Abstract
Hypersensitive response (HR) is a programmed cell death that is commonly associated with disease resistance in plants. Among the different HR-related early induced genes, the AtMYB30 gene is specifically, rapidly, and transiently expressed during incompatible interactions between Arabidopsis and bacterial pathogens. Its expression was also shown to be deregulated in Arabidopsis mutants affected in the control of cell death initiation. Here, we demonstrate that overexpression in Arabidopsis and tobacco of AtMYB30 (i) accelerates and intensifies the appearance of the HR in response to different avirulent bacterial pathogens, (ii) causes HR-like responses to virulent strains, and (iii) increases resistance against different bacterial pathogens, and a virulent biotrophic fungal pathogen, Cercospora nicotianae. In antisense AtMYB30 Arabidopsis lines, HR cell death is strongly decreased or suppressed in response to avirulent bacterial strains, resistance against different bacterial pathogens decreased, and the expression of HR- and defense-related genes was altered. Taken together, these results strongly suggest that AtMYB30 is a positive regulator of hypersensitive cell death.
In response to pathogen attack, plants have developed complex signaling and defense mechanisms to protect themselves. One of the most efficient and immediate resistance reactions is the hypersensitive response (HR), which is characterized by the rapid death of the plant cells directly in contact with, or in close proximity to, the pathogen. The HR thus confines the pathogen by stopping it from spreading from the site of the attempted infection. Our understanding of the molecular mechanisms underlying the HR is progressing rapidly (1). Numerous data suggest that the HR is a form of programmed cell death, as its initiation depends on active plant metabolism (2, 3) and can be triggered by elicitor molecules from bacteria or fungi (4–6). The overexpression of certain transgenes in plants can lead to the appearance of necrotic lesions resembling those observed during an HR (7, 8).‖ In addition, a number of plant mutants develop spontaneous necrotic lesions in the absence of any pathogen attack and other hallmarks of the HR, such as the expression of pathogenesis-related (PR) genes and/or an enhanced resistance to pathogen infection (9–11). However, the nature of many of the components involved in the hypersensitive cell death program remains a black box.
Genetic analysis of lesion mimic mutants has permitted the identification of certain actors and/or regulators of this program, including the LSD1 (lesion-simulating disease resistance 1) gene, which encodes a putative zinc finger protein (12). We have previously identified genes that are specifically expressed during the HR in tobacco (13) and Arabidopsis (14) and shown for one of them a role of negative regulator of the HR (15). Among them, an R2R3 MYB-related gene in Arabidopsis thaliana, AtMYB30, was isolated on the basis of its specific, rapid, and transient transcriptional activation during the very first steps of the HR in response to the avirulent strain 147 of Xanthomonas campestris pv. campestris and to other bacterial pathogens (16). The use of Arabidopsis lsd mutants (10) and their corresponding suppressor phx (phoenix) mutants (17) showed that AtMYB30 expression is closely associated with the initiation of hypersensitive cell death, rather than its propagation (16). AtMYB30 may thus be a component of a cell death pathway conditioning the HR. However, the exact role of this putative transcriptional factor remained unknown. Here, by the analysis of transgenic plants expressing sense or antisense AtMYB30 transcripts constitutively, we show that overexpression of AtMYB30 accelerates the appearance of the HR in response to avirulent bacterial pathogens and causes HR-like responses to virulent bacterial pathogens. In addition, it increases resistance against different bacterial pathogens and a biotrophic fungal pathogen, Cercospora nicotianae. In antisense AtMYB30 Arabidopsis lines, HR cell death is strongly decreased or suppressed in response to an avirulent pathogen. We propose that AtMYB30 is a positive regulator of the programmed cell death associated with plant disease resistance.
Materials and Methods
DNA Constructions.
The coding region of AtMYB30 (16) was cloned into a derivative of pBI221 (CLONTECH), which contained the cauliflower mosaic virus 35S promoter fragment. The recombinant plasmids were named pBIM131 for sense constructions, pBIW13 for antisense constructions, and pBI111 for the control vector.
Plant Growth Conditions and Transformation.
A. thaliana plants, accession Wassilewskaija (Ws-4), were used in these experiments and transformed according to Bechtold et al. (18). The cultivar of Nicotiana tabacum L. used in this study was Bottom Special. Plant growth conditions, transformation, selection of the transformants, and determination of the T2 generation genotype were performed as described by Pontier et al. (13). These transgenic plants were in all respects phenotypically similar to untransformed Arabidopsis and tobacco plants, except during senescence (data not shown).
Pathogen Inoculation of Transgenic Plants.
All of the infection experiments were performed on kanamycin-resistant T2 plants, as described (13, 16). For C. nicotianae, 8-week-old tobacco plants were inoculated locally on the leaves with a syringe, with 100 μl of a suspension containing 2.4 × 105 spores per ml.
Quantification of the Transcript Levels.
Total RNA was isolated from leaves, as described by Lacomme and Roby (19). cDNAs were synthesized by using the SuperScript II RT system of Life Technologies (Cergy Pontoise, France). For reverse transcription–PCR, β-tubulin was used as internal constitutive control. For specific AtMYB30 cDNA, two primers were used during the same PCR (5′-GGCGAAAAAGGCTCTCTCTG and 5′-ATCACCAATCTGTCCACCAG). All of the PCRs were performed on Robocycler gradient 96 (Stratagene). Experiments were carried out in duplicate, in two independent experiments.
Detection of Cell Death.
Cell death was quantified by monitoring the uptake of Evans blue by leaf discs from inoculated or healthy plants, as described by Baker and Mock (20). The assay was performed with six leaf discs (1.5 cm2) punched out with a cork borer from the inoculated zones of 2–3 plants. Three replicates per point were performed.
Lipid Peroxydation Analysis.
Hydroxy and hydroperoxy fatty acids and components derived from the lipid peroxydation metabolism were analyzed as described by Rustérucci et al. (21). In the HPLC analyses, the 12- and 16-hydroxy isomers of 18:3 [12-hydroxy-9,13,15,(Z,E,Z)-octadecatrienoic acid and 16-hydroxy-9,12,14,(Z,Z,E)-octadecatrienoic acid, respectively] were reassigned in the present work, using authentic samples (a gift from I. Feussner, Institute for Plant Genetics and Crop Plant Research, Gatersleben, Germany).
β-Glucuronidase (GUS) Activity.
GUS activity was measured as described (22).
RNA Gel Blot Analysis.
Total RNA was isolated from leaves and Northern analysis was performed as described by Lummerzheim et al. (23). DNA fragments corresponding to the full-length cDNA clones of hypersensitivity-related gene 203 (hsr203), hsr515, PR-1a, and PR-5 in the case of tobacco, Athsr3 and PR-1 in the case of Arabidopsis, or the ribosomal intergenic spacer (24) of rDNA, were used as probes. Transcript levels for each gene were quantified by scanning the RNA gel blot and measuring the relative intensity of the corresponding bands with imagequant software (Molecular Dynamics).
Results
Overexpression and Antisense-Mediated Depletion of AtMYB30 Lead to Altered Responses to Avirulent and Virulent Pathogen Attack.
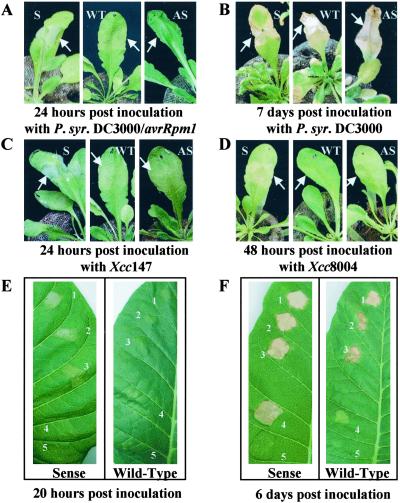
To address the possible role of AtMYB30 in the HR, we examined the effects of overexpression and antisense-mediated depletion of this MYB-like gene in Arabidopsis and tobacco, an heterologous system and an amenable model for molecular and biochemical study of plant–pathogen interactions. We obtained several transgenic Arabidopsis and tobacco plants expressing AtMYB30 either in sense orientation (pBIM131) or antisense orientation (pBIW13), or containing the vector (pBI111). The phenotype of these lines was visually compared with that of the wild type, after inoculation with either avirulent or virulent bacterial pathogens. When challenged with Pseudomonas syringae pv. tomato strain DC3000(avrRpm1) (DC3000/avrRpm1) or X. campestris pv. campestris strain 147 (Xcc147), Arabidopsis wild-type plants show a typical HR, characterized by the rapid (24–48 h under our conditions) collapse and drying of the inoculated tissues (Fig. 1 A and C). The same observation can be made in tobacco in response to an 5.107 colony-forming unit (cfu)/ml inoculation of Ralstonia solanacearum GMI1000 strain. This response is less intense and/or less rapid when the inoculum is reduced (Fig. 1F). In sense lines and antisense lines (Fig. 1 A and C), the appearance of the HR symptoms was accelerated and retarded, respectively. In addition, an intensification in the sense lines was clearly seen in tobacco for inocula under 5.107 cfu/ml. These observations were repeatedly made for 4–6 representative transgenic lines (data not shown) when compared alongside wild-type and control lines.
Figure 1.
Phenotypes of AtMYB30 sense and antisense Arabidopsis and tobacco lines in response to inoculation with avirulent or virulent bacteria. Symptoms observed in Arabidopsis sense, wild-type, and antisense lines: (A) 24 h postinoculation with DC3000/avrRpm1 (107 cfu/ml), (B) 7 days postinoculation with DC3000 (107 cfu/ml), (C) 24 h postinoculation with Xcc147 (108 cfu/ml), and (D) 48 h postinoculation with Xcc8004 (108 cfu/ml). (E and F) Phenotypes of tobacco transgenic lines in response to inoculation with R. solanacearum. Different inocula were used for the avirulent strain (GMI1000), ranging from 5.107 cfu/ml (no. 1), 107 cfu/ml (no. 2), to 5.106 cfu/ml (no. 3); the virulent strain (K60) was inoculated at 5.107 cfu/ml (no. 4), and a control inoculation was performed with water (no. 5).
Similar experiments were undertaken with virulent strains of the same pathogens. In the wild-type plants, a spreading chlorosis on the leaf, starting from the inoculated zone 2–3 days after inoculation was observed in Arabidopsis (Fig. 1 B and D) and tobacco (Fig. 1F). Surprisingly, sense lines showed HR-like responses, and these symptoms did not spread out of the inoculated zone (Fig. 1 B, D, and E). In the case of the antisense lines, no significant differences could be observed with the wild-type or control lines.
AtMYB30 Is a Positive Regulator of Cell Death Occurring in Response to Avirulent and Virulent Pathogen Attack.
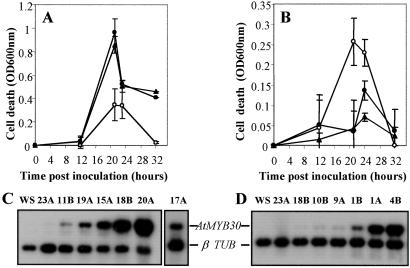
To address the relationship between AtMYB30 overexpression and HR cell death after bacterial inoculation, the exact timing and appearance of lethality of the inoculated cells in control or transformed plants were monitored with the Evans blue leaf disk assay (20). These data are reported in Fig. 2, together with an evaluation by reverse transcription–PCR of the expression level of the transgenes in these lines. When challenged with an avirulent strain of P. syringae, cells of the wild-type line located at the inoculation site show an increase of Evans blue uptake 12–24 h after inoculation, indicative of cell death (Fig. 2A). The dye being no longer retained after tissue collapse (20), cell death rate decreases between 24 and 30 h after inoculation and is hardly detectable 32 h after inoculation. In the overexpressing lines, a strikingly different pattern of the accumulation of the Evans blue was observed, the cell death rate being amplified as much as three times over the control and the process being accelerated. In contrast, in the antisense lines, accumulation of the Evans blue is hardly detected (Fig. 2B). These results, together with the evaluation of transgene transcript levels in the transgenic lines, are consistent with a direct role for AtMYB30 in promoting the HR phenotype. They are also in agreement with previous data indicating that AtMYB30 is expressed at very low levels (16), because sense or antisense lines expressing low levels of AtMYB30 present a clear altered phenotype. Similar data were obtained in Arabidopsis and tobacco, in response to other pathogens (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org).
Figure 2.
Effect of overexpression and antisense depletion of AtMYB30 on cell death occurring in response to bacterial avirulent pathogen attack. (A and B) Evaluation of cell death in transgenic Arabidopsis leaves after inoculation with the avirulent strain DC3000/avrRpm1 of P. syringae at 107 cfu/ml. Uptake of Evans blue by leaves from wild-type plants (○), sense transgenic plants (A) (pBIM131–20A, ●; pBIM131–17A, ▴) and antisense transgenic plants (B) (pBIW13–1A, ●; pBIW13–1B, ▴). Data are expressed as OD units, and means and SD from three replicates in two independent experiments are given. (C and D) Reverse transcription–PCR analysis of AtMYB30 and β-tubulin mRNA accumulation in plant extracts from wild-type (WS) plants, control plants (23A), sense lines (11B, 19A, 15A, 18B, 20A, and 17A), and antisense lines (18B, 10B, 9A, 1B, 1A, and 4B).
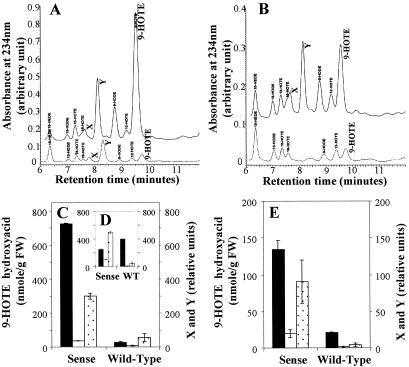
Besides a quantitative evaluation of cell death in transgenic AtMYB30 lines, the analysis of typical biochemical cell death markers in plants, such as oxylipins, might help to determine the nature of cell death that is modulated in these lines. Recently, the HR in cryptogein-elicited tobacco leaves was shown to depend on an intense 9S-LOX (lipoxygenase) metabolism, and the products of this metabolism, i.e., free fatty acid hydroperoxides, were proposed to participate in the execution of HR cell death (21). In this context, HPLC analysis of hydroxy fatty acids obtained after NaBH4 reductive extraction and NaOH hydrolysis of total lipids was conducted on the sense line pBIM131–40A and compared with the wild type, in response to avirulent and virulent strains of R. solanacearum. Typical chromatograms of hydroxy fatty acids are shown in Fig. 3 A and B. As previously reported, all positional isomers of 18:2 and 18:3 fatty acids were identified and quantified with reference to an internal standard, 15-hydroxy-11,13(Z,E) eicosadienoic acid, the major isomers being 9-hydroxy-10,12(Z,E) octadecadienoic acid (9-HODE) and 9-hydroxy-10,12,15(E,Z,Z) octadecatrienoic acid (9-HOTE). In addition, two early eluting-compounds, not identified as hydroxy fatty acids and appearing only in extracts from leaves undergoing HR, designated X and Y (21), were detected. After inoculation with the HR-inducing pathogen, the two profiles are similar (Fig. 3A), but in the sense line, lipid peroxidation increased markedly in the early time points (22 h after inoculation), as compared with the wild type. This finding is especially clear for the 9-HOTE isomer and the X and Y compounds (Fig. 3C). At later time points (72 h), the difference between the two lines is attenuated for all of the isomers, including the 9-HOTE isomer, X and Y levels remaining higher in the sense line (Fig. 3D). In contrast, in response to the virulent pathogen, the HPLC chromatographic profiles obtained from the sense line versus the wild type are clearly different (Fig. 3B). While a chromatogram typical of a compatible interaction (low rates of lipid peroxidation, no preeminence of 9-HODE and 9-HOTE, absence of X and Y compounds) was observed for the wild type whatever the stage of the infection process, the sense line exhibited a profile very similar to that observed during an HR (high lipid peroxidation, preeminence of 9-HODE and 9-HOTE, detection of X and Y compounds) (Fig. 3E). This profile is observed specifically at the appearance of the lesions, 72 h after inoculation. These data are consistent with the HR-like phenotype previously observed for the sense lines in response to virulent pathogens.
Figure 3.
Effect of overexpression of AtMYB30 on hydroxy fatty acid production in response to bacterial pathogen attack. HPLC analysis of hydroxy fatty acids extracted from wild-type and sense transgenic plants (pBIM131–40A). (A and B) HPLC traces of wild-type leaf extracts (Lower) and sense transgenic leaf extracts (Upper), 22 h after inoculation with an avirulent strain (5.107 cfu/ml) (A) or 76 h after inoculation with a virulent strain (5.107 cfu/ml) (B) of R. solanacearum. The various hydroxy fatty acids isomers of 18:2 and 18:3 fatty acids were identified as described (21) and quantified with reference to the internal standard 15-hydroxy-11,13(Z,E) eicosadienoic acid. (C–E). Changes in 9-HOTE (filled column), X (empty column), and Y (dotted column) compound levels of sense and wild-type tobacco leaves 22 h (C) and 72 h (D) after inoculation with an avirulent strain, or 76 h after inoculation with a virulent strain (E) of R. solanacearum. Mean and SD from three replicates of the same experiment are given.
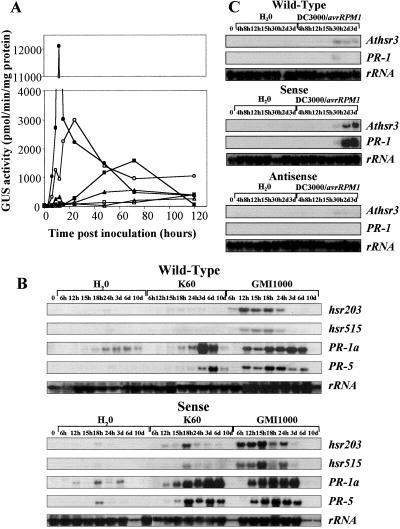
Another way to elucidate the basis for the observed regulation of HR cell death by AtMYB30 is to examine the expression of genes that are either closely associated with HR, such as the hsr genes (13, 26), or related to resistance, such as PR genes (27, 28). For this purpose, we generated a cross between one of the tobacco sense lines (pBIM131–40A) and a reference line expressing a transcriptional fusion between the hsr203J promoter and the GUS reporter gene. GUS activity was then measured after inoculation with avirulent and virulent strains of R. solanacearum (Fig. 4A). In the reference line, the hsr203J gene shows an expression pattern specific for the incompatible interaction, the expression being maximal in response to the HR-inducing strain, early and transient. In the line resulting from the cross, the hsr203J promoter activation was about four times higher and could be detected earlier, 6 h postinoculation with the avirulent strain. In the compatible interaction with the virulent strain, the expression of the hsr203J gene was also increased as compared with the reference line, again consistent with the HR-like phenotype displayed by the sense lines in response to virulent pathogen.
Figure 4.
Effects of AtMYB30 overexpression on hsr and defense-related gene expression. (A) Time course of hsr203J promoter activation in transgenic tobacco plants resulting (or not, control) from a cross (F2 plants whose genotype has been determined) between an AtMYB30 sense line (pBIM131–40A) and a line containing the hsr203J promoter-GUS fusion (13), after inoculation with different isolates of R. solanacearum (5.107 cfu/ml) or water. GUS activity was measured in extracts of two leaves (equivalent of four replicates) after inoculation with the avirulent strain (○), virulent strain (□), and water (▵) from the control line (open symbols) or the F2 line (filled symbols). (B and C) Alterations in the expression of hsr genes and PR genes in transgenic sense and control tobacco plants (B) and in transgenic sense, antisense, and control Arabidopsis plants (C). Total RNA was isolated at different times after inoculation with Ralstonia (5.107 cfu/ml) (B) or P. syringae DC3000/avrRpm1 (107 cfu/ml) (C) from the leaves.
The expression of genes associated with HR cell death in tobacco, hsr203 and hsr515 (13, 26), and Arabidopsis, Athsr3 (14), was examined by Northern blot analysis (Fig. 4 B and C). Expression of these genes was highly activated in tobacco, especially at early time points in the line overexpressing AtMYB30 during the incompatible interaction, as compared with the wild-type lines (Fig. 4B and Fig. 8A, which is published as supporting information on the PNAS web site). Their transcripts were also easily detectable in the sense tobacco line in response to the virulent pathogen, in contrast to the wild-type control line. The PR-1 and PR-5 genes, two genes related to disease resistance in plants, also showed increased transcript levels in the sense lines in response to both virulent and avirulent strains of Ralstonia (Figs. 4B and 8A) as compared with the wild-type lines. In Arabidopsis, Athsr3 and PR-1 gene expression was also clearly enhanced in the sense line and decreased in the antisense line in response to P. syringae DC3000/avrRpm1, as compared with the wild-type line (Figs. 4C and 8B). However, the kinetics of expression were similar in all cases, probably because of the rapidity of the HR process triggered by P. syringae harboring the avrRpm1 gene.
Taken together, these results indicate that overexpression of AtMYB30 enhances and accelerates hypersensitive cell death in response to an HR-inducing strain, initiates an HR-like response to a virulent bacterial pathogen, and alters the expression of HR- and defense-related genes.
Overexpression of AtMYB30 Confers Enhanced Resistance to Pathogens.
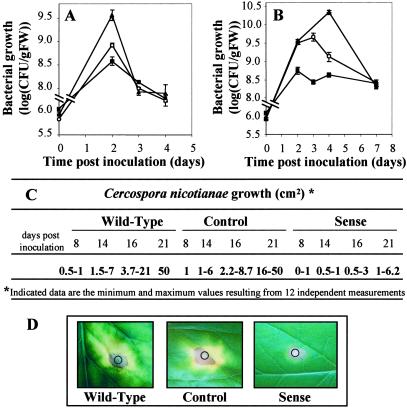
The above results suggested that overexpression of AtMYB30 in Arabidopsis and tobacco should lead to enhanced disease resistance. We therefore monitored the in planta growth of virulent and avirulent P. syringae strains in the wild-type, sense (pBIM131–20A), or antisense (pBIW13–1A) lines. In response to the avirulent strain, a slightly reduced growth of bacteria was observed 2 days after inoculation in the sense line, whereas in the antisense line at the same time point, bacterial growth was higher than the control (Fig. 5A). In a similar way, use of a virulent strain of P. syringae (Fig. 5B) showed that bacterial growth during the first three or four days after inoculation is 11 times lower in the sense line and 12 times higher in the antisense line than in the wild-type line. Thus, the overexpression of AtMYB30 restricts the growth of both avirulent and virulent strains of P. syringae and conversely for AtMYB30 underexpression by antisense depletion. Similar results have been obtained with other bacterial pathogens of Arabidopsis and tobacco. In addition, the overexpression of AtMYB30 protected tobacco from infection by a fungal pathogen, C. nicotianae. The sense line showed increased resistance to this pathogen, as measured by colonization distance from the inoculated zone, when compared with wild-type controls (Fig. 5 C and D). Taken together, these results strongly suggest that AtMYB30 plays a role in the resistance against diverse pathogens.
Figure 5.
Disease resistance of transgenic Arabidopsis and tobacco expressing constitutively AtMYB30 to different virulent and avirulent pathogens. (A and B) Measurement of in planta growth of P. syringae pv. tomato in wild-type Arabidopsis plants (○), sense transgenic plants (pBIM131–20A, ■), or antisense plants (pBIW13–1A, ▴), inoculated (A) with the avirulent strain DC3000/avrRpm1 (107 cfu/ml) or (B) with the virulent strain DC3000 (107 cfu/ml). Data points represent the mean of three replicate experiments and SDs are shown. (C) Growth of C. nicotianae in wild-type tobacco plants, control plants (pBI111–23A), or sense transgenic plants (pBIM131–40A). Twelve to 20 independent measurements of fungal growth were made on 10 leaves from five plants at each time point, and the maximal and minimal data are indicated in cm2. Statistically significant differences have been measured between the sense line and the control line [t test, 0.01 < P < 0.05 (14 days postinoculation, dpi); P < 0.0001 (16 dpi), P < 0.0001 (21 dpi)] and between the sense line and the wild-type line [t test, P < 0.0001 (14 dpi); P < 0.0001 (16 dpi); P < 0.0001 (21 dpi)]. (D) Disease symptoms caused by C. nicotianae 14 days after inoculation in the wild-type, control, and sense lines. The black circle indicates the inoculated zone.
Discussion
Although the HR is thought to occur via a cell death program, very little is known about the plant genes involved in this particular program. Among them, transcription factors probably play key roles in the regulation of an array of genes whose protein products lead to the cell death per se. We have recently identified a gene, AtMYB30, encoding an R2R3 MYB protein in A. thaliana, whose expression pattern is specific, early, and transient during the HR (16). We also showed that the lsd4 and lsd5 mutations, both responsible for an uncontrolled initiation of hypersensitive cell death (10, 16), also cause increased expression of AtMYB30. In this article, we report that the overexpression of AtMYB30 in Arabidopsis and tobacco leads to an acceleration and intensification of hypersensitive cell death in response to an avirulent pathogen attack and to the induction of an HR-like response upon virulent pathogen attack. In addition, resistance to pathogens and HR molecular markers such as products of the oxylipin metabolism, hsr and PR gene expression, are enhanced in the constitutively expressing lines. Conversely, the antisense-mediated depletion of AtMYB30 in transgenic plants led to a strong decrease or suppression of the HR. These results identify AtMYB30 as a positive regulator of hypersensitive cell death in response to pathogen attack.
AtMYB30 Is a Component of the HR Regulatory Pathway.
In contrast to lesion mimic genes, overexpression of AtMYB30 in transgenic plants does not lead to spontaneous formation of HR-like lesions in the absence of pathogen attack (32), but instead confers accelerated and enhanced hypersensitive cell death. It is not clear whether the phenotypes associated with the lesion mimic genes are caused by an alteration of the cellular metabolism induced by their overexpression, or whether these genes intervene directly or indirectly in the HR signaling pathway. In the case of AtMYB30, overexpression leads to the very specific effects described above. Only a few other genes have also been reported to act similarly: Pti1 overexpression accelerates the HR in tobacco in response to P. syringae pv. tabaci strain expressing avrPto (33). Pti1 is a serine/threonine kinase that interacts with the Pto protein conferring resistance to bacterial speck disease in tomato and that is phosphorylated by Pto. Thus, this protein is probably a component of a phosphorylation cascade involved in gene-for-gene plant disease resistance. In contrast, decreased production of a chloroplastic protease (FtsH) in tobacco (34) led to an acceleration of the HR induced by tobacco mosaic virus. More recently, antisense expression of hsr203 was also shown to confer an acceleration of the HR in tobacco in response to diverse pathogens (15). All of these genes have a potential regulatory function in the HR, although their exact mode of action remains unknown.
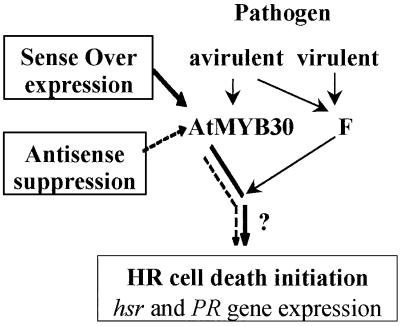
AtMYB30 overexpression does not cause spontaneous HR-like lesions. We hypothesize (Fig. 6) that the expression of AtMYB30 is not sufficient by itself to induce the hypersensitive cell death program and that AtMYB30 probably acts in cooperation with other factor(s) for initiation of the program. Indeed, other MYB domain-containing proteins have been shown to display protein–protein interactions that greatly influence their biological activities (25, 35–39). In maize, the activity of the MYB factor C1 strictly depends on R presence, a basic helix–loop–helix transcription factor (35): the expression of genes that are regulated by the C1/R tandem is turned on only when both C1 and R genes are expressed (36). Such a tightly controlled mechanism may be crucial in the initiation of plant cell death and is consistent with our results. This situation can be compared with the overexpression of c-myb (37), which does not lead to apoptotic cell death by itself, but rather enhances the lethal effect of the proapoptotic agent, transforming growth factor β1. A search for proteins interacting with AtMYB30 could provide new information about the role of this protein on the initiation of programmed cell death upon pathogen attack.
Figure 6.
Proposed model for the role of AtMYB30 in cell death initiation. AtMYB30 gene is specifically, rapidly, and transiently expressed in response to avirulent pathogens. Overexpression of AtMYB30 leads to an acceleration and enhancement of HR cell death, whereas in antisense AtMYB30 lines, HR cell death is strongly decreased or suppressed. AtMYB30 overexpression does not confer a lesion mimic phenotype, so the expression of AtMYB30 is not sufficient by itself to induce the hypersensitive cell death program. AtMYB30 probably acts in cooperation with other factor(s) (F) for initiation of the program.
Essential Role for AtMYB30 in Plant Disease Resistance.
AtMYB30 overexpression not only intensifies the HR to avirulent bacteria, but also mimics the HR in response to virulent pathogens. The observed necrotic response, although slightly delayed as compared with a bona fide HR, is accompanied by the induction of multiple defense mechanisms and the induction of enhanced resistance, and importantly, possesses the hydroxy fatty acid production profile that is the signature of a typical HR. This finding suggests that in the compatible interaction the putative interacting partner of AtMYB30 is produced. In addition, although it is not known whether cell death associated with various diseases is a genetically programmed process, our results suggest that it is distinct from the HR. However, it cannot be excluded that the primary function of this gene might be to control other function(s) influencing cell death rate in the plant cell. The identification of the target genes of this transcriptional factor should elucidate this point in the future.
AtMYB30 is not the sole example of a plant gene whose overexpression enhances resistance to biotrophic pathogens. Tobacco overexpressing Pti1 from tomato (33) are more resistant to P. syringae pv. tabaci harboring the avirulence gene avrPto, i.e., the HR appears earlier than in the wild-type line. However, the corresponding Pseudomonas virulent strain does not provoke HR-like symptoms on tobacco overexpressing Pti1, in contrast to our system. The same comment can be made for engineered hsr203 transgenic plants (15). This finding suggests that AtMYB30, whatever its role in HR development, might act in a critical signaling step preceding cell death.
Cell death has a central role in incompatible plant–pathogen interactions (30), and study of its function in resistance has recently been initiated (31). Although HR is one of the most important factors impeding growth of biotrophic pathogens, it promotes the disease caused by some necrotrophs. Increased resistance of tobacco to several necrotrophic pathogens has been observed upon expression of negative regulators of apoptosis (32). Work presented here concerns only biotrophic pathogens. In this respect, our data are in accordance with these previous studies.
In conclusion, we have identified AtMYB30 as an important actor in the hypersensitive cell death program. It will be used as a starting point for the identification of other actors in this program. It will be of particular importance, in view of our results, to identify the proteins that interact with AtMYB30 in the initiation of hypersensitive cell death. The nature of the target genes of AtMYB30 that may be found by using the DNA microarray technology or more focused approaches should permit a better understanding of the role of this transcriptional factor in the control of the HR. Finally, the possible use of this gene for the production of disease-resistance crop plants should be tested.
Supplementary Material
Acknowledgments
We thank Claudine Balagué for stimulating discussions, Alain Toppan and Bruno Grezes-Besset for providing the C. nicotianae strain and their help for inoculation tests, and Jean Pierre Agnel for technical assistance. We are also grateful to Nigel Grimsley for reviewing the manuscript. This work was supported by a grant from the French Ministry of National Education and Research (to X.D. and F.V.).
Abbreviations
- HR
hypersensitive response
- PR
pathogenesis-related
- GUS
β-glucuronidase
- cfu
colony-forming unit
- 9-HOTE
9-hydroxy-10,12,15(E,Z,Z) octadecatrienoic acid
- hsr
hypersensitivity-related gene
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Repetti, P. P. & Staskawicz, B., Ninth International Conference on Arabidopsis Research, June 24–29, 1998, Madison, WI, abstract 537.
References
- 1.Richberg M H, Aviv D H, Dangl J L. Curr Opin Plant Biol. 1998;1:480–485. doi: 10.1016/s1369-5266(98)80039-3. [DOI] [PubMed] [Google Scholar]
- 2.Keen N T, Ersek T, Long M, Bruegger B, Holliday M. Physiol Plant Pathol. 1981;18:325–337. [Google Scholar]
- 3.Levine A, Pennell R I, Alvarez M E, Palmer R, Lamb C. Curr Biol. 1996;6:427–437. doi: 10.1016/s0960-9822(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 4.Blein J-P, Milat M-L, Ricci P. Plant Physiol. 1991;95:486–491. doi: 10.1104/pp.95.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He S Y, Huang H C, Collmer A. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 6.Arlat M, Van Gijsegem F, Huet J C, Pernollet J C, Boucher C. EMBO J. 1994;13:543–553. doi: 10.1002/j.1460-2075.1994.tb06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittler R, Shulaev V, Lam E. Plant Cell. 1995;7:29–42. doi: 10.1105/tpc.7.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacomme C, Santa Cruz S. Proc Natl Acad Sci USA. 1999;96:7956–7961. doi: 10.1073/pnas.96.14.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg J T, Klessig D F, Ausubel F M. Plant J. 1993;4:327–341. doi: 10.1046/j.1365-313x.1993.04020327.x. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich R A, Delaney T P, Uknes S J, Ward E R, Ryals J A, Dangl J L. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg J T, Guo A, Klessig D F, Ausubel F M. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich R A, Richberg M H, Shmidt R, Dean C, Dangl J L. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- 13.Pontier D, Godiard L, Marco Y, Roby D. Plant J. 1994;5:507–521. doi: 10.1046/j.1365-313x.1994.5040507.x. [DOI] [PubMed] [Google Scholar]
- 14.Lacomme C, Roby D. FEBS Lett. 1999;459:149–153. doi: 10.1016/s0014-5793(99)01233-8. [DOI] [PubMed] [Google Scholar]
- 15.Tronchet M, Ranty B, Marco Y, Roby D. Plant J. 2001;27:115–127. doi: 10.1046/j.1365-313x.2001.01072.x. [DOI] [PubMed] [Google Scholar]
- 16.Daniel X, Lacomme C, Morel J-B, Roby D. Plant J. 1999;20:57–66. doi: 10.1046/j.1365-313x.1999.00578.x. [DOI] [PubMed] [Google Scholar]
- 17.Morel J-B, Dangl J L. Genetics. 1999;151:305–319. doi: 10.1093/genetics/151.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- 19.Lacomme C, Roby D. Plant Mol Biol. 1996;30:995–1008. doi: 10.1007/BF00020810. [DOI] [PubMed] [Google Scholar]
- 20.Baker C J, Mock N M. Plant Cell Tissue Organ Cult. 1994;39:7–12. [Google Scholar]
- 21.Rustérucci C, Montillet J L, Agnel J P, Battesti C, Alonso B, Knoll A, Bessoule J J, Etienne P, Suty L, Blein J P, Triantaphylidès C. J Biol Chem. 1999;274:36446–36455. doi: 10.1074/jbc.274.51.36446. [DOI] [PubMed] [Google Scholar]
- 22.Roby D, Broglie K, Cressman R, Biddle P, Chet I, Broglie R. Plant Cell. 1990;2:999–1007. doi: 10.1105/tpc.2.10.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lummerzheim M, de Olivera D, Castresana C, Miguens F C, Louzada E, Roby D, Van Montagu M, Timmerman B. Mol Plant–Microbe Interact. 1993;6:532–544. [Google Scholar]
- 24.Grellet F, Delcasso-Tremousaygue D, Delseny M. Plant Mol Biol. 1989;12:695–706. doi: 10.1007/BF00044160. [DOI] [PubMed] [Google Scholar]
- 25.Ying G G, Proost P, Vazn Damne J, Bruschi M, Introna M, Golay J. J Biol Chem. 2000;275:4152–4158. doi: 10.1074/jbc.275.6.4152. [DOI] [PubMed] [Google Scholar]
- 26.Czernic P, Huang H C, Marco Y. Plant Mol Biol. 1996;31:255–265. doi: 10.1007/BF00021788. [DOI] [PubMed] [Google Scholar]
- 27.Ward E R, Uknes S J, Williams S C, Dincher S S, Wiederhold D L, Alexander D C, Ahl-Goy P, Metraux J-P, Ryals J. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kazan K, Murray F R, Goulter K C, Llewellyn D J, Manners J M. Mol Plant–Microbe Interact. 1998;11:555–562. [Google Scholar]
- 29.Huang H C, Schuuring R, Denny T P, Atkinson M M, Baker C J, Yucell T, Hutchinson S W, Collmer A. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenberg J T. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:525–545. doi: 10.1146/annurev.arplant.48.1.525. [DOI] [PubMed] [Google Scholar]
- 31.Govrin E M, Levine A. Curr Biol. 2000;10:751–757. doi: 10.1016/s0960-9822(00)00560-1. [DOI] [PubMed] [Google Scholar]
- 32.Dickman M B, Park Y K, Oltersdorf T, Clemente T, French R. Proc Natl Acad Sci USA. 2001;98:6957–6962. doi: 10.1073/pnas.091108998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Loh Y-T, Bressan R A, Martin G B. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 34.Seo S, Okamoto M, Iwai T, Iwano M, Fukui K, Isogai A, Nakajima N, Ohashi Y. Plant Cell. 2000;12:917–932. doi: 10.1105/tpc.12.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth B A, Goff S A, Klein T M, Fromm M E. Plant Cell. 1991;3:317–325. doi: 10.1105/tpc.3.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grotewold E, Chamberlain M, St Claire G, Swenson J, Siame B A, Butler L G, Snook M, Bowen B. Plant Cell. 1998;10:727–740. [PMC free article] [PubMed] [Google Scholar]
- 37.Selvakumaran M, Lin H-K, Sjin R T T, Reed J C, Lieberman D A, Hoffman B. Mol Cell Biol. 1994;14:2352–2360. doi: 10.1128/mcb.14.4.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganter B, Fu S I, Lipsick J S. EMBO J. 1998;17:255–268. doi: 10.1093/emboj/17.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedge S P, Kumar A, Kurschner C, Shapiro L H. Mol Cell Biol. 1998;18:2729–2737. doi: 10.1128/mcb.18.5.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.