adopt a conformation similar to microtubule polymer or intermediate between dimer and polymer. In support of this idea, CLIP-170 dissociates from tubulin dimer with kinetics similar to its dissociation from microtubule polymer when measurements are performed by fluorescence anisotropy. Conversely, in SPR experiments where precautions have been taken to ensure that tubulin remains in a dimeric state, the dissociation of the CLIP-170-tubulin complex is much slower. An additional piece of evidence supporting this explanation is that EB1 binding to tubulin is consistent whether measured by fluorescence anisotropy or SPR (Folker and Goodson, unpublished results). Therefore it seems likely that tubulin oligomerization is complicating the fluorescence anisotropy measurements and that the SPR measurements reflect the true characteristics of the CLIP-170-tubulin dimer interaction.
The preassociation, copolymerization, and regulated release mechanism is consistent with previous fluorescence microscope characterization of interactions between CLIP-170 and microtubules in vitro. These experiments showed that when CLIP-170 is added to a system of dynamically polymerizing microtubules, it associates specifically with newly polymerized microtubules, but exhibits no preference for the plus-ends of these microtubules. Similarly, CLIP-170 binds along the length of preformed microtubules, showing no preference for ends (Diamantopoulos et al., 1999). The sum of these observations indicate that the plus-end tracking activity of CLIP-170 should be observed only when two conditions are met: 1) free tubulin is present and can polymerize dynamically onto microtubule plus-ends; and 2) a regulatory system is present facilitating CLIP-170 dissociation from older regions of the polymer. Consistent with this prediction, it has not been possible to observe CLIP-170 plus-end tracking behavior with purified systems of dynamic microtubules in vitro, although negative evidence must always be interpreted with caution (Diamantopoulos et al., 1999; Folker and Goodson, unpublished results).
Comparison between CLIP-170 and Its Relatives Other Systems
Although our evidence indicates that CLIP-170 tracks microtubule plus-ends by end-loading and does so by virtue of its own interactions with tubulin and microtubules, genetic and cell biological experiments suggest that CLIP-170 relatives in yeast (S. pombe and S. cerevisiae) move on microtubules via kinesin motors (Busch and Brunner, 2004; Busch et al., 2004; Carvalho et al., 2004) and may require EB1 homologues to bind microtubules effectively(Butner and Kirschner, 1991; Busch et al., 2004). Why would orthologous yeast and vertebrate proteins differ in such fundamental ways?
One answer is that even though CLIP-170, Bik1, and Tip1p appear to be orthologues, they have significant structural differences that may reflect fundamental functional differences. Most obviously, CLIP-170 and its metazoan orthologues have two CAP-Gly domains, whereas relatives from single-celled organisms sequenced to date have only one (Gregoretti and Goodson, unpublished results). Alternatively, the yeast and vertebrate proteins may not be as different as they seem. The web of interactions between proteins at microtubule plus-ends is complex. Perhaps CLIP-170 and its fungal relatives engage in a set of common interactions, but the interactions that are dominant differ between organisms. In light of this reasoning, is interesting to consider the possibility that CLIP-170 interacts with kinesin superfamily proteins and EB1. Indeed, evidence already exists for a direct interaction between CLIP-170 and EB1 (Goodson et al., 2003; Folker and Goodson, unpublished results). Finally, it is important to remember that tubulin itself has very different properties in mammalian and yeast cells. Yeast microtubules grow at one tenth the speed of mammalian microtubules, contain significantly more GTP tubulin, and are (by virtue of cell size) much shorter (Dougherty et al., 1998, 2001). Perhaps these differences in microtubule dynamics dictate differences in the necessary functional characteristics of microtubule plus-end tracking proteins.
Relationship between the CLIP-170 Plus-End Tracking Mechanism and CLIP-170 Function
Understanding how CLIP-170 achieves its dynamic localization to microtubule plus-ends is critical to elucidating its function. For example, it has been suggested that +TIP behavior can be a mechanism for delivering proteins to the cell cortex (Schuyler and Pellman, 2001; Maekawa et al., 2003; Busch et al., 2004). This is correct only if there is net transport of the protein in question. Instead, as discussed above, the bulk of CLIP-170 appears to remain stationary with respect to the microtubule. If CLIP-170 plus-end tracking does not function to deliver CLIP-170 to targets such as the cell cortex, then what is the function of this behavior, and how does it relate to the function of CLIP-170? This question remains unresolved, in part because the complex network of interactions between plus-end tracking proteins is only now becoming apparent (Schuyler and Pellman, 2001; Carvalho et al., 2003; Vaughan, 2004). CLIP-170 function may be meaningful only in terms of its interactions with other members of the network.
However, one important observation is that CLIP-170 promotes microtubule growth both in vivo and in vitro (Diamantopoulos et al., 1999; Komarova et al., 2002). The mechanism of growth promotion is under investigation, but the observation that CLIP-170 has stronger affinity for GTP-like than GDP-like microtubules is likely significant. Additionally, previous analyses have indicated that CLIP-170 associates with and increases the concentration of tubulin oligomers (Diamantopoulos et al., 1999; Arnal et al., 2004). These observations lead to the intriguing speculation that CLIP-170 stabilizes the GTP conformation, cross-links protofilaments, or perhaps promotes the addition of a preformed cap. These activities, which are not mutually exclusive, provide attractive explanations for the ability of CLIP-170 and its relatives to promote processive microtubule growth (Brunner and Nurse, 2000; Komarova et al., 2002; Carvalho et al., 2004).
Supplementary Material
Acknowledgments
We thank Elena Lastochkin for careful preparation of tubulin and CLIP-170 fragments, Jose Chaverri for help with cloning, and Becky Davis for training in use of the Biacore machine. We are grateful to Yulia Komarova, Kevin Vaughan, Crislyn D'Souza-Schorey, Mary Prorok, and Gergana Ugrinova and members of the Goodson laboratory for insightful discussions and comments on the manuscript. We also thank the anonymous reviewers for their detailed and constructive criticism. This work was supported by the following grants to H.V.G.: a Scientist Development grant from the American Heart Association, and 1R01 GM065420 from the National Institutes of Health.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-12-1106) on August 24, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
Footnotes
Although regions that started out dim normally remained so, it is interesting to note that bright spots sometimes fluctuated in fluorescence intensity. These variations may represent binding of CLIP-170 to other CLIP-170 molecules as can occur via CLIP-170 head-tail interactions (Lansbergen et al., 2004).
It is interesting to note that in the SPR experiments the “rapid phase” of the dissociation of CLIP-170 from GTP tubulin (which was in fact very slow) was similar to the single phase dissociation of GDP tubulin from CLIP-170 (the half-lives were 170 and 190 s, Figure 4D). This similarity, together with the observation that most of the GTP tubulin does not dissociate during the time course of the experiment, suggests that the rapid phase of “GTP tubulin dissociation” may in fact represent the dissociation of tubulin that has hydrolyzed its GTP to GDP. This issue requires further investigation, but it does not alter our conclusion that the dissociation of CLIP-170 from both tubulin and microtubules is slow.
References
- Arnal, I., Heichette, C., Diamantopoulos, G. S., and Chretien, D. (2004). CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 14, 2086-2095. [DOI] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695-704. [DOI] [PubMed] [Google Scholar]
- Bulinski, J. C., Odde, D. J., Howell, B. J., Salmon, T. D., and Waterman-Storer, C. M. (2001). Rapid dynamics of the microtubule binding of ensconsin in vivo. J. Cell Sci. 114, 3885-3897. [DOI] [PubMed] [Google Scholar]
- Busch, K. E., and Brunner, D. (2004). The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14, 548-559. [DOI] [PubMed] [Google Scholar]
- Busch, K. E., Hayles, J., Nurse, P., and Brunner, D. (2004). Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev. Cell 6, 831-843. [DOI] [PubMed] [Google Scholar]
- Butner, K. A., and Kirschner, M. W. (1991). Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115, 717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, P., Gupta, M. L., Jr., Hoyt, M. A., and Pellman, D. (2004). Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell 6, 815-829. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., Tirnauer, J. S., and Pellman, D. (2003). Surfing on microtubule ends. Trends Cell Biol. 13, 229-237. [DOI] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T. J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117. [DOI] [PubMed] [Google Scholar]
- Diamantopoulos, G. S., Perez, F., Goodson, H. V., Batelier, G., Melki, R., Kreis, T. E., and Rickard, J. E. (1999). Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 144, 99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty, C. A., Himes, R. H., Wilson, L., and Farrell, K. W. (1998). Detection of GTP and Pi in wild-type and mutated yeast microtubules: implications for the role of the GTP/GDP-Pi cap in microtubule dynamics. Biochemistry 37, 10861-10865. [DOI] [PubMed] [Google Scholar]
- Dougherty, C. A., Sage, C. R., Davis, A., and Farrell, K. W. (2001). Mutation in the beta-tubulin signature motif suppresses microtubule GTPase activity and dynamics, and slows mitosis. Biochemistry 40, 15725-15732. [DOI] [PubMed] [Google Scholar]
- Faulkner, N. E., Dujardin, D. L., Tai, C. Y., Vaughan, K. T., O'Connell, C. B., Wang, Y., and Vallee, R. B. (2000). A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2, 784-791. [DOI] [PubMed] [Google Scholar]
- Goodson, H. V., Skube, S. B., Stalder, R., Valetti, C., Kreis, T. E., Morrison, E. E., and Schroer, T. A. (2003). CLIP-170 interacts with dynactin complex and the APC-binding protein EB1 by different mechanisms. Cell Motil. Cytoskelet. 55, 156-173. [DOI] [PubMed] [Google Scholar]
- Griparic, L., and Keller, T. C., 3rd. (1999). Differential usage of two 5′ splice sites in a complex exon generates additional protein sequence complexity in chicken CLIP-170 isoforms. Biochim. Biophys. Acta 1449, 119-124. [DOI] [PubMed] [Google Scholar]
- Gundersen, G. G., Gomes, E. R., and Wen, Y. (2004). Cortical control of microtubule stability and polarization. Curr. Opin. Cell Biol. 16, 106-112. [DOI] [PubMed] [Google Scholar]
- Hill, T. L., and Carlier, M. F. (1983). Steady-state theory of the interference of GTP hydrolysis in the mechanism of microtubule assembly. Proc. Natl. Acad. Sci. USA 80, 7234-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, G., and Weber, K. (1978). Radioimmunoassay for tubulin: a quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell 14, 795-804. [DOI] [PubMed] [Google Scholar]
- Hyman, A., Drechsel, D., Kellogg, D., Salser, S., Sawin, K., Steffen, P., Wordeman, L., and Mitchison, T. (1991). Preparation of modified tubulins. Methods Enzymol. 196, 478-485. [DOI] [PubMed] [Google Scholar]
- Hyman, A. A., Salser, S., Drechsel, D. N., Unwin, N., and Mitchison, T. J. (1992). Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolyzable analogue, GMPCPP. Mol. Biol. Cell 3, 1155-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, K. B., Burds, A. A., Swedlow, J. R., Bekir, S. S., Sorger, P. K., and Nathke, I. S. (2001). A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat. Cell Biol. 3, 429-432. [DOI] [PubMed] [Google Scholar]
- Kirschner, M., and Mitchison, T. (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell 45, 329-342. [DOI] [PubMed] [Google Scholar]
- Komarova, Y. A., Akhmanova, A. S., Kojima, S., Galjart, N., and Borisy, G. G. (2002). Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159, 589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen, G., Komarova, Y., Modesti, M., Hoogenraad, C. C., Goodson, H. V., Grosveld, F., Galjart, N., Borisy, G. G., and Akhmanova, A. (2004). Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J. Cell Biol. 166, 1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos, D., Kusch, J., Grava, S., Vogel, J., and Barral, Y. (2003). Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112, 561-574. [DOI] [PubMed] [Google Scholar]
- Maekawa, H., Usui, T., Knop, M., and Schiebel, E. (2003). Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22, 438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, E. E., Wardleworth, B. N., Askham, J. M., Markham, A. F., and Meredith, D. M. (1998). EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene 17, 3471-3477. [DOI] [PubMed] [Google Scholar]
- Perez, F., Diamantopoulos, G. S., Stalder, R., and Kreis, T. E. (1999). CLIP-170 highlights growing microtubule ends in vivo. Cell 96, 517-527. [DOI] [PubMed] [Google Scholar]
- Pierre, P., Scheel, J., Rickard, J. E., and Kreis, T. E. (1992). CLIP-170 links endocytic vesicles to microtubules. Cell 70, 887-900. [DOI] [PubMed] [Google Scholar]
- Rickard, J. E., and Kreis, T. E. (1990). Identification of a novel nucleotide-sensitive microtubule-binding protein in HeLa cells. J. Cell Biol. 110, 1623-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard, J. E., and Kreis, T. E. (1991). Binding of pp170 to microtubules is regulated by phosphorylation. J. Biol. Chem. 266, 17597-17605. [PubMed] [Google Scholar]
- Rodionov, V., Nadezhdina, E., and Borisy, G. (1999). Centrosomal control of microtubule dynamics. Proc. Natl. Acad. Sci. USA 96, 115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel, J., Pierre, P., Rickard, J. E., Diamantopoulos, G. S., Valetti, C., van der Goot, F. G., Haner, M., Aebi, U., and Kreis, T. E. (1999). Purification and analysis of authentic CLIP-170 and recombinant fragments. J. Biol. Chem. 274, 25883-25891. [DOI] [PubMed] [Google Scholar]
- Schuyler, S. C., and Pellman, D. (2001). Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell 105, 421-424. [DOI] [PubMed] [Google Scholar]
- Severin, F. F., Sorger, P. K., and Hyman, A. A. (1997). Kinetochores distinguish GTP from GDP forms of the microtubule lattice. Nature 388, 888-891. [DOI] [PubMed] [Google Scholar]
- Terpetschnig, E., Szmacinski, H., Malak, H., and Lakowicz, J. R. (1995). Metal-ligand complexes as a new class of long-lived fluorophores for protein hydrodynamics. Biophys. J. 68, 342-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, K. T. (2004). Surfing, regulating and capturing: are all microtubule-tip-tracking proteins created equal? Trends Cell Biol. 14, 491-496. [DOI] [PubMed] [Google Scholar]
- Vaughan, K. T., Tynan, S. H., Faulkner, N. E., Echeverri, C. J., and Vallee, R. B. (1999). Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J. Cell Sci. 112(Pt 10), 1437-1447. [DOI] [PubMed] [Google Scholar]
- Vaughan, P. S., Miura, P., Henderson, M., Byrne, B., and Vaughan, K. T. (2002). A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J. Cell Biol. 158, 305-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.